Recognition of non-canonical purine•purine RNA motifs by hnRNP A2 mediates targeted delivery of neuronal RNAs to dendrites.
Abstract
In neurons, regulation of gene expression occurs in part through translational control at the synapse. A fundamental requirement for such local control is the targeted delivery of select neuronal mRNAs and regulatory RNAs to distal dendritic sites. The nature of spatial RNA destination codes, and the mechanism by which they are interpreted for dendritic delivery, remain poorly understood. We find here that in a key dendritic RNA transport pathway (exemplified by BC1 RNA, a dendritic regulatory RNA, and protein kinase M ζ [PKMζ] mRNA, a dendritic mRNA), noncanonical purine•purine nucleotide interactions are functional determinants of RNA targeting motifs. These motifs are specifically recognized by heterogeneous nuclear ribonucleoprotein A2 (hnRNP A2), a trans-acting factor required for dendritic delivery. Binding to hnRNP A2 and ensuing dendritic delivery are effectively competed by RNAs with CGG triplet repeat expansions. CGG repeats, when expanded in the 5′ untranslated region of fragile X mental retardation 1 (FMR1) mRNA, cause fragile X–associated tremor/ataxia syndrome. The data suggest that cellular dysregulation observed in the presence of CGG repeat RNA may result from molecular competition in neuronal RNA transport pathways.
Introduction
RNA transport and localization are important means in the control of gene expression in eukaryotic cells (Dahm and Kiebler, 2007). In neurons, RNA transport to dendritic and axonal domains is considered an essential underpinning of neuronal functionality and plasticity (Mohr et al., 2001; Smith, 2004; Dahm et al., 2007; Miyashiro et al., 2009). Fundamental to neuronal RNA transport mechanisms are cis-acting spatial codes that specify intracellular delivery destinations and trans-acting factors that decode such information for the cellular transport machinery (Blichenberg et al., 1999, 2001; Mohr and Richter, 2003; Shan et al., 2003; Muslimov et al., 2006; Bramham and Wells, 2007; Jambhekar and Derisi, 2007). Although the information contained in RNA spatial destination codes is derived from genomic nucleotide sequences, decoding requires recognition of the code “gestalt” (the overall RNA motif structure representing the code) by trans-acting protein factors (Smith, 2004).
Eukaryotic RNA targeting motifs are diverse and cooperate with various trans-acting factors for decoding (Eberwine et al., 2002; Bramham and Wells, 2007; Jambhekar and Derisi, 2007). RNA motif structures that specify intracellular targeting are known as spatial destination codes, zip codes, or targeting elements. In neurons, the synapto-dendritic delivery of select mRNAs and regulatory RNAs is a prerequisite for translation at the synapse, and thus an important determinant of local protein synthetic output (Kindler et al., 2005; Dahm et al., 2007). One of the key trans-acting factors that recognize dendritic targeting elements (DTEs) in neuronal RNAs is heterogeneous nuclear ribonucleoprotein A2 (hnRNP A2; Shan et al., 2003; Smith, 2004; Muslimov et al., 2006; Gao et al., 2008). Interactions of trans-acting factors with DTE spatial codes are the principal determinants of neuronal RNA transport specificities. As a consequence, they define synaptic RNA repertoires and thus control the spectrum of local protein synthetic capacity.
Fragile X–associated tremor/ataxia syndrome (FXTAS) is precipitated by CGG triplet repeat expansion in the 5′ untranslated region (UTR) of fragile X mental retardation 1 (FMR1) mRNA (Jacquemont et al., 2007; Brouwer et al., 2009; Hagerman et al., 2010). At 55–200 repeats (referred to as premutation), CGG triplets appear to cause gain-of-function RNA toxicity that results in cellular dysregulation (Jacquemont et al., 2007; Brouwer et al., 2009; Hagerman et al., 2010). CGG premutations may cause FXTAS, a late-onset neurodegenerative disorder (Jin et al., 2003) characterized by cognitive decline and motor disorders, and may also give rise to neurodevelopmental impairments with features resembling mild fragile X syndrome (FXS) or autism (Jacquemont et al., 2007; Hagerman et al., 2010). We noted that analogous to DTEs of the GA motif subtype (Muslimov et al., 2006), CGG RNA repeats form stem loops that interact with hnRNP A2 (Sofola et al., 2007; Swanson and Orr, 2007). In further analogy, both GA-type DTEs and CGG repeat stem loops are characterized by noncanonical (non–Watson Crick [non-WC]) purine•purine base interactions (Napierala et al., 2005; Muslimov et al., 2006; Tiedge, 2006; Zumwalt et al., 2007; Sobczak et al., 2010). Given their potentially equivalent noncanonical, hnRNP A2–interacting RNA motif content, we hypothesized that GA targeting motifs and CGG repeat stem-loops compete with each other for binding to trans-acting factor hnRNP A2.
Here we dissect the molecular basis of GA motif recognition by hnRNP A2, and the consequences of CGG versus GA motif competition in A2 recognition and dendritic targeting. Working with a representative of dendritic regulatory RNAs, BC1 RNA (Iacoangeli et al., 2010), and with a representative of dendritic mRNAs, PKMζ mRNA (Pastalkova et al., 2006; Serrano et al., 2008), we show that noncanonical structural motifs are critical for recognition and targeting. We found that CGG repeats compete with the BC1 GA motif for an essential targeting resource, hnRNP A2, resulting in impaired dendritic delivery. The data implicate RNA competition in disease-relevant neuronal transport mechanisms.
Results
Interaction of hnRNP A2 with targeted RNAs
In this work, we sought to establish principles and the significance of hnRNP A2 interactions with spatial codes in dendritically targeted neuronal RNAs. To this end, we first examined binding of hnRNP A2 to prototypical dendritic RNAs using quantitative electrophoretic mobility shift assays (EMSAs) with recombinant protein (Ryder et al., 2008; Chao et al., 2010). We focused on RNAs with spatial codes that we had previously predicted to be recognized by hnRNP A2 (Muslimov et al., 2006; Tiedge, 2006).
Regulatory BC1 RNA, a non-protein-coding small cytoplasmic RNA (scRNA), is an effector of translational control in neurons (Wang et al., 2002, 2005; Kondrashov et al., 2005; Lin et al., 2008; Iacoangeli et al., 2010). BC1 RNA is specifically targeted to distal dendritic domains (Muslimov et al., 1997, 2006), and we have previously shown that a noncanonical GA motif in the apical part of the 5′ BC1 stem loop domain is responsible for distal dendritic targeting (Muslimov et al., 2006). The defining attributes of GA motifs are a core of tandem noncanonical G•A base pairs, an adjacent internal loop of variable geometry, and flanking standard WC base pairs, typically of the GC type (Lescoute et al., 2005; Muslimov et al., 2006). GA motifs are dimorphic and can alternate between an extended and a kinked (kink-turn or K-turn [KT]) conformation (Goody et al., 2004; Tiedge, 2006; Schroeder et al., 2010). Secondary structures of GA motifs used in this work are shown in Fig. S1.
We performed EMSA analysis with recombinant hnRNP A2 to probe interactions of this trans-acting factor with RNA targeting motifs. Fig. 1 shows that wild type (WT) BC1 RNA was shifted to lower mobility by recombinant hnRNP A2. In clear contrast, a mutant BC1 RNA in which noncanonical G•A base pairs in the apical GA motif had been converted into standard WC pairs failed to interact with hnRNP A2 (Fig. 1 A; see Fig. S1 for GA motif mutants used.) These data indicate that noncanonical nucleotide interactions in the 5′ BC1 GA motif are indispensable for recognition by hnRNP A2.
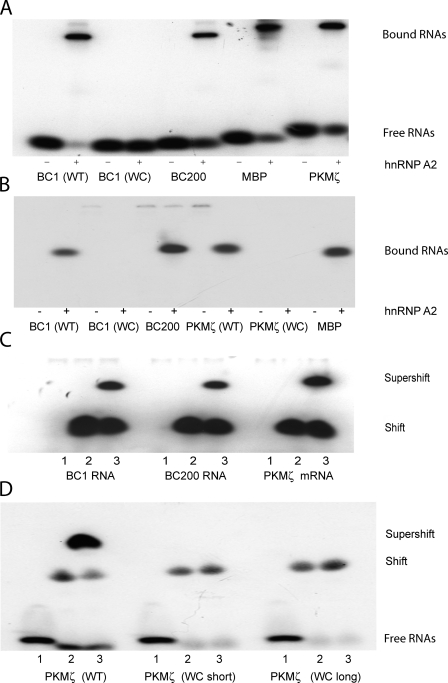
Figure 1.
Dendritically transported BC1 RNA and PKMζ mRNA interact with hnRNP A2. EMSA experiments were performed using native PAGE. (A and B) BC1 RNA and PKMζ mRNA bind to recombinant hnRNP A2. EMSA experiments revealed shifts to lower mobility as a result of binding of radiolabeled RNAs to the protein. (A) Gel shifts induced by hnRNP A2 were observed with BC1 RNA, BC200 RNA, PKMζ mRNA, and MBP mRNA. For BC1 RNA and PKMζ mRNA, derivatives were used in which the noncanonical GA motif had been converted to canonical WC base pairing. Such WC mutants, in contrast to the respective WT species, failed to interact with hnRNP A2 (A and B; only bound RNAs are shown in B). For secondary structures of WT and WC mutant motifs in BC1 RNA and PKMζ mRNA, see Fig. S1. The PKMζ mRNA WC mutant used in B was variant long. (C and D) BC1 RNA, BC200 RNA, and PKMζ mRNA are recognized by hnRNP A2 in brain extracts. (C) An antibody specific for hnRNP A2 (Ma et al., 2002) produced a supershift with BC1 RNA, BC200 RNA, and PKMζ mRNA. Shifted and supershifted bands are shown. (D) Recognition of PKMζ mRNA by hnRNP A2 in brain extracts depends on a noncanonical motif structure in a 3′ UTR stem loop. WT PKMζ mRNA was supershifted by antibodies against hnRNP A2; in contrast, WC mutant PKMζ mRNAs were not. Loading (C and D): 1, RNAs; 2, RNAs plus brain extract; 3, RNAs plus brain extract plus anti-A2 antibody.
We next examined interactions of hnRNP A2 with PKMζ mRNA. PKMζ is a persistently active synaptic kinase that plays an essential role in long-term memory maintenance (Pastalkova et al., 2006; Serrano et al., 2008). We have previously reported that a 44-nucleotide stem-loop structure in the 3′ UTR of PKMζ mRNA is required for distal dendritic targeting (Muslimov et al., 2004). We subsequently hypothesized, on the basis of structural similarities, that this stem-loop harbors a targeting-relevant noncanonical GA motif (Muslimov et al., 2006; Tiedge, 2006). We now show that PKMζ mRNA binds specifically to hnRNP A2 (Fig. 1). We generated two PKMζ mRNA mutants in which the 44-nucleotide stem-loop structure in the 3′ UTR was altered such that the tandem G•A pairs and the adjacent asymmetric internal loop motif were converted to canonical WC base pairs (WC mutant long and WC mutant short; Fig. S1). Neither of these canonical conversion mutants displayed any detectable binding to hnRNP A2 (WC mutant long shown in Fig. 1 B; WC mutant short data not depicted). The results indicate that a noncanonical GA motif in a 3′ UTR stem-loop structure of PKMζ mRNA is specifically recognized by hnRNP A2.
BC200 RNA is the primate functional analogue of rodent BC1 RNA (Tiedge et al., 1993; Lin et al., 2008). The two RNA genes are not orthologues, as their respective phylogenetic pedigrees are distinct (Martignetti and Brosius, 1993a,b). For this reason, it was necessary to examine BC200 RNA, next to BC1 RNA, for its capacity to interact with hnRNP A2. Fig. 1 shows that hnRNP A2 binding capacities to BC200 and to BC1 RNAs were indistinguishable. (Analogously to BC1 RNA, BC200 RNA is also transported to distal dendritic domains; Fig. S2).
As a positive control in our EMSA analysis, we included myelin basic protein (MBP) mRNA, as it has previously been shown to interact with hnRNP A2 (Hoek et al., 1998; Munro et al., 1999; Shan et al., 2000). This finding was replicated in our assay (Fig. 1). In a negative control, we probed binding of BC1 RNA and PKMζ mRNA to eukaryotic initiation factor 4G (eIF4G), an RNA-binding protein that interacts with mRNAs during the initiation phase of translation (Gingras et al., 1999; Pestova et al., 2007). For this purpose, we used the central, RNA-binding domain of eIF4G, as described previously (Pestova et al., 1996). The results show that eIF4G failed to produce a shift of BC1 RNA and PKMζ mRNA to lower mobilities, in contrast to hnRNP A2, which clearly did (Fig. S3).
For independent corroboration of the EMSA data, we performed supershift experiments with antibodies specific for hnRNP A2 (Ma et al., 2002) using established techniques with rat brain extracts (Wang et al., 2002). As shown in Fig. 1 (C and D), BC1 RNA, BC200 RNA, and PKMζ mRNA were supershifted to lower mobilities by anti-hnRNP A2 antibodies in rat brain extracts. We conclude, on the basis of the combined data, that trans-acting factor hnRNP A2 specifically recognizes targeting-determinant noncanonical GA motifs in the dendritic RNAs examined.
High-affinity binding of hnRNP A2 to the BC1 GA motif
What is the binding affinity of hnRNP A2 for BC1 RNA? To establish equilibrium binding constants, we used quantitative EMSA analysis to titrate the binding partners, measuring the fraction of bound BC1 RNA as a function of hnRNP A2 protein concentration (Fig. 2 A). The equilibrium dissociation constant (Kd) was then calculated by fitting the data to the Hill equation, as described previously (Ryder et al., 2008; Chao et al., 2010). This analysis revealed a Kd of 8.2 nM, which indicates that BC1 RNA binds with high affinity to hnRNP A2.
Figure 2.
High-affinity binding of dendritic BC1 RNA to hnRNP A2 is modulated by Mg2+ and Ca2+ ions. Binding affinities were established by quantitative EMSA using native PAGE. WT BC1 RNA (A) and WC mutant BC1 RNA (B) were titrated with increasing concentrations of hnRNP A2 in the presence of 10 mM MgCl2. Although a Kd of 8.2 nM was obtained for WT BC1 RNA, no equilibrium constant could be established for WC mutant BC1 RNA, as no binding was observed even at maximal hnRNP A2 concentrations. (C) Binding affinity of BC1 RNA to hnRNP A2 is dependent on the concentration of Mg2+ ions. The following equilibrium dissociation constants were obtained: 25 mM MgCl2, Kd = 3.5 nM; 10 mM MgCl2, Kd = 8.2 nM; 5 mM MgCl2, Kd = 520 nM; 2.5 mM MgCl2, Kd = 980 nM. (D) Binding of BC1 RNA to hnRNP A2 is modulated by Ca2+ ion concentrations. Dissociation constants: 10 mM CaCl2, Kd = 0.6 nM; 5 mM CaCl2, Kd = 1.0 nM; 1 mM CaCl2, Kd = 210 nM; 0.5 mM CaCl2, Kd = 250 nM; 0.1 mM CaCl2, Kd = 380 nM.
We next used BC1 RNA in which the 5′ GA motif has been converted to canonical base pairing (Fig. S1). Titration with hnRNP A2 did not reveal any indication of binding, even if excessively high protein concentrations (up to 50 µM) were used (Fig. 2 B). We conclude that an intact noncanonical GA motif in the 5′ BC1 domain is required for high-affinity binding to hnRNP A2.
Previous work has indicated that at sufficiently high concentrations of Mg2+ ions, the apical 5′ BC1 GA motif adopts the conformation of a KT motif (Muslimov et al., 2006). KT motifs are sharply bent conformations of GA motifs that serve as recognition sites for protein interactions (Goody et al., 2004; Lescoute et al., 2005). We therefore hypothesized that the affinity for binding of BC1 RNA to hnRNP A2 may be a function of Mg2+ ion concentrations. Titration curves in the presence of varying Mg2+ concentrations confirmed that this was indeed the case (Fig. 2 C). At Mg2+ concentrations of 10 mM or above, the Kd for BC1 RNA–hnRNP A2 binding was in the nanomolar concentration range (Kd = 8.2 nM at 10 mM Mg2+, Kd = 3.5 nM at 25 mM Mg2+). In contrast, at Mg2+ concentrations <10 mM, the BC1 RNA–hnRNP A2 binding affinity was weaker by two orders of magnitude (Kd = 520 nM at 5 mM Mg2+; Kd = 980 nM at 2.5 mM Mg2+; Fig. 2 C). Thus, we observed a substantial increase in binding affinity going from 5 mM to 10 mM Mg2+.
An earlier study indicated that Mg2+ and Ca2+ ions are equivalent in their abilities to support the kinked conformation of ribosomal RNA KT motif KT-38 (Matsumura et al., 2003). We therefore asked whether the affinity for binding of BC1 RNA to hnRNP A2 is also modulated by Ca2+ ions. Titration curves shown in Fig. 2 D show that the Kd for BC1 RNA–hnRNP A2 binding underwent a shift of more than two orders of magnitude between 1 mM Ca2+ (Kd = 210 nM) and 5 mM Ca2+ (Kd = 1.0 nM). Thus, although increasing Mg2+ and Ca2+ concentrations both shifted BC1 RNA–hnRNP A2 equilibrium constants to higher affinities, this shift was induced at lower relative concentrations of Ca2+ compared to Mg2+.
In conclusion, the data show that high-affinity binding of BC1 RNA to hnRNP A2 requires a noncanonical 5′ GA motif, and they suggest that interactions of the GA-type targeting element with hnRNP A2 can be modulated in a Mg2+/Ca2+-dependent manner.
Requirement of hnRNP A2 for dendritic targeting
We have previously shown that the apical noncanonical GA motif in the 5′ BC1 domain is required for distal dendritic targeting (Muslimov et al., 2006). With the results presented in this paper, we now show that this motif is specifically recognized by hnRNP A2, resulting in high-affinity, Mg2+/Ca2+-modulated binding. These data raise the question of whether interactions with hnRNP A2 are in fact necessary for the delivery of BC1 RNA to distal dendritic domains.
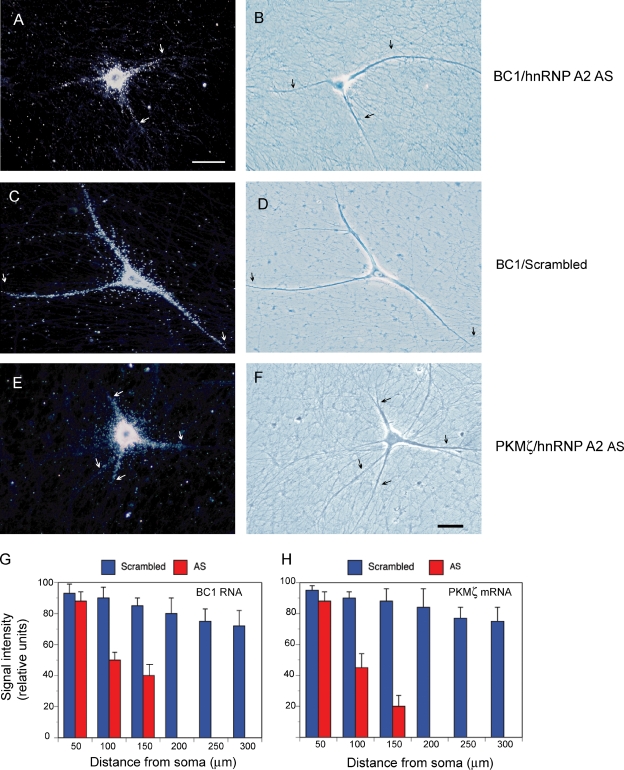
We chose an antisense oligonucleotide approach to address this question. We used antisense oligonucleotides directed against the translational start site region of hnRNP A2, as described previously (Kwon et al., 1999; Munro et al., 1999). Exposure of sympathetic neurons in culture to such oligonucleotides resulted in a specific reduction of cellular levels of hnRNP A2 by >80% (Fig. S4). Cultured neurons exposed to A2 antisense oligonucleotide were then examined for dendritic targeting of BC1 RNA, using our established microinjection paradigm (Muslimov et al., 1997, 2002, 2004, 2006). We observed a significant reduction in distal dendritic targeting competence in the presence of A2 antisense oligonucleotides, but not in the presence of scrambled-sequence control oligonucleotides (Fig. 3, A–D and G). The data indicate that hnRNP A2 is required for distal dendritic targeting of BC1 RNA.
Figure 3.
Dendritic transport of BC1 RNA and PKMζ mRNA is inhibited by hnRNP A2 antisense oligonucleotides. Photomicrographs show subcellular localization of injected full-length BC1 RNA (A and B) and PKMζ mRNA (E and F) in neurons pretreated with hnRNP A2 antisense oligonucleotides. BC1 RNA and PKMζ mRNA were detectable in proximal but not distal dendritic domains. In contrast, BC1 RNA (C and D) and PKMζ mRNA (not depicted) were transported along the entire dendritic extent in neurons pretreated with scrambled-sequence oligonucleotides. Dark-field (left) and phase-contrast (right) images are shown. RNA signal appears as white silver grains in dark field images. Arrows indicate the extent of dendritic labeling. Number of cells analyzed: (A and B) 10 neurons, 34 dendrites; (C and D) 10 neurons, 30 dendrites; (E and F) 9 neurons, 31 dendrites. Bars, 50 µm. (G and H) Quantitative data are shown in the format of mean ± SEM of relative signal intensities along the dendritic extent for BC1 RNA (G) and PKMζ mRNA (H). One-way ANOVA, Dunnett’s post-hoc analysis (comparison of all interval points between A2 antisense and scrambled-sequence): P > 0.5 for interval points 50 µm, P < 0.001 for interval points 100 µm and beyond (for both BC1 RNA and PKMζ mRNA). Error bars indicate SEM.
We also examined whether application of A2 antisense oligonucleotides impacted the somato-dendritic localization of endogenous BC1 RNA. We first performed in situ hybridization experiments to confirm that endogenous BC1 RNA is located to somato-dendritic domains of sympathetic neurons in culture (Fig. 4, A and B), which is analogous to the previously reported localization in hippocampal neurons in culture (Muslimov et al., 1998). Subsequent experiments, shown in Fig. 4 (C and D), revealed no significant changes in dendritic localization of endogenous BC1 RNA after a 1-h exposure of sympathetic neurons to A2 antisense oligonucleotides. In contrast, a substantial decrease of dendritic (and an increase of somatic) levels of endogenous BC1 RNA was observed after 24 h in the presence of A2 antisense oligonucleotides (Fig. 4, E–G). The data thus suggest that interference with dendritic transport of endogenous BC1 RNA does not seem to result in immediate reductions of dendritic RNA levels, as local reservoirs of preexisting RNAs are not rapidly depleted. Over time, however, dendritic levels decline as local preexisting RNAs are not being replenished.
Figure 4.
A2 antisense oligonucleotides cause long-term decrease in dendritic localization of endogenous BC1 RNA. (A and B) In situ hybridization experiments reveal somato-dendritic localization of endogenous BC1 RNA in sympathetic neurons in culture. Dark field (A) and phase contrast (B) photomicrographs show significant labeling for BC1 RNA along the dendritic extent. Dendrites are indicated by arrows, axons by red arrowheads. 8 neurons and 26 dendrites were analyzed. In situ hybridization experiments using sense-strand control RNA probes did not result in any significant cellular labeling (not depicted). Subcellular distributions of other endogenous molecular components have been published earlier as follows: PKMζ mRNA (somato-dendritic; Muslimov et al., 2004), α-tubulin mRNA (restricted to soma; Bruckenstein et al., 1990; Mohr et al., 2001), hnRNP A2 (somato-dendritic; Shan et al., 2003; Smith, 2004). (C and D) After 1 h of exposure to A2 antisense oligonucleotides, dendritic localization of BC1 RNA remained indistinguishable from neurons maintained in the absence of oligonucleotides (A and B). Dendritic labeling is indicated by arrows. 9 neurons and 35 dendrites were analyzed. (E and F) After 24 h of exposure to A2 antisense oligonucleotides, a significant reduction was observed in dendritic (but not somatic) levels of endogenous BC1 RNA. Arrows indicate extent of the labeling signal. Red arrowheads in F indicate a morphologically identifiable axon. 12 neurons and 36 dendrites were analyzed. Scrambled-sequence oligonucleotides were not effective in reducing dendritic localization (not depicted). Bars, 50 µm. (G) Quantitative analysis of in situ hybridization data confirmed a significant reduction in the dendritic localization of BC1 RNA after 24 h (but not after 1 h) of treatment with A2 antisense oligonucleotides. The analysis also revealed that somatic levels of BC1 RNA increased after 24 h, though not after 1 h of A2 antisense treatment. One-way ANOVA, Dunnett’s post-hoc analysis: comparison of all interval points between 1 h A2 antisense and scrambled-sequence, P > 0.5 for all interval points. Comparison of all interval points between 24 h A2 antisense and scrambled-sequence: P < 0.0001 for interval points 50 µm and beyond, P < 0.01 for interval points of 0 µm. Error bars indicate SEM.
For PKMζ mRNA, a 44-nt stem-loop structure in the 3′ UTR is required for distal dendritic targeting (Muslimov et al., 2004). We have now shown that a noncanonical GA motif within this structure is specifically recognized by hnRNP A2 (Fig. 1). Is this motif, and recognition by hnRNP A2, required for distal dendritic delivery? We performed two sets of experiments to address these questions. First, we probed dendritic transport of PKMζ mRNA in cultured neurons exposed to A2 antisense oligonucleotides. In the presence of such oligonucleotides, but not in the presence of scrambled-sequence oligonucleotides, distal dendritic delivery of PKMζ mRNA was significantly reduced (Fig. 3, E, F, and H). Second, we examined dendritic transport of PKMζ mRNA in which the 3′ UTR noncanonical GA motif had been converted to canonical WC base pairing (Fig. S1). Fig. 5 shows that such mutants failed to be delivered to distal dendritic domains, in contrast to the WT PKMζ mRNA which clearly was. These results indicate that dendritic transport of PKMζ mRNA is mediated by hnRNP A2 and requires a noncanonical GA motif in a 3′ UTR stem-loop structure. The combined results suggest that the molecular mechanisms underlying dendritic transport of BC1 RNA and PKMζ mRNA are remarkably similar.
Figure 5.
A noncanonical GA motif in the 3′ UTR is requisite for dendritic delivery of PKMζ mRNA. (A and B) WT PKMζ mRNA was transported along the entire dendritic extent, reaching distal dendritic tips. (C–F) In contrast, WC mutant PKMζ mRNA entered only proximal dendritic segments. Two different WC mutants were used (see Fig. S1). Arrows indicate the extent of dendritic labeling. Number of cells analyzed: (A and B) 9 neurons, 30 dendrites; (C and D) 10 neurons, 32 dendrites; (E and F) 8 neurons, 29 dendrites. Bar, 50 µm. (G) Quantitative data were subjected to one-way ANOVA, Dunnett’s post-hoc analysis. Comparison of all interval points between WT and WC mutants: P > 0.5 for interval points 50 µm, P < 0.001 for interval points 100 µm and beyond (for both WC mutants, in comparison with WT). Error bars indicate SEM.
CGG triplet repeat competition
CGG triplets, expanded to 55–200 repeats, in the 5′ UTR of FMR1 mRNA are causative of human premutation disorders including FXTAS (Jacquemont et al., 2007; Brouwer et al., 2009; Hagerman et al., 2010). Such repeats form stem-loop structures that are stabilized by noncanonical G•G base pairs (Napierala et al., 2005; Zumwalt et al., 2007). Recent evidence suggests that CGG repeat stem-loop structures bind to hnRNP A2 (Sofola et al., 2007; Swanson and Orr, 2007; Brouwer et al., 2009). We therefore hypothesized that noncanonical CGG repeat structures may compete with noncanonical GA motifs that serve as DTEs. Such competition would be expected to cause diminished GA motif binding to hnRNP A2 and, consequently, reduced dendritic delivery.
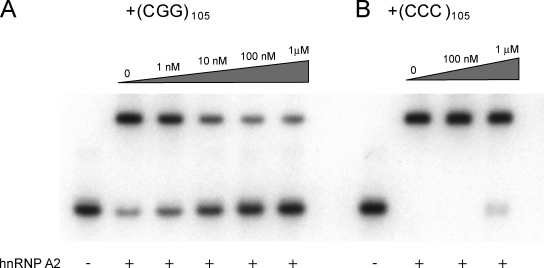
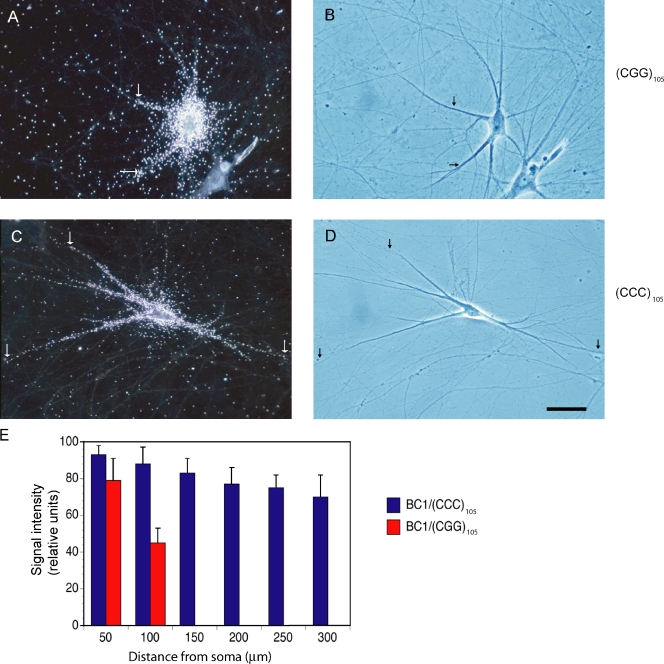
We performed two sets of experiments to test this hypothesis. First, we examined binding of BC1 RNA to hnRNP A2 in the presence of (CGG)105 repeat RNA. A repeat length of 105 CGG units was chosen as premutation representative, which is consistent with previous work in the field (Jin et al., 2007; Sofola et al., 2007). We observed that (CGG)105 repeat RNA, but not (CCC)105 repeat RNA, effectively competed with binding of BC1 RNA to hnRNP A2 (Fig. 6). Second, we investigated the impact of (CGG)105 repeat RNA on dendritic targeting of BC1 RNA. In these experiments, we applied (CGG)105 repeat RNA at levels that we estimated were comparable to FMR1 mRNA levels in premutation disease cells (see Materials and methods). We found that under these conditions, distal dendritic delivery of BC1 RNA was significantly reduced in the presence of (CGG)105 RNA (Fig. 7). In contrast, (CCC)105 RNA had no effect on dendritic BC1 targeting (Fig. 7).
Figure 6.
(CGG)105 RNA competes with BC1 RNA for binding to hnRNP A2. EMSA experiments were performed using native PAGE. (A) Increasing concentrations of (CGG)105 RNA reduced formation of BC1 RNA–hnRNP A2 complexes. (B) In contrast, (CCC)105 RNA showed no effect on BC1 RNA–hnRNP A2 interactions. hnRNP A2 was used at 100 nM, MgCl2 at 10 mM.
Figure 7.
Dendritic delivery of BC1 RNA is significantly reduced in the presence of (CGG)105 RNA. (A and B) After coinjection with (CGG)105 RNA, transport of BC1 RNA along the dendritic extent was reduced. 6 neurons and 17 dendrites were analyzed. (C and D) In contrast, after coinjection with (CCC)105 RNA, BC1 RNA was delivered along the dendritic extent, to distal-most dendritic segments and tips, in a manner indistinguishable from dendritic BC1 transport in the absence of repeat RNA. 11 neurons and 35 dendrites were analyzed. Concentrations of (CGG)105 RNA and (CCC)105 RNA in the injection pipette were 1 µM. Arrows indicate the extent of dendritic labeling. Note that higher amounts of labeled RNA were injected in A and B (as compared with C and D) to verify that subcellular distribution (here somatic restriction) is independent of injection amounts (see also Materials and methods). Bar, 50 µm. (E) Data were examined by one-way ANOVA, Dunnett’s post-hoc analysis (comparison of all interval points between (CGG)105 RNA and (CCC)105 RNA): P > 0.5 for interval points 50 µm, P < 0.001 for interval points 100 µm and beyond. Error bars indicate SEM.
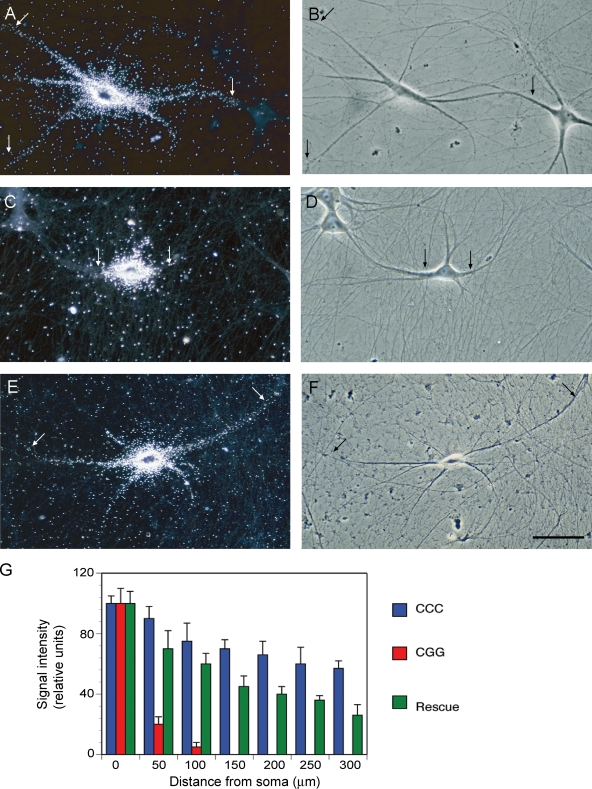
To corroborate that reduced dendritic targeting in the presence of (CGG)105 RNA was the result of competition with hnRNP A2, we performed rescue experiments in which recombinant hnRNP A2 was introduced into sympathetic neurons in culture by microinjection. At a molar ratio of 3:1 of injected hnRNP A2 to coinjected (CGG)105 RNA, dendritic transport of BC1 RNA was restored to a significant degree (Fig. 8). The data support the notion that CGG repeat RNA interferes with dendritic delivery of BC1 RNA by sequestering an essential shared component, hnRNP A2. It is also noted that although restoration of dendritic delivery by hnRNP A2 was significant, it was not complete (Fig. 8 G). It is possible that in addition to hnRNP A2, other trans-acting factors that are required for the dendritic targeting of BC1 RNA are sequestered by CGG repeat RNA.
Figure 8.
Dendritic delivery of BC1 RNA is restored by hnRNP A2. (A and B) BC1 RNA was transported to dendritic domains after microinjection into sympathetic neurons in culture. 12 neurons and 36 dendrites were analyzed. (C and D) In the presence of (CGG)105 RNA, distal dendritic delivery of BC1 RNA was significantly reduced (see also Fig. 7). 10 neurons and 29 dendrites were analyzed. (E and F) After coinjection with recombinant hnRNP A2 (at a 3:1 molar ratio to (CGG)105 RNA), dendritic delivery of BC1 RNA was restored to a significant degree. 11 neurons and 38 dendrites were analyzed. Arrows indicate the extent of dendritic labeling. Bar, 50 µm. (G) Quantitative analysis confirmed that coinjection of hnRNP A2 with (CGG)105 RNA restored dendritic delivery of BC1 RNA to a significant degree. One-way ANOVA, Dunnett’s post-hoc analysis (comparison of all interval points between (CGG)105 RNA and (CGG)105 RNA coinjected with hnRNP A2): P > 0.05 for interval points 50 µm, P < 0.0001 for interval points 100 µm and beyond. Error bars indicate SEM.
In conclusion, our results suggest that molecular competition of CGG triplet repeat RNA with the BC1 RNA GA targeting motif results in diminished hnRNP A2 binding and reduced distal dendritic delivery.
Role of CGG triplet repeats in RNA targeting
Given that CGG repeats in the 5′ UTR of the FMR1 gene have the potential to expand and thus cause disease (Jacquemont et al., 2007; Brouwer et al., 2009), the question arises of why, despite this adverse potential, normal-size repeats (< 55 CGG units) have been retained in the course of phylogenetic development. Are they indispensable because they perform an essential function?
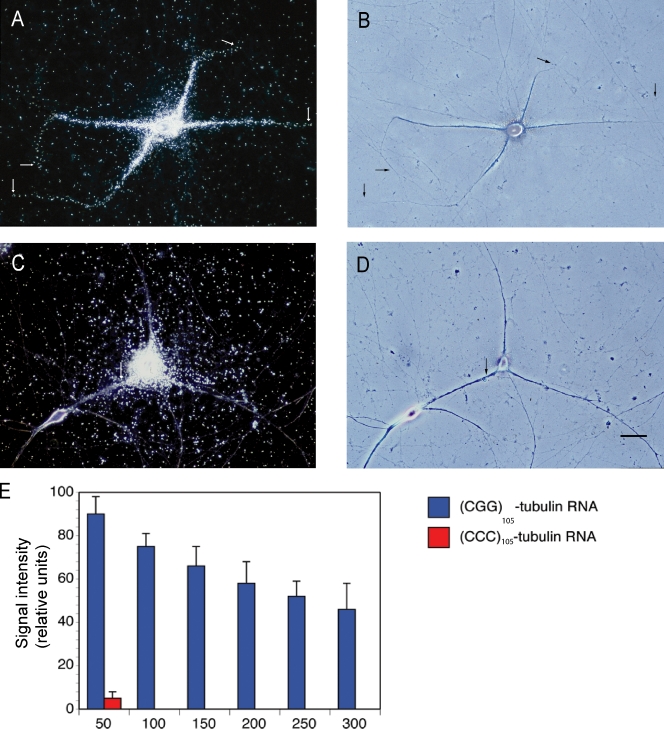
We speculated that CGG repeats may in fact be determinants of dendritic targeting. This speculation also takes into consideration the observation that CGG repeats, analogous to GA targeting motifs, feature noncanonical purine•purine pairs that are flanked by standard WC GC pairs. We addressed this question by generating chimeric RNAs in which a (CGG)24 segment was inserted into the 5′ UTR of α-tubulin mRNA. This mRNA is normally nondendritic; i.e., it remains restricted to neuronal somata (Bruckenstein et al., 1990; Mohr et al., 2001). A 24-unit repeat was chosen to represent the subthreshold, “healthy” range of CGG repeats in FMR1 mRNA that is found in the normal population (Brouwer et al., 2009). Fig. 9 shows that (CGG)24 α-tubulin RNA, but not (CCC)24 α-tubulin RNA, was delivered to distal dendritic domains. The results indicate that CGG repeat stem loops can serve as spatial codes to specify dendritic RNA targeting.
Figure 9.
Chimeric (CGG)24 α-tubulin RNA is transported to dendrites. (A and B) Microinjected chimeric (CGG)24 α-tubulin RNA was specifically delivered to dendrites, reaching distal dendritic tips. 10 neurons and 33 dendrites were analyzed. Arrows indicate the extent of dendritic labeling. (C and D) In contrast, microinjected chimeric (CCC)24 α-tubulin RNA failed to be transported along dendrites. 12 neurons and 36 dendrites were analyzed. Higher amounts of labeled RNA were injected in C and D to verify that somatic restriction is independent of injection amounts (see also Materials and methods). Bar, 50 µm. (E) Quantitative analysis: one-way ANOVA, Dunnett’s post-hoc analysis (comparison of all interval points between (CGG)24 α-tubulin RNA and (CCC)24 α-tubulin RNA): P < 0.0001 for interval points 50 µm and beyond. Error bars indicate SEM.
RNA stability
A crucial determinant in RNA biology, specifically including RNA transport, is RNA stability. For this reason, we have in this and previous work taken extensive precautions to monitor the relative stability of transcripts (Materials and methods; Fig. S5; Muslimov et al., 1997, 2004, 2006). Such precautions are of utmost importance, as any introduced mutation, for instance in a DTE, may carry the risk of differentially affecting RNA stability. This risk exists irrespective of the technique used to express nonnative RNAs in cells (i.e., injection, transfection of RNA or DNA constructs, or other). If a mutant RNA is inherently less stable than its WT counterpart, unreliable results will be obtained no matter how the RNAs were introduced or expressed in cells. We therefore find it advisable that relative RNA stabilities be rigorously monitored in any RNA transport study.
In a similar vein, caution is urged when localization is investigated after expressing nonnative double-stranded RNAs over prolonged periods of time, as for instance in transgenic animals (Khanam et al., 2007). Cellular defense mechanisms may be activated in such cases (Kaufman, 2000; Pe’ery and Mathews, 2000), and tandem GA pairs within double-stranded RNA contexts are known to be particularly prone to elicit such responses (Bevilacqua et al., 1998).
Discussion
Translational control is a key mechanism in the regulation of gene expression in eukaryotic cells (Mathews et al., 2007). In neurons, the local translation of select mRNAs at synapto-dendritic sites plays an important role in the implementation of long-term neuronal plasticity (Job and Eberwine, 2001; Kindler et al., 2005; Jambhekar and Derisi, 2007; Miyashiro et al., 2009). A major determinant of local protein-synthetic capacity, for example at a synapse, is the availability of a specific set of RNAs at that site. Therefore, a fundamental question in neuronal gene expression is presented by the need for RNAs to be selectively transported to their subcellular destination sites (Smith, 2004; Bramham and Wells, 2007; Dahm et al., 2007). How is spatial information encoded, and how is such information decoded through specific RNA motif recognition? Central to this question is the recognition of RNA spatial destination codes by cognate trans-acting factors (Smith, 2004).
Noncanonical motif recognition
We report here that in one neuronal RNA transport pathway, such recognition is mediated by interactions of the trans-acting protein hnRNP A2 with RNA motifs that feature noncanonical attributes. These motifs contain cores of noncanonical purine•purine base pairs. Tandem G•A/A•G pairs are often contiguous with a variable internal loop featuring unpaired nucleotides; in turn, this GA pair–internal loop element is flanked by standard WC pairs, often of the GC type. For the purpose of our analysis, we focused on one representative of dendritic regulatory RNAs (BC1 RNA) and on one representative of dendritic mRNAs (PKMζ mRNA).
We found that hnRNP A2 was required for the distal dendritic targeting of BC1 RNA and PKMζ mRNA. Conversion of GA motif noncanonical base pairs into standard WC base pairs prevented binding to hnRNP A2 and abolished distal dendritic targeting. Binding of BC1 RNA to hnRNP A2 was of high affinity, with a Kd in the nanomolar concentration range. A similar Kd has been reported for binding of hnRNP A2 to MBP mRNA, an mRNA that is localized in oligodendrocytes (Shan et al., 2000).
Remarkably, the binding affinity was exquisitely sensitive to concentrations of Mg2+ and Ca2+ ions. Relatively small concentration shifts—from 5 mM to 10 mM Mg2+ or from 1 mM to 5 mM Ca2+—resulted in an increase in binding affinity by more than two orders of magnitude. We posit that such affinity shifts may have functional relevance. GA motifs exist in a dynamic equilibrium between an extended and a tightly kinked conformation (Goody et al., 2004; Dennis and Omer, 2005; Lescoute et al., 2005). The kinked KT motif conformation is adopted at sufficiently high Mg2+ or Ca2+ concentrations in vitro (typically >5 mM; Matsumura et al., 2003; Goody et al., 2004; Schroeder et al., 2010), and the 5′ BC1 domain is known to be kinked under these conditions (Muslimov et al., 2006). The data therefore suggest that the BC1 GA motif conformation is dimorphic and dependent on divalent cation concentrations, and that the kinked conformation is recognized by hnRNP A2 with higher affinity.
The GA motif transition from the extended to the kinked conformation appears to proceed via a hinge-like motion on a fast timescale (Cojocaru et al., 2005a,b; Rázga et al., 2005, 2006). This “molecular-elbow” transition is promoted by the presence of divalent cations and by interactions with cognate proteins (Matsumura et al., 2003; Goody et al., 2004; Schroeder et al., 2010), two determinants that may act in concert (Turner et al., 2005). We suggest that brief, transient increases in local divalent cation availability may induce a rapid shift in the extended-kinked conformation equilibrium. Such a shift would allow cognate proteins to engage the motif at higher affinity, and possibly to lock the kinked conformation (Turner et al., 2005). Although it remains to be established in future work how such conformational changes, induced by divalent cations, may impact dendritic targeting, recent data indicate that chelation of intracellular Ca2+ (using BAPTA-AM) results in diminished distal dendritic delivery of BC1 RNA (unpublished data). The possibility is thus raised that Ca2+ may signal to dendritic RNA transport.
Ca2+ transients, including intracellular Ca2+ waves that originate in synaptic domains and propagate along the dendrites toward the soma (Larkum et al., 2003; Berridge, 2009), are obvious candidates for such signaling, especially as local transient Ca2+ increases can be significantly higher than global levels (Neher, 1998). Intracellular Mg2+ surges that occur after depolarization (Kato et al., 1998; Gotoh et al., 1999) may also modulate hnRNP A2 interactions with cargo RNAs. In the intracellular milieu, it is likely that additional proteins engage the targeting motif–hnRNP A2 complex (e.g., Purα; Johnson et al., 2006; Swanson and Orr, 2007), and that such assembly of RNA–protein ensembles will further fine-tune ion-dependent binding interactions and affinities.
CGG competition
In the RNA stem-loop GA motifs discussed here, noncanonical purine•purine pairings are requisite determinants for binding to hnRNP A2. Binding to hnRNP A2 has also been reported for CGG triplet repeat RNA motifs (Sofola et al., 2007; Swanson and Orr, 2007; Brouwer et al., 2009). CGG repeats form stem loops in which noncanonical G•G base pairs are flanked by standard WC GC pairs, resulting in stable RNA motif structures (Napierala et al., 2005; Zumwalt et al., 2007; Sobczak et al., 2010). Collectively, these observations suggested that spatial code GA motifs and CGG repeat stem-loops share relevant structural features (i.e., GC-flanked noncanonical purine•purine pairs). We therefore hypothesized that such GA motifs and CGG repeat motifs may engage in a dynamic competition for an essential cellular resource, hnRNP A2. This hypothesis was corroborated by experiments showing that (CGG)105 RNA, but not (CCC)105 RNA, effectively competed with BC1 RNA for binding to hnRNP A2, and that such CGG competition resulted in significantly reduced dendritic delivery.
CGG triplet repeats in the 5′ UTR of the FMR1 gene are polymorphic (Jacquemont et al., 2007; Brouwer et al., 2009). In the normal population, CGG repeat lengths range from 5 up to 55, with an average length of 30 repeats (Fu et al., 1991). Intergenerational repeat instability may result in triplet expansion to >55 CGG units. Repeat lengths of 55–200 CGG triplets are causative of FXTAS and other premutation disorders, whereas repeat lengths of >200 CGG triplets, referred to as full mutation, are causative of FXS. In the first case, FMR1 mRNA with the 5′ UTR CGG repeat expansion is expressed; in the second case, hypermethylation of the gene results in transcriptional silencing (Brouwer et al., 2009). Thus, although FXS is a protein loss-of-function disease (resulting from functional absence of FMRP), FXTAS has been referred to as an RNA gain-of-function disease (Jacquemont et al., 2007; Brouwer et al., 2009; Hagerman et al., 2010). Expanded CGG repeats in the premutation range appear to precipitate “RNA toxicity” (Jacquemont et al., 2007; Hagerman et al., 2010), and it has been suggested that partial sequestration of cellular factors (including hnRNP A2) by CGG repeat RNA may be an underlying cause of such toxicity (Iwahashi et al., 2006; Sofola et al., 2007; Swanson and Orr, 2007; Oostra and Willemsen, 2009). However, supporting experimental evidence was not available (Brouwer et al., 2009).
Our data support the CGG-premutation RNA gain-of-function hypothesis. GC-embedded noncanonical purine•purine RNA motifs are recognized by hnRNP A2, and such noncanonical attributes are displayed both by CGG repeat stem loops and targeting-determinant GA motifs. We suggest that competition for hnRNP A2 establishes a dynamic equilibrium between FMR1 mRNA CGG stem loops and dendritic RNA GA motifs. At normal repeat lengths of ∼30 CGG units, this equilibrium would be “healthy,” whereas beyond the 55 CGG unit threshold, hnRNP A2 binding would become unbalanced and skewed in favor of CGG repeat FMR1 mRNA. Increased CGG repeat lengths, coupled with significantly elevated levels of FMR1 mRNA in premutation cells, appear to be sufficient to cause co-sequestration of FMR1 mRNA and hnRNP A2 in nuclear inclusions that are observed in FXTAS (Tassone et al., 2000, 2004; Iwahashi et al., 2006; Hagerman et al., 2010). As a result of these alterations, we suggest that transport-competent neuronal RNAs are becoming increasingly deprived of a trans-acting factor that is essential for their delivery to dendritic domains. We therefore put forward the hypothesis—and invite its further experimental scrutiny—that cellular RNA transport deficiencies are underlying CGG repeat premutation disorders.
Non-protein-coding neuronal brain cytoplasmic (BC) RNAs are mediators of translational control of gene expression and have been implicated in the local regulation of protein synthesis at the synapse (Kindler et al., 2005; Wang and Tiedge, 2009; Iacoangeli et al., 2010). Impaired dendritic delivery of BC200 RNA has been observed in affected cortical areas of Alzheimer’s disease (AD) brains (Mus et al., 2007). It may therefore be tempting to speculate that BC RNA transport defects may be causally related to long-term neurodegenerative consequences that are observed in AD and FXTAS. However, there is one caveat, as dendritic transport not only of BC RNAs but also of relevant neuronal mRNAs may be negatively affected if levels of hnRNP A2 are reduced as a result of CGG repeat sequestration. Several neuronal mRNAs that bind to hnRNP A2 have been identified or predicted on the basis of structural features (Fig. 1; Munro et al., 1999; Muslimov et al., 2006; Tiedge, 2006; Gao et al., 2008). Currently, however, we are lacking a comprehensive understanding of the complement of dendritic mRNAs that rely on hnRNP A2 for targeting. Impaired dendritic delivery of some of such hnRNP A2–dependent neuronal mRNAs may contribute to phenotypical manifestations in FXTAS and related premutation diseases. For example, we show that hnRNP A2 is required for dendritic delivery of PKMζ mRNA, and that a GA motif with internal loop and flanking GC pairs in the 3′ UTR of the mRNA is requisite for recognition by hnRNP A2 and for dendritic transport. Future work will address the question of whether dendritic delivery of any such mRNAs is in competition with CGG repeat RNA.
Role of CGG motifs
An enigmatic aspect in trinucleotide repeat research has been the question of why such repeats, troublesome once expanded, have been retained over the course of recent evolution (Brouwer et al., 2009). What functional benefits may they offer that make their continued presence indispensable? Prompted by our finding that CGG repeat motifs compete with GA targeting motifs for binding to RNA transport factor hnRNP A2, we subsequently realized that such CGG RNA motifs can in fact function as spatial destination codes for dendritic delivery. These data further corroborate our hypothesis of a common denominator in CGG repeat codes and GA motif codes: WC-embedded noncanonical purine•purine pair motifs. We posit that standard size (∼30 units) CGG RNA stem-loop motifs and GA-type RNA targeting motifs are equal partners in dynamic competition for dendritic transport resources. CGG repeats that exceed a threshold (i.e., >55 CGG units) will become dominant and may outcompete relevant other neuronal RNAs for access to the essential transport resource hnRNP A2.
Materials and methods
Plasmids and RNA preparation
The following plasmids were used in this work: (1) pBCX607, to generate full-length WT BC1 RNA (Cheng et al., 1996; Muslimov et al., 1997); (2) pBC1IL-A:WC, to generate BC1 RNA in which noncanonical base pairs in the apical 5′ domain have been replaced with standard WC pairs (Fig. S1; Muslimov et al., 2006); (3) pPKMζ-(48–1982), to generate transport-competent segment nt 48–1982 of WT PKMζ mRNA (Muslimov et al., 2004); (4) pPKMζL-WC, equivalent to pPKMζ-(48–1982) except that noncanonical base pairs and unpaired nucleotides have been replaced with standard WC base pairs (mutant WC long, see Fig. S1); (5) pPKMζS-WC, equivalent to pPKMζ-(48–1982) except that noncanonical base pairs in the 3′ UTR have been replaced with standard WC base pairs, and unpaired nucleotides have been eliminated (mutant WC short, see Fig. S1); (6) pBC200, to generate full-length human BC200 RNA (Kondrashov et al., 2005); (7) pBSP, to generate a transport-competent segment of MBP mRNA (the plasmid was a gift from K. Ainger and J. Carson, University of Connecticut Health Center, Farmington, CT) and was constructed as a rat MBP SalI 666 to PvuII 953 insert in pBluescriptII (SK+); (8) p(CGG)105, to generate (CGG)105 RNA; (9) p(CCC)105, to generate (CCC)105 RNA ((CGG)105 or (CCC)105 DNA was inserted into pBluescript SK (+) between the KpnI and DraII sites); (10) pSL300, to generate irrelevant random-sequence RNA (Muslimov et al., 1997); (11) p(CGG)24-tubulin, to generate (CGG)24 α-tubulin chimeric RNA; (12) p(CCC)24-tubulin, to generate (CCC)24 α-tubulin chimeric RNA: both DNA fragments ((CGG)24 α-tubulin and (CCC)24 α-tubulin) were cloned into pBluescript SK (+) between the XbaI and Asp718 sites; (13) pET-9c, to express recombinant full-length hnRNP A2 (plasmid was a gift from R. Smith, University of Queensland, Brisbane, Australia; Munro et al., 1999).
Plasmids were verified by sequencing. Plasmids 3, 4, 5, 11, and 12 were linearized with XbaI, plasmid 7 with PvuII, plasmids 8 and 9 with DraII, plasmid 10 with KpnI, and all other plasmids with DraI. 35S-labeled RNAs were generated by in vitro transcription, and size and integrity of all transcripts were ascertained by PAGE (Muslimov et al., 1997, 2002, 2004, 2006).
Expression of recombinant proteins
Recombinant hnRNP A2 was expressed and purified as described previously (Munro et al., 1999). In brief, recombinant protein was purified by chromatography on diethylaminoethyl cellulose (GE Healthcare), followed by chromatography on Sephacryl S-300 (GE Healthcare). Samples were further purified on a C4 reverse-phase HPLC column (Vydac) using a linear 10–50% acetonitrile gradient in 0.1% trifluoroacetic acid. Protein was lyophilized and finally dissolved in water. Identity and purity of hnRNP A2 was established by SDS-PAGE and by electrospray mass spectrometry on a Sciex 165 spectrometer (PerkinElmer).
Recombinant eIF4G (central RNA-binding domain eIF4G737-1116) was generated from plasmid pET28(His6-eIF4GI737-1116) and purified by FPLC using a Mono S column (GE Healthcare). Linear gradients of 100 mM to 1 M KCl were used to elute eIF4G737–1116 at ∼290 mM KCl (Lin et al., 2008).
Native PAGE and EMSA
Native PAGE was performed in 8% polyacrylamide gels (19:1 acrylamide/bisacrylamide) in 90 mM Tris-borate, pH 8.3, in the presence of 15 mM MgCl2 at room temperature for 12 h at 15 V (Goody et al., 2004; Muslimov et al., 2006). EMSA and supershift experiments were performed as described previously (Wang et al., 2002; Muslimov et al., 2006; Lin et al., 2008). In brief, 32P-labeled RNA probes (50,000 cpm per reaction) were heated for 10 min at 70°C, cooled for 5 min at room temperature, and incubated together with proteins in binding buffer (300 mM KCl, 5 mM MgCl2, 2 mM DTT, 5% glycerol, and 20 mM Hepes, pH 7.6) for 20 min at room temperature. Results were quantified using a Storm 860 phosphorimaging system with ImageQuant software (GE Healthcare). Antibodies directed against hnRNP A2 (Ma et al., 2002) were provided by R. Smith (University of Queensland, Brisbane, Australia).
To establish binding affinities, fractions of free versus bound RNA were measured by phosphorimaging. The data were fitted to a modified version of the Hill equation to determine equilibrium constants (Ryder et al., 2008; Chao et al., 2010).
Cell culture, RNA microinjection, and in situ hybridization
Primary cultures of sympathetic neurons were generated and maintained as described previously (Higgins et al., 1991; Muslimov et al., 1997, 2004, 2006). In brief, superior cervical ganglia from embryonic day 19–21 Sprague Dawley rat embryos were dissociated, and neurons were maintained on glass coverslips. Dendritic growth was induced by supplementing the medium with basement membrane extract on the third day in vitro. 1 µM cytosine arabinofuranoside was used on the second and fifth days after plating to minimize nonneuronal proliferation. Transport experiments were performed with neurons maintained in culture for 4 wk. Characteristics of sympathetic neurons in primary culture have been described previously (Higgins et al., 1991). Work with animals was approved by the Institutional Animal Care and Use Committee.
Microinjection of radiolabeled RNA has been described previously (Muslimov et al., 1997, 2004, 2006). Our continued preference for this methodology has been motivated by the following considerations. (a) Radiolabeled RNA was used for injection because it has been our experience that with nonradioactive labels (e.g., fluorescent and digoxigenin labels), the differential character of dendritic transport could not always be as clearly and reproducibly documented as with radiolabels. We believe that this may be caused by the fact that in complex RNA motifs (such as GA/KT motifs), most nucleotide interactions, particularly noncanonical ones, are indispensable and therefore exquisitely sensitive to steric hindrance by nonnative side groups. (b) Microinjection was preferred over other delivery methods because it allows unparalleled control over amounts of RNA introduced into cells (Muslimov et al., 2006). Over the past several years, microinjection has increasingly and routinely been used to introduce RNA into nerve cells (Shan et al., 2003; Schratt et al., 2006; Tübing et al., 2010).
RNAs were 35S-radiolabeled at 3 × 106 cpm/µl and were microinjected into the perikaryal regions of sympathetic neurons in culture at volumes of a few femtoliters per pulse (Muslimov et al., 1997, 2006). We occasionally performed nuclear injections to check if nuclear experience of a transcript was targeting-relevant. No such case was observed. As before (Muslimov et al., 1997, 2006), we varied injection amounts over a concentration range of at least one order of magnitude to ensure that observed dendritic targeting patterns were concentration-independent. (CGG)105 and (CCC)105 RNAs were introduced at intracellular concentrations that were comparable to levels of FMR1 mRNA in premutation cells. Previous work (Tassone et al., 2000) has shown that normal expression levels of FMR1 mRNA are comparable to levels of β-glucuronidase (GUS) mRNA, a commonly used PCR standard. In premutation cells, transcription of FMR1 mRNA is strongly increased, resulting in expression levels that are elevated by a mean factor of eight for CGG repeat numbers of 100–200 (Tassone et al., 2000; Hagerman et al., 2010). With this information, we estimate that relative intracellular levels of (CGG)105 or (CCC)105 repeat RNAs, after microinjection into sympathetic neurons, were as follows. Low-amount injection routine: repeat RNA levels equivalent to or lower than (by a factor of at most four) premutation FMR1 mRNA levels. Medium-amount injection routine: repeat RNA levels equivalent to or higher than (by a factor of at most two) premutation FMR1 mRNA levels. High-amount injection routine: repeat RNA levels higher than (by a factor of at most six) premutation FMR1 mRNA levels. Data from high-amount injection routine experiments are not presented in the figures and were not entered into quantitative analyses.
To monitor RNA stability, we assessed average integrated total signal intensities per injected cell for each transcript, as described previously (Muslimov et al., 2006). This intensity, i.e., the average sum of somatic and dendritic signal intensities, was used as an indicator of intracellular RNA stability after injection and over the period of incubation (Muslimov et al., 2006). No significant differences were observed for a microinjected RNA or its derivatives under the experimental conditions used in this work (including hnRNP A2 antisense, CGG competition).
In addition, RNA stability was monitored by PAGE (Muslimov et al., 1997, 2006). Fig. S5 shows that (CGG)105 RNA and (CCC)105 RNA did not significantly degrade when incubated with rat brain extract for up to 4 h, the maximal incubation time used in RNA transport experiments. The data also show that (CGG)105 RNA and (CCC)105 RNA did not differ from each other in their relative stabilities when incubated with brain extract.
In situ hybridization with cultured neurons was performed with 35S-labeled RNA probes (Muslimov et al., 1998, 2004).
Treatment of neurons with hnRNP A2 antisense oligonucleotides
Antisense oligodeoxyribonucleotides were complementary to a region containing the translational start site of hnRNP A2 (Kwon et al., 1999; Munro et al., 1999). Sympathetic neurons in culture were incubated in medium containing 8 µM oligonucleotides (antisense or scrambled-sequence) for at least 18 h (Kwon et al., 1999; Munro et al., 1999; Shan et al., 2003). Oligonucleotides were obtained from Sigma-Aldrich. hnRNP A2 levels in treated neurons were established by Western blot analysis (Shan et al., 2003). Proteins were resolved by SDS-PAGE and were blotted onto Millipore Immobilon-P PVDF membranes (GenHunter). For development, a mouse antibody against a peptide segment in Exon 7 of hnRNP A2 (Ma et al., 2002) was used at 1:10,000 dilution. A rabbit antibody against α-tubulin (Sigma Aldrich) was used as a control (1:2,000 dilution).
Microscopy and image analysis
A Microphot-FXA microscope (Nikon) was used for data acquisition (Muslimov et al., 2006). Neurons were imaged at room temperature using dark-field and phase-contrast optics. The following objective lenses were used: Plan-Fluor 10×/0.30, 160 mechanical tube length/0.17 cover glass thickness; PhC Plan 20×/0.50, DL 160/0.17; Plan 20×/0.50, differential interference contrast (DIC) 160/0.17; and Ph3 DL Plan 40×/0.65, 160/0.17. Digital images were acquired with a 5-megapixel charge-coupled device (CCD) camera (Digital Sight DS-Fi1; Nikon), or with a cooled CCD camera (CoolSNAP HQ, Roper Scientific). Image analysis was performed using MetaMorph software (Molecular Devices), and illustrations were generated using Photoshop and Illustrator software (Adobe). For further details, see Muslimov et al. (2006).
To establish RNA distribution profiles along dendrites, silver grain densities were measured along dendritic shafts at 50-µm interval points, up to a maximal distance of 300 µm (Muslimov et al., 2006). Signal intensities at 0-µm interval points (base of dendrites) were assigned a relative value of 100; mean absolute signal intensities did not substantially differ at this interval point between wild-type and respective mutant RNAs. Two to four dendrites were analyzed per neuron (typically ≥10 neurons per experiment).
Statistical analyses were performed using SPSS software. All experiments described in this work were performed at least four times, unless noted otherwise in figure legends. Error bars represent SEM.
Online supplemental material
Fig. S1 shows secondary structures of targeting-relevant domains in BC1 RNA and PKMζ mRNA, and respective derivatives used in this work. Fig. S2 documents distal dendritic delivery of human BC200 RNA. Fig. S3 provides additional confirmation that interactions of BC1 RNA and PKMζ mRNA with hnRNP A2 are specific. Fig. S4 verifies that application of hnRNP A2 antisense oligonucleotides results in reduced levels of hnRNP A2 in sympathetic neurons in culture. Fig. S5 documents comparable relative stabilities of (CGG)105 and (CCC)105 RNAs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010027/DC1.
Acknowledgments
We thank Dr. Jeremy Weedon (State University of New York Brooklyn Scientific Computing Center) for statistical consultation, and members of the Tiedge, Wong, and Fenton laboratories for advice and discussion.
This work was supported in part by National Institutes of Health grants NS046769 and DA026110 (to H. Tiedge).
Footnotes
Abbreviations used in this paper:
- BC
- brain cytoplasmic
- DTE
- dendritic targeting element
- eIF4G
- eukaryotic initiation factor 4G
- EMSA
- electrophoretic mobility shift assay
- FMR
- fragile X mental retardation
- FXS
- fragile X syndrome
- FXTAS
- fragile X–associated tremor/ataxia syndrome
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- KT
- K-turn
- MBP
- myelin basic protein
- UTR
- untranslated region
- WC
- Watson-Crick
- WT
- wild type
References
- Berridge M.J. 2009. Inositol trisphosphate and calcium signalling mechanisms. Biochim. Biophys. Acta. 1793:933–940 10.1016/j.bbamcr.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Bevilacqua P.C., George C.X., Samuel C.E., Cech T.R. 1998. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 37:6303–6316 10.1021/bi980113j [DOI] [PubMed] [Google Scholar]
- Blichenberg A., Schwanke B., Rehbein M., Garner C.C., Richter D., Kindler S. 1999. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J. Neurosci. 19:8818–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg A., Rehbein M., Müller R., Garner C.C., Richter D., Kindler S. 2001. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur. J. Neurosci. 13:1881–1888 10.1046/j.0953-816x.2001.01565.x [DOI] [PubMed] [Google Scholar]
- Bramham C.R., Wells D.G. 2007. Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8:776–789 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- Brouwer J.R., Willemsen R., Oostra B.A. 2009. Microsatellite repeat instability and neurological disease. Bioessays. 31:71–83 10.1002/bies.080122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckenstein D.A., Lein P.J., Higgins D., Fremeau R.T., Jr 1990. Distinct spatial localization of specific mRNAs in cultured sympathetic neurons. Neuron. 5:809–819 10.1016/0896-6273(90)90340-L [DOI] [PubMed] [Google Scholar]
- Chao J.A., Patskovsky Y., Patel V., Levy M., Almo S.C., Singer R.H. 2010. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 24:148–158 10.1101/gad.1862910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.G., Tiedge H., Brosius J. 1996. Identification and characterization of BC1 RNP particles. DNA Cell Biol. 15:549–559 10.1089/dna.1996.15.549 [DOI] [PubMed] [Google Scholar]
- Cojocaru V., Klement R., Jovin T.M. 2005a. Loss of G-A base pairs is insufficient for achieving a large opening of U4 snRNA K-turn motif. Nucleic Acids Res. 33:3435–3446 10.1093/nar/gki664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V., Nottrott S., Klement R., Jovin T.M. 2005b. The snRNP 15.5K protein folds its cognate K-turn RNA: a combined theoretical and biochemical study. RNA. 11:197–209 10.1261/rna.7149605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm R., Kiebler M.A. 2007. RNA localization: new roles for an evolutionarily ancient mechanism. Semin. Cell Dev. Biol. 18:161–162 10.1016/j.semcdb.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Dahm R., Kiebler M., Macchi P. 2007. RNA localisation in the nervous system. Semin. Cell Dev. Biol. 18:216–223 10.1016/j.semcdb.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Dennis P.P., Omer A. 2005. Small non-coding RNAs in Archaea. Curr. Opin. Microbiol. 8:685–694 10.1016/j.mib.2005.10.013 [DOI] [PubMed] [Google Scholar]
- Eberwine J., Belt B., Kacharmina J.E., Miyashiro K. 2002. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem. Res. 27:1065–1077 10.1023/A:1020956805307 [DOI] [PubMed] [Google Scholar]
- Fu Y.H., Kuhl D.P., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S., Verkerk A.J., Holden J.J., Fenwick R.G., Jr, Warren S.T., et al. 1991. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 67:1047–1058 10.1016/0092-8674(91)90283-5 [DOI] [PubMed] [Google Scholar]
- Gao Y., Tatavarty V., Korza G., Levin M.K., Carson J.H. 2008. Multiplexed dendritic targeting of α calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol. Biol. Cell. 19:2311–2327 10.1091/mbc.E07-09-0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Raught B., Sonenberg N. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963 10.1146/annurev.biochem.68.1.913 [DOI] [PubMed] [Google Scholar]
- Goody T.A., Melcher S.E., Norman D.G., Lilley D.M. 2004. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 10:254–264 10.1261/rna.5176604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh H., Kajikawa M., Kato H., Suto K. 1999. Intracellular Mg2+ surge follows Ca2+ increase during depolarization in cultured neurons. Brain Res. 828:163–168 10.1016/S0006-8993(99)01298-6 [DOI] [PubMed] [Google Scholar]
- Hagerman R., Hoem G., Hagerman P. 2010. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol Autism. 1:12 10.1186/2040-2392-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D., Lein P.J., Osterhout D.J., Johnson M.I. 1991. Tissue culture of mammalian autonomic neurons. Culturing Nerve Cells. Banker G., Goslin K., MIT Press, Cambridge: 177–205 [Google Scholar]
- Hoek K.S., Kidd G.J., Carson J.H., Smith R. 1998. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 37:7021–7029 10.1021/bi9800247 [DOI] [PubMed] [Google Scholar]
- Iacoangeli A., Bianchi R., Tiedge H. 2010. Regulatory RNAs in brain function and disorders. Brain Res. 1338:36–47 10.1016/j.brainres.2010.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi C.K., Yasui D.H., An H.J., Greco C.M., Tassone F., Nannen K., Babineau B., Lebrilla C.B., Hagerman R.J., Hagerman P.J. 2006. Protein composition of the intranuclear inclusions of FXTAS. Brain. 129:256–271 10.1093/brain/awh650 [DOI] [PubMed] [Google Scholar]
- Jacquemont S., Hagerman R.J., Hagerman P.J., Leehey M.A. 2007. Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol. 6:45–55 10.1016/S1474-4422(06)70676-7 [DOI] [PubMed] [Google Scholar]
- Jambhekar A., Derisi J.L. 2007. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 13:625–642 10.1261/rna.262607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Zarnescu D.C., Zhang F., Pearson C.E., Lucchesi J.C., Moses K., Warren S.T. 2003. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 39:739–747 10.1016/S0896-6273(03)00533-6 [DOI] [PubMed] [Google Scholar]
- Jin P., Duan R., Qurashi A., Qin Y., Tian D., Rosser T.C., Liu H., Feng Y., Warren S.T. 2007. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 55:556–564 10.1016/j.neuron.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C., Eberwine J. 2001. Localization and translation of mRNA in dendrites and axons. Nat. Rev. Neurosci. 2:889–898 10.1038/35104069 [DOI] [PubMed] [Google Scholar]
- Johnson E.M., Kinoshita Y., Weinreb D.B., Wortman M.J., Simon R., Khalili K., Winckler B., Gordon J. 2006. Role of Pur α in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J. Neurosci. Res. 83:929–943 10.1002/jnr.20806 [DOI] [PubMed] [Google Scholar]
- Kato H., Gotoh H., Kajikawa M., Suto K. 1998. Depolarization triggers intracellular magnesium surge in cultured dorsal root ganglion neurons. Brain Res. 779:329–333 10.1016/S0006-8993(97)01232-8 [DOI] [PubMed] [Google Scholar]
- Kaufman R.J. 2000. The double-stranded RNA-activated protein kinase PKR. Translational Control of Gene Expression. Sonenberg N., Hershey J.W.B., Mathews M.B., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 503–527 [Google Scholar]
- Khanam T., Raabe C.A., Kiefmann M., Handel S., Skryabin B.V., Brosius J. 2007. Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PLoS ONE. 2:e961 10.1371/journal.pone.0000961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S., Wang H., Richter D., Tiedge H. 2005. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21:223–245 10.1146/annurev.cellbio.21.122303.120653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A.V., Kiefmann M., Ebnet K., Khanam T., Muddashetty R.S., Brosius J. 2005. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J. Mol. Biol. 353:88–103 10.1016/j.jmb.2005.07.049 [DOI] [PubMed] [Google Scholar]
- Kwon S., Barbarese E., Carson J.H. 1999. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J. Cell Biol. 147:247–256 10.1083/jcb.147.2.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum M.E., Watanabe S., Nakamura T., Lasser-Ross N., Ross W.N. 2003. Synaptically activated Ca2+ waves in layer 2/3 and layer 5 rat neocortical pyramidal neurons. J. Physiol. 549:471–488 10.1113/jphysiol.2002.037614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A., Leontis N.B., Massire C., Westhof E. 2005. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 33:2395–2409 10.1093/nar/gki535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Pestova T.V., Hellen C.U., Tiedge H. 2008. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell. Biol. 28:3008–3019 10.1128/MCB.01800-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A.S., Moran-Jones K., Shan J., Munro T.P., Snee M.J., Hoek K.S., Smith R. 2002. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J. Biol. Chem. 277:18010–18020 10.1074/jbc.M200050200 [DOI] [PubMed] [Google Scholar]
- Martignetti J.A., Brosius J. 1993a. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc. Natl. Acad. Sci. USA. 90:11563–11567 10.1073/pnas.90.24.11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti J.A., Brosius J. 1993b. Neural BC1 RNA as an evolutionary marker: guinea pig remains a rodent. Proc. Natl. Acad. Sci. USA. 90:9698–9702 10.1073/pnas.90.20.9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews M.B., Sonenberg N., Hershey J.W.B. 2007. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 934 pp [Google Scholar]
- Matsumura S., Ikawa Y., Inoue T. 2003. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 31:5544–5551 10.1093/nar/gkg760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro K.Y., Bell T.J., Sul J.Y., Eberwine J. 2009. Subcellular neuropharmacology: the importance of intracellular targeting. Trends Pharmacol. Sci. 30:203–211 10.1016/j.tips.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Mohr E., Richter D. 2003. Molecular determinants and physiological relevance of extrasomatic RNA localization in neurons. Front. Neuroendocrinol. 24:128–139 10.1016/S0091-3022(03)00011-6 [DOI] [PubMed] [Google Scholar]
- Mohr E., Prakash N., Vieluf K., Fuhrmann C., Buck F., Richter D. 2001. Vasopressin mRNA localization in nerve cells: characterization of cis-acting elements and trans-acting factors. Proc. Natl. Acad. Sci. USA. 98:7072–7079 10.1073/pnas.111146598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro T.P., Magee R.J., Kidd G.J., Carson J.H., Barbarese E., Smith L.M., Smith R. 1999. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 274:34389–34395 10.1074/jbc.274.48.34389 [DOI] [PubMed] [Google Scholar]
- Mus E., Hof P.R., Tiedge H. 2007. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 104:10679–10684 10.1073/pnas.0701532104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Santi E., Homel P., Perini S., Higgins D., Tiedge H. 1997. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 17:4722–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Banker G., Brosius J., Tiedge H. 1998. Activity-dependent regulation of dendritic BC1 RNA in hippocampal neurons in culture. J. Cell Biol. 141:1601–1611 10.1083/jcb.141.7.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Titmus M., Koenig E., Tiedge H. 2002. Transport of neuronal BC1 RNA in Mauthner axons. J. Neurosci. 22:4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Nimmrich V., Hernandez A.I., Tcherepanov A., Sacktor T.C., Tiedge H. 2004. Dendritic transport and localization of protein kinase Mzeta mRNA: implications for molecular memory consolidation. J. Biol. Chem. 279:52613–52622 10.1074/jbc.M409240200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov I.A., Iacoangeli A., Brosius J., Tiedge H. 2006. Spatial codes in dendritic BC1 RNA. J. Cell Biol. 175:427–439 10.1083/jcb.200607008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napierala M., Michalowski D., de Mezer M., Krzyzosiak W.J. 2005. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 33:451–463 10.1093/nar/gki186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher E. 1998. Vesicle pools and Ca2+ microdomains: new tools for understanding their roles in neurotransmitter release. Neuron. 20:389–399 10.1016/S0896-6273(00)80983-6 [DOI] [PubMed] [Google Scholar]
- Oostra B.A., Willemsen R. 2009. FMR1: a gene with three faces. Biochim. Biophys. Acta. 1790:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastalkova E., Serrano P., Pinkhasova D., Wallace E., Fenton A.A., Sacktor T.C. 2006. Storage of spatial information by the maintenance mechanism of LTP. Science. 313:1141–1144 10.1126/science.1128657 [DOI] [PubMed] [Google Scholar]
- Pe’ery T., Mathews M.B. 2000. Viral translation strategies and host defense mechanisms. Translational Control of Gene Expression. Sonenberg N., Hershey J.W.B., Mathews M.B., Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory: 400–424 [Google Scholar]
- Pestova T.V., Shatsky I.N., Hellen C.U. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T., Lorsch J.R., Hellen C.U.T. 2007. The mechanism of translation initiation in eukaryotes. Translational Control in Biology and Medicine. Mathews M.B., Sonenberg N., Hershey J.W.B., Cold Spring Harbor Laboratory Press, Cold Spring Harbor: 87–128 [Google Scholar]
- Rázga F., Koca J., Sponer J., Leontis N.B. 2005. Hinge-like motions in RNA kink-turns: the role of the second a-minor motif and nominally unpaired bases. Biophys. J. 88:3466–3485 10.1529/biophysj.104.054916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rázga F., Zacharias M., Réblová K., Koca J., Sponer J. 2006. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure. 14:825–835 10.1016/j.str.2006.02.012 [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky T.S., Kopylov A.M., Brosius J., Hüttenhofer A. 2001. Neuronal BC1 RNA structure: evolutionary conversion of a tRNA(Ala) domain into an extended stem-loop structure. RNA. 7:722–730 10.1017/S1355838201002485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder S.P., Recht M.I., Williamson J.R. 2008. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 488:99–115 10.1007/978-1-60327-475-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. 2006. A brain-specific microRNA regulates dendritic spine development. Nature. 439:283–289 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- Schroeder K.T., McPhee S.A., Ouellet J., Lilley D.M. 2010. A structural database for k-turn motifs in RNA. RNA. 16:1463–1468 10.1261/rna.2207910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Friedman E.L., Kenney J., Taubenfeld S.M., Zimmerman J.M., Hanna J., Alberini C., Kelley A.E., Maren S., Rudy J.W., et al. 2008. PKMzeta maintains spatial, instrumental, and classically conditioned long-term memories. PLoS Biol. 6:2698–2706 10.1371/journal.pbio.0060318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J., Moran-Jones K., Munro T.P., Kidd G.J., Winzor D.J., Hoek K.S., Smith R. 2000. Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J. Biol. Chem. 275:38286–38295 10.1074/jbc.M007642200 [DOI] [PubMed] [Google Scholar]
- Shan J., Munro T.P., Barbarese E., Carson J.H., Smith R. 2003. A molecular mechanism for mRNA trafficking in neuronal dendrites. J. Neurosci. 23:8859–8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. 2004. Moving molecules: mRNA trafficking in Mammalian oligodendrocytes and neurons. Neuroscientist. 10:495–500 10.1177/1073858404266759 [DOI] [PubMed] [Google Scholar]
- Sobczak K., Michlewski G., de Mezer M., Kierzek E., Krol J., Olejniczak M., Kierzek R., Krzyzosiak W.J. 2010. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 285:12755–12764 10.1074/jbc.M109.078790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofola O.A., Jin P., Qin Y., Duan R., Liu H., de Haro M., Nelson D.L., Botas J. 2007. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 55:565–571 10.1016/j.neuron.2007.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M.S., Orr H.T. 2007. Fragile X tremor/ataxia syndrome: blame the messenger! Neuron. 55:535–537 10.1016/j.neuron.2007.07.032 [DOI] [PubMed] [Google Scholar]
- Tassone F., Hagerman R.J., Taylor A.K., Gane L.W., Godfrey T.E., Hagerman P.J. 2000. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 66:6–15 10.1086/302720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F., Iwahashi C., Hagerman P.J. 2004. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS). RNA Biol. 1:103–105 10.4161/rna.1.2.1035 [DOI] [PubMed] [Google Scholar]
- Tiedge H. 2006. K-turn motifs in spatial RNA coding. RNA Biol. 3:133–139 10.4161/rna.3.4.3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H., Chen W., Brosius J. 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 13:2382–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tübing F., Vendra G., Mikl M., Macchi P., Thomas S., Kiebler M.A. 2010. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J. Neurosci. 30:4160–4170 10.1523/JNEUROSCI.3537-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B., Melcher S.E., Wilson T.J., Norman D.G., Lilley D.M. 2005. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 11:1192–1200 10.1261/rna.2680605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Tiedge H. 2009. Dendrites: localized translation. Encyclopedia of Neuroscience. Squire Larry R., editor Academic Press, San Diego: 431–435 [Google Scholar]
- Wang H., Iacoangeli A., Popp S., Muslimov I.A., Imataka H., Sonenberg N., Lomakin I.B., Tiedge H. 2002. Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 22:10232–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Iacoangeli A., Lin D., Williams K., Denman R.B., Hellen C.U.T., Tiedge H. 2005. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 171:811–821 10.1083/jcb.200506006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumwalt M., Ludwig A., Hagerman P.J., Dieckmann T. 2007. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 4:93–100 10.4161/rna.4.2.5039 [DOI] [PubMed] [Google Scholar]