Abstract
Aquaporins are multifunctional membrane channels which belong to the family of major intrinsic proteins (MIPs) and are best known for their ability to facilitate the movement of water. In the present study, earlier results from microarray experiments were followed up. These experiments had suggested that, in barley (Hordeum vulgare L.), aquaporin family members are expressed in distinct patterns during leaf development. Real-time PCR and in situ hybridization were used to analyse the level and tissue-distribution of expression of candidate aquaporins, focusing on plasma membrane and tonoplast intrinsic proteins (PIPs, TIPs). Water channel function of seven aquaporins, whose transcripts were the most abundant and the most variable, was tested through expression in yeast and, in part, through expression in oocytes. All PIP1 and PIP2 subfamily members changed in expression during leaf development, with expression being much higher or lower in growing compared with mature tissue. The same applied to those TIPs which were expressed at detectable levels. Specific roles during leaf development are proposed for particular aquaporins.
Keywords: barley (Hordeum vulgare L.), Casparian bands, cell expansion, leaf growth, major intrinsic protein, transpiration stream
Introduction
Aquaporins belong to the family of major intrinsic proteins (MIPs). In higher plants, they group into plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), Nod26-like proteins (NIPs), small, basic intrinsic proteins (SIPs), and the recently classified X-intrinsic proteins (XIPs) (Johanson et al., 2001; Danielson and Johanson, 2008). Aquaporins are multifunctional membrane channels, but they are best known for their ability to facilitate the movement of water across membranes (Johansson et al., 2000; Kaldenhoff and Fischer, 2006; Maurel, 2007; Maurel et al., 2008). Water channel activity is displayed in particular by members of the PIP2 subgroup of PIPs and TIPs.
The physiological role of individual aquaporin isoforms in plants is still a matter of debate, (Flexas et al., 2006; Hachez et al., 2006b; Ma et al., 2006; Takano et al., 2006; Katsuhara et al., 2008; Heinen et al., 2009). Most studies which have investigated the water channel function of aquaporins have focused on root water uptake or the response of plants to environmental stresses which cause a shortage in water supply (for reviews see Maurel, 2007; Maurel et al., 2008). Few studies have addressed the role of aquaporins in growth, particularly leaf cell expansion (Hukin et al., 2002; Sakurai et al., 2005; Wei et al., 2007; Hachez et al., 2008; Obroucheva and Sin'kevich, 2010). The rather diffuse arrangement of the developmental stages in leaves of dicotyledonous plants (e.g. Arabidopsis) makes such an analysis difficult, in contrast to the leaves of grasses where developmental stages occur within spatially well-defined zones. In addition, the leaf cells of grasses develop while they are enclosed by sheaths of older leaves and experience a micro-climate and light environment which is very different from that experienced by cells of the mature blade. This may provide a clue as to the regulation of expression of MIPs during leaf development. Hachez et al. (2008), studying PIPs in maize (Zea mays) observed that expression generally increased during leaf development. By contrast, Wei et al. (2007) observed that a PIP1 isoform was expressed almost exclusively in growing, but not in mature leaf tissue of barley, whereas Schünmann et al. (1994) reported similar developmental changes in the expression of a TIP. Apart from the latter studies, there exists little information about the development- and growth-dependent expression in leaves of TIPs and how it relates to the expression of PIPs (Chaumont et al., 1998; Frangne et al., 2001). PIPs, being localized at the plasma membrane facilitate the movement of water into and out of cells, whereas TIPs, being localized at the tonoplast, facilitate rapid osmotic equilibration between the vacuole and cytosol (Maurel et al., 1993). Regardless of whether water traverses cells by passing plasma membrane and tonoplast in series or bypasses the vacuole along the cytosol, one would expect overlapping patterns of expression for some of the PIP and TIP family members.
As part of a previous study on cuticle development in barley leaves (Richardson et al., 2007a) microarray analyses were carried out in which the expression of genes between growing and non-growing leaf regions was compared. This study also provided information on the expression of MIPs. The data suggested that most MIPs are expressed differentially during leaf development. The aim of the present study was to follow up these observations through a more detailed and quantitative analysis of expression in leaf zones and through functional characterization and determination of tissue-specificity of expression of a selected group of candidate MIPs. The focus was on PIPs and TIPs since these are most likely to have a role in controlling the water flow associated with cell elongation growth and transpiration. Those candidate genes which had shown the largest differences in expression between leaf regions were selected for detailed analyses. To obtain some information about the leaf specificity of expression and function of candidate MIPs, expression was also analysed in mature and growing root tissue. Together, these analyses made it possible to allocate particular roles during leaf development to specific aquaporin isoforms and to propose factors which influence the expression of these aquaporins in barley. Most of the previous studies on barley aquaporins have been carried out by Katsuhara and colleagues (Hanba et al., 2004; Katsuhara et al., 2002, 2003a; Katsuhara and Hanba, 2008). These studies focused on the water channel HvPIP2;1, while two further barley aquaporins, HvPIP1;3 and HvPIP1;5 were found not to display water channel activity. Wei et al. (2007) observed that the barley PIP1 HvPIP1;6 (which is identical to the barley MIP annotated as HvPIP1;1) displayed some water channel activity. Since the water channel function of these four barley PIPs had been tested and published previously, these genes were not selected for functionality tests in the present study, regardless of the pattern of expression.
Materials and methods
Plant growth
Barley (Hordeum vulgare L. cv. Golf (Svalöf Weibull AB, Svalöv, Sweden)) plants were grown hydroponically in a growth chamber as described previously under a 16/8 h light/dark period (Knipfer and Fricke, 2010). Plants were analysed when they were 14–16-d-old. At this developmental stage, the major root system consisted of seminal roots, with adventitious roots starting to develop. Leaf three was elongating at 2–3 mm h−1 and was 12–19 cm long, of which 7–8 cm were enclosed in and 5–11 cm emerged from the sheath of leaf two.
Plant harvest
Work by Katsuhara et al. (2003b) and Katsuhara (2007) has shown that expression of barley aquaporins in roots can differ between day and night. It was possible that this would also be the case for leaves. Trying to apply all of the present analyses to both time periods would have been beyond the scope of the present study and would have compromised the range of approaches taken and the number of candidate aquaporins analysed. Also, most of the leaf growth (>80%) and water uptake (>95%) of barley plants occurred during the day period and it was therefore decided to restrict analyses and harvest of plants to a 2 h-time period, 4–6 h into the photoperiod. Leaf elongation and transpiration rate were at a steady and (near-) maximum value during this period (not shown).
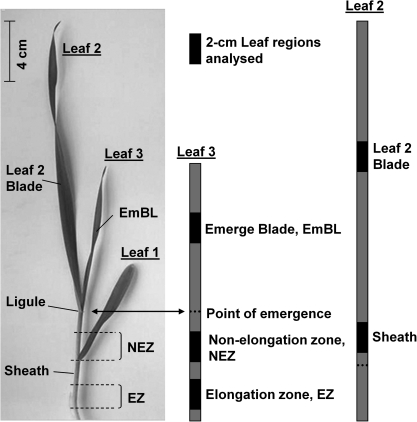
Leaf samples were taken from up to three developmental zones along the elongating leaf three of barley, the elongation zone (EZ), the non-elongation zone (NEZ), and the emerged blade (EmBL). In addition, a leaf sample was taken from the mature blade and from midway along the sheath of leaf two (L2 and Sh). Leaf samples consisted of 2-cm-long segments which were taken from the centre region of the respective zone (Fig. 1). In some experiments, root samples were analysed. These were taken from mature and growing tissue and from seminal and adventitious roots (Boscari et al., 2009; Knipfer et al., 2011). Although adventitious roots contributed less than 10% to water uptake in the barley plants studied, they were also analysed since it could not be ruled out that some aquaporins were expressed specifically in this type of root. Samples were frozen in liquid nitrogen and pooled from 3–4 plants before being stored at –80 °C.
Fig. 1.
Leaf regions analysed in the present study. Leaf samples were taken from up to three developmental zones along the elongating leaf three of barley, the elongation zone (EZ), the non-elongation zone (NEZ) and the emerged blade (EmBL). In addition, a leaf sample was taken from the mature blade and from midway along the sheath of leaf two. Leaf samples consisted of 2-cm-long segments which were taken from the centre region of the respective zone. For qPCR analyses of the elongation zone of leaf three, the younger leaf four which was wrapped within this region was removed.
Expression analyses
RNA was extracted from plant samples and cDNA synthesized as described previously (Boscari et al., 2009). Expression of candidate genes was analysed by real-time PCR (qPCR), using a Stratagene rapid cycler Mx3000P and SYBR-Green as reagent (Takara Bio Inc, Otsu, Shiga 520-2193, Japan) on 96-well plates, following the manufacturer's protocol. cDNA samples from three independent batches of plants were analysed and qPCR data processed as described previously (Boscari et al., 2009) using the ΔCt method (Pfaffl, 2001). Primers used for qPCR analyses were designed using Primer3 (http://frodo.wi.mit.edu/) and the less conserved 3' untranslated region of candidate genes. Sequences of primers are listed in Supplementary File S1 at JXB online. Each reaction contained 40 ng of cDNA. Three genes were chosen as expression references, these were ubiquitin, H+-ATPase, and GAPDH (glycerin-aldehyde-phosphate dehydrogenase) (Boscari et al., 2009). In experiments where expression was compared between five leaf regions, HvSIP2;1 was used instead of GADPH, since preliminary analyses showed that HvSIP2;1 was suited even better for this particular type of experiment. Expression data for a range of reference genes is shown in Supplementary File S5 at JXB online.
Isolation and cloning of candidate genes
Primer sequences used for the isolation of candidate genes are listed in Supplementary File S1 at JXB online. BarleyBase (http://www.barleybase.orgShen et al., 2005) was screened for contig sequences using keywords such as ‘aquaporin’, ‘water channel’, ‘TIP’, and ‘PIP’ (see also Richardson et al., 2007a). Sequences were screened for the existence of the NPA/NPX motif characteristic of MIPs. Full-length sequences were obtained from the NCBI (http://www.ncbi.nlm.nih.gov/) database, and primers were designed to clone a selection of aquaporins, using cDNA which had been synthesized from RNA extracted from 2-week-old barley seedlings. The open reading frame was amplified by PCR using proof reading polymerase KOD Hot Start DNA Polymerase (Merck, Darmstadt, Germany). The PCR product was cloned into the Gateway entry vector pCR®8/GW/TOPO® (pCR®8/GW/TOPO® TA Cloning® Kit, Invitrogen), and the insert verified by sequencing.
Nucleotide sequence alignments and analysis were conducted as described by Richardson et al. (2007a). Phylogenetic trees were constructed using the Bayesian inference in BEAST (Drummond and Rambaut, 2007). Annotations and sequences used for the construction of trees are given in Supplementary File S2 at JXB online.
Functionality test of candidate genes in yeast
The open reading frame was cloned into the modified yeast expression vector pYeDP60u (Hamann and Møller, 2007), which is derived from pYeDP60, using a uracil-excision-based cloning system as described by Nour-Eldin et al. (2006) (USER™ Friendly Cloning Kit, Biolabs). Sequences were amplified using specific uracil-containing primers (see Supplementary File S1 at JXB online). The resulting vector was verified by sequencing.
Strain 31019b (Marini et al., 1997) transformed with pYeDP60u containing selected aquaporins was used to study water transport activity. Spheroplasts were isolated and subjected to swelling assays as described previously by Bienert et al. (2007). Swelling was recorded by a fast kinetic instrument (SFM-300, Biologic) equipped with a spectrofluorometer (MOS-250, Biologic). The instrument was loaded with spheroplasts expressing a particular aquaporin or empty vector (negative control) and 10–20 successive runs were carried out and averaged to yield a trace recording for that particular measurement. The rate constant of the decrease in scattered light intensity is proportional to the water permeability coefficient (Calamita et al., 2005). Rate constants were calculated by fitting a single exponential to the traces of control spheroplasts and a double exponential to the aquaporin-expressing spheroplasts (Liu et al., 2006; Bienert et al., 2007). Several experiments were carried out involving independently transformed yeast strains.
Functionality test of candidate genes in oocytes
cRNA was synthesized using an mMESSAGE mMACHINE kit (Ambion, Austin, Texas, USA) and injected into Xenopus laevis stage V and VI oocytes, which were incubated in modified Barth's saline, as described previously (Wei et al., 2007). Two to four days following injection, oocytes were tested for functionality of expressed candidate genes using swelling assays (Wei et al., 2007). Three batches of oocytes were analysed, with similar results.
In situ hybridization
Tissue preparation and in situ hybridization was performed as described by Jabnoune et al. (2009). Hybridization experiments were conducted on the ‘Plate-Forme d'Histocytologie et Imagerie Cellulaire Végétale (PHIV platform)’ (http://phiv.cirad.fr/). Primers for cRNA probes were designed in the 3' UTR cDNA region of candidate genes; an 18S ribosome cDNA was used as positive control (for primer sequences, see Supplementary File S1 at JXB online). Hybridization signals were detected with VectorBlue KIT III (Vector laboratories, Burlingame, CA) and observed with a microscope (Leica DM6000, Leica, Germany). Three independent batches of plants were analysed, with similar results.
Transient expression in onion epidermis
The full-length coding sequence of HvPIP2;2, HvPIP2;7, and HvTIP1;1 was cloned downstream of a coding sequence for the Enhanced Yellow Fluorescence Protein (EYFP) under the control of the CaMV 35S constitutive promoter in the vector pSAT6-EYFP-N1 (accession number: AY818378), kindly provided by the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). Candidate MIP genes were co-expressed with the empty pSAT6-DsRed2-N1 (accession number: AY818373) targeting the cytoplasm and the plasma membrane-mcherry marker pm-rk (http://www.bio.utk.edu/cellbiol/markers/) (Nelson et al., 2007).
Clones were sequenced to ensure that they were in-frame with the EYFP reporter gene. Biolistic transformation of onion (Allium cepa L.) epidermal cells was performed as described by Pata et al. (2008) using 1 μg of plasmid DNA for each transformation. After 48 h of incubation, the subcellular localization of expression was observed using an Olympus FV1000 confocal laser scanning microscope.
Detection of Casparian bands
For the detection of Casparian bands, leaf cross-sections, which were either cut by hand using fresh material or were spare sections prepared from embedded material for in situ hybridizsation, were stained for 30 min with 0.1% berberine-hemisulphate (bright yellow signal) and counterstained for 1–3 min with 0.5% toluidine blue. Sections were viewed under fluorescence light using a UV filter with an excitation wavelength of 390–420 nm (Brundrett et al., 1988; Hachez et al., 2006a). Sections were observed with a LEICA microscope (LEICA DM IL, Wetzlar, Germany) and captured with a digital camera (LEICA DFC300 FX, Wetzlar, Germany).
Statistical analyses
Student's t test and ANOVA (Excel) were used for statistical analyses.
Results
Phylogenetic analysis of barley MIPs
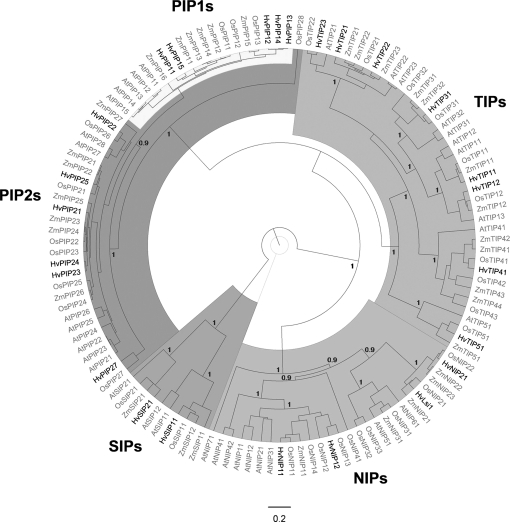
Sequence alignment (ClustalW) and research in the NCBI sequence database identified 22 unique aquaporins among the 35 microarray MIP probe set, five HvPIP1s; four HvPIP2s; seven HvTIPs; four HvNIPs, and two HvSIPs (for details see Supplementary File S2 at JXB online). Two additional aquaporins HvPIP2;4 and HvLsi1 (HvNIP2;1) were not included on the microarray but were found on the NCBI databases. Surprisingly, the silicon transporters HvLsi1 and HvLsi6, which belong to the aquaporin family, were both annotated as HvNIP2;1. For this study, HvNIP2;1 was referred to as HvLsi6 since this was the annotation which had been given first.
After close analysis, HV_CEb0007N06r2_at, HF03B07r_at, and Contig19630_at were the only probe sets presenting no significant similarities with the sequences available on the NCBI database. Unlike the other two sequences, Contig19630_at showed significant homology with the SIP2 subgroup of aquaporins (top hit for ZmSIP2;1). Contig19630_at was the only SIP2 isoform identified in barley.
All sequences had previously been annotated except two SIPs (Contig6339_s_at/Contig6340_at and Contig19630_at) and one PIP2 (Contig19393_at). Having cloned the full-length coding sequence of Contig19393_at in the present study, the gene was annotated as HvPIP2;7 (accession number 314622; GU584120) according to its closest homologue OsPIP2;7 from Oryza sativa, as there existed no PIP2;7 isoform in Arabidopsis thaliana. By contrast, the barley SIPs were not cloned and were named according to their closest homologue in Arabidopsis thaliana, AtSIP1;1 and AtSIP2;1 [HvSIP1;1 (Contig6339_s_at/Contig6340_at), HvSIP2;1 (Contig19630_at)] (see Supplementary Table S3.2 at JXB online).
Phylogenetic analysis of barley MIPs, together with MIP sequences of Arabidopsis, rice, and maize, showed that all genes grouped into known MIP subfamilies (Fig. 2). The total number of MIPs identified in barley, so far, was 25 (five PIP1s, six PIP2s, eight TIPs, four NIPs, and two SIPs; see Supplementary File S2 at JXB online). This exceeded the number of barley MIP genes as proposed by Katsuhara and Hanba (2008).
Fig. 2.
Bayesian phylogenetic analysis of barley major intrinsic proteins (MIPs). Barley MIP protein sequences were aligned with MIPs of Arabidopsis, maize, and rice using ClustalW. Barley MIPs analysed are highlighted. Annotations and protein sequences are given in Supplementary File S2 at JXB online. Concerning HvNIP2;1, Fig. 2 refers to the gene annotated first (accession number BA166444) and not to the gene referred to by Schnurbusch et al. (2010) as a boron transporter (HvNIP2;1). The latter gene is identical in sequence to the silicon transporter (HvLsi1, shown) reported by Chiba et al. (2009). HvSIP2;1 is not annotated; only a partial sequence is known, which shows the highest homology to AtSIP2;1 among Arabidopsis SIPs. The distance scale represents the evolutionary distance, expressed as the number of substitutions per amino acid. The figures displayed on the main nodes reflect the posterior probability.
Identification of candidate genes
Microarray experiments, using a barley Affymetrix chip (Close et al., 2004), were carried out during 2004/2005 as part of a study on cuticle development in barley (Richardson et al., 2007a). The plants used for microarray analyses had been grown under similar, controlled conditions and were of the same developmental age as the plants analysed in the present study. The entire set of microarray raw data (Richardson et al., 2007a) following analyses with the RankProduct (RP) method (Breitling et al., 2004, Breitling and Herzyk, 2005) can be found in the supplementary material of Richardson et al. (2007a). Screening of BarleyBase showed that the Affymetrix chip contained 35 MIP-related sequences (see Supplementary File S2 at JXB online); 22 of the sequences were identified as unique aquaporins (see previous paragraph). Their pattern of expression between the leaf developmental regions is summarized in Supplementary File S3 at JXB online.
Contig19393_at (HvPIP2;7) was the third most differentially expressed gene between the emerged blade and elongation zone (lower expression) of all >21 000 sequences on the chip; four more MIPs were among the 140 most differentially expressed genes (HVSMEf0019H18r2_at, RP18, HvTIP1;2; Contig1223_at, RP 88, HvPIP2;3; Contig1309_at, RP121, closest to HvTIP1;2; and Contig1219_s_at, RP137, HvPIP1;4). The two contigs which were expressed highest in the elongation zone compared with the emerged blade were Contig14229_at (RP 129, HvNIP1;1) and Contig1216_s_at (RP 384, HvPIP2;2) (not shown).
Nineteen of the 35 aquaporin-related sequences were expressed differentially between leaf regions. Based on the level of difference (low RP value) and likeliness of the candidate sequence encoding an aquaporin with water channel activity (e.g. PIP2s), Contig19393_at (HvPIP2;7), Contig1216_s_at (HvPIP2;2), Contig1315_s_at (HvTIP2;3) and HVSMEf0019H18r2_at (HvTIP1;2) were selected for detailed analyses. Contig1222_s_at (HvPIP2;5) was also selected due to its high level of expression and HvTIP1;1 (accession number: X80266), a TIP which was not included on the chip and had been shown to be expressed particularly in the leaf elongation zone (Schünmann et al., 1994). The present study was carried out in close association with a study on the role of aquaporins in barley root water uptake (Knipfer et al., 2011). Therefore, water channel activity was also tested for HvPIP1;2, which was expressed almost exclusively in root compared with leaf tissue (Table 1).
Table 1.
Expression of major intrinsic proteins in root and leaf tissue of barley
| Gene | Roota Average | SD | Leaf Average | SD |
| HvPIP1;1 | 0.7952 | 0.2963 | 1.2038 | 0.8911 |
| HvPIP1;2 | 0.6542* | 0.3153 | 0.0007 | 0.0004 |
| HvPIP1;3 | 1.7418* | 0.8452 | 0.5693 | 0.6482 |
| HvPIP1;4 | 0.0313* | 0.0138 | 0.0024 | 0.0026 |
| HvPIP1;5 | 0.0084 | 0.0015 | 0.0472 | 0.0307 |
| HvPIP2;1 | 0.1024* | 0.0477 | 0.0167 | 0.0187 |
| HvPIP2;2 | 0.0627 | 0.0237 | 0.3046 | 0.2428 |
| HvPIP2;3 | 0.0752* | 0.0260 | 0.0149 | 0.0075 |
| HvPIP2;4 | 0.2412* | 0.1156 | 0.0379 | 0.0479 |
| HvPIP2;5 | 2.2871 | 0.4679 | 2.3250 | 1.9264 |
| HvPIP2;7 | 0.0157 | 0.0267 | 0.0091 | 0.0139 |
| HvTIP1;1 | 5.0038 | 1.8757 | 6.6860 | 5.2284 |
| HvTIP1;2 | 0.1463** | 0.0749 | 0.0438 | 0.0623 |
| HvTIP2;1 | LD | LD | ||
| HvTIP2;3 | 0.3914 | 0.2262 | 0.3203 | 0.1062 |
| HvTIP3;1 | 0.00006 | 0.00005 | 0.00004 | 0.00002 |
| HvNIP1;1 | LD | LD | ||
| HvNIP2;1 | LD | LD |
Expression was analysed by qPCR. Average 2–(ΔCt) values for (n=) three batches of plants are shown, together with standard deviations (SD). In each batch the average value of expression in adventitious and seminal roots ((AR+SR)/2) and the average expression in leaf 2 and the elongation zone of leaf three ((L2+EZ)/2) was used to represent expression in root and shoot, respectively; LD, limit of detection.
*, **, Statistically significant (paired t test, Excel) difference between root and shoot at P <0.05 and P <0.01.
qPCR expression analyses
Expression in leaf tissues:
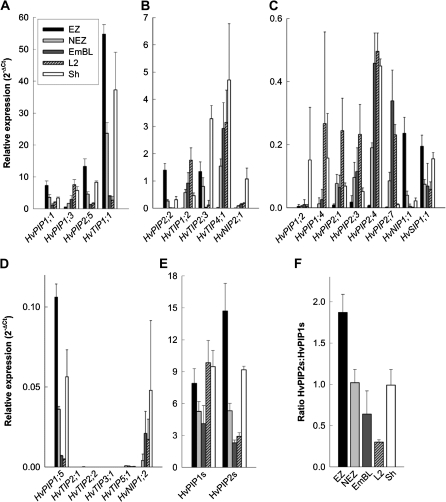
qPCR data generally supported microarray data. The expression of 23 barley MIPs, including all known members of the PIP1 and PIP2 family, was analysed (Fig. 3A–D). Five genes showed hardly any or no expression in leaf tissue. These genes were almost exclusively TIPs (HvTIP2;1, HvTIP2;2, HvTIP3;1, HvTIP5;1, and HvPIP1;2). Of the remaining 18 genes, seven (HvPIP1;1, HvPIP1;5, HvPIP2;2, HvPIP2;5, HvTIP1;1, HvTIP2;3, and HvNIP1;1) were expressed higher and ten (HvPIP1;3, HvPIP1;4, HvPIP2;1, HvPIP2;3, HvPIP2;4, HvPIP2;7, HvTIP1;2, HvTIP4;1, HvNIP1;2, and HvNIP2;1) were expressed lower in the elongation zone compared to mature and emerged leaf tissue. Differences in the expression level of a particular candidate aquaporin between leaf regions were mostly statistically significant (all PIP2s, HvPIP1;1, 1;3 and 1;5; HvTIP1;1, 1;2, 2;3, and 4;1; see Supplementary File S4 at JXB online), and expression differed by orders of magnitude. As a result, most of the genes which showed the highest expression in the elongation zone showed much lower or hardly detectable expression in the emerged blade, while the opposite applied to genes which showed the highest expression in emerged and mature leaf tissue. Expression in the non-elongation zone was at an intermediate level to that in the emerged blade and elongation zone. In those cases where expression was high or highest in the sheath, it was close to the level of expression in either elongating or mature leaf tissue. The exception being HvPIP1;2, HvTIP2;3, and HvNIP2;1. One gene, namely HvSIP2;1, was expressed uniformly between leaf tissues, to an extent that it could be used as a reference gene of expression. The other SIP analysed (HvSIP1;1) also showed comparatively minor differences in expression between leaf regions.
Fig. 3.
Real-time (qPCR) expression analyses of major intrinsic proteins in leaf regions of barley. The elongation zone (EZ), adjacent non-elongation zone (NEZ), and emerged-blade portion (EmBL) of the growing leaf three were analysed, together with the blade and sheath of the mature leaf two (L2 and Sh, respectively). (A, B, C, D) Values of relative expression show how many times higher or lower the expression of a candidate gene was compared with that of the reference genes (ubiquitin, H+-ATPase, HvSIP2;1), using the 2–(ΔCt) method. (E) Total expression of the PIP1 and PIP2 subfamily, together with (F) ratio of expression. Results are averages ±SD (error bars) of three experiments. Statistical significance of difference in expression between leaf regions is summarised in Supplementary File S4 at JXB online.
Most of the PIP genes, which were expressed highest in the elongation zone, were expressed statistically lower in all other leaf regions tested (see Supplementary File S4 at JXB online). Similarly, all TIPs tested were expressed significantly higher or lower in the elongation zone compared with the non-elongation zone, emerged blade, and leaf two. HvTIP1;1 was expressed almost as high in sheath tissue as in the elongation zone (see Supplementary File S4 at JXB online). The fewest differences in expression were observed between the emerged blade of the growing leaf three and the mature blade of leaf two. This applied to both PIPs and TIPs (see Supplementary File S4 at JXB online).
Plant PIP1s and PIP2s form functional subgroups in that most PIP2s show substantial water channel activity, whereas PIP1s either facilitate the movement of other, small neutral solutes or increase water channel activity when co-expressed with PIP2s (Fetter et al., 2004; Maurel et al., 2008). To obtain a measure of the ‘investment’ into or ‘usage’ of each subgroup during leaf developmental stages, the sum of expression of PIP1s and PIP2s was calculated. Total expression of PIP1s was comparable between leaf regions, whereas PIP2 expression was up to six times higher in the elongation zone (Fig. 3E). This was due to much higher expression of HvPIP2;5 in elongating leaf tissue. As a result, the ratio of expression of PIP2:PIP1 decreased continually during leaf cell development, from 1.9 in the elongation zone to 0.3 in the mature blade (Fig. 3F).
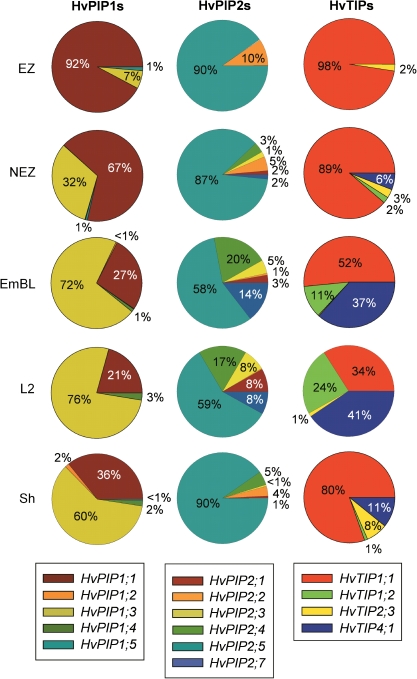
In the leaf elongation zone, a single PIP isoform accounted for more than 90% of expression of PIP1s (HvPIP1;1) and PIP2s (HvPIP2;5), respectively, with a second PIP accounting for the bulk of remaining expression within each group (HvPIP1;3 and HvPIP2;2; Fig. 4). As leaf tissues matured, expression of HvPIP1;3 became dominant among PIP1s, and almost all PIP2s contributed significantly to expression. The percentage distribution for sheath tissue was, in some ways, similar to that of the elongation zone (PIP2s) and that of the mature blade (PIP1s).
Fig. 4.
The percentage contribution of individual family members to total expression of PIP1s, PIP2s, and TIPs in leaf regions of barley. Data were calculated from the expression values shown in Fig. 3. The TIPs analysed should represent the bulk of expression of TIPs (Alexandersson et al., 2005; Sakurai et al., 2005). HvTIP2;1, HvTIP2;2, HvTIP3;1, and HvTIP5;1 together accounted for less than 1% of the expression of TIPs and are not included in the pie charts. Numbers give the mean percentage contribution calculated from three experiments. Standard deviations are given in Supplementary File S4 at JXB online.
Expression in leaf and root:
Expression was compared between leaves (combined expression in growing and mature tissue) and roots (combined expression in seminal and adventitious roots; growing and mature regions) (Table 1). Four of the genes tested showed very low expression (HvTIP3;1) or expression near the limit of detection (HvTIP2;1, HvNIP1;1, HvNIP2;1) in leaves and roots. One MIP (HvPIP1;2) was expressed almost exclusively in roots, while two genes (HvPIP1;5, HvPIP2;2) were expressed several-fold higher in leaf tissue. Seven genes were expressed significantly differently between the root and leaf. In all cases, expression was higher in roots. None of the MIPs tested was expressed exclusively in leaves.
Functionality tests of candidate genes
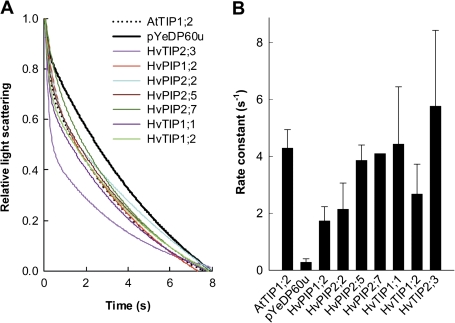
Spheroplast swelling was measured as a reduction in light scattering in a stopped-flow spectrophotometer and was completed within seconds following transfer of spheroplasts to hypo-osmotic medium (Fig. 5A). Expression of barley aquaporins in spheroplasts increased the initial rate of swelling compared with the empty vector control (Fig. 5A). Increases ranged from 7–20-fold (Fig. 5B). The effect of preincubation of spheroplasts with the aquaporin inhibitor Ag on the aquaporin-dependent rate constant was tested for HvPIP2;5 and HvTIP1;1. In both cases, the rate constant was reduced by 65% (not shown, means of three experiments).
Fig. 5.
Test of water channel function of selected barley MIPs through expression in yeast and subsequent swelling assays of isolated spheroplasts. (A) Swelling kinetics of a representative batch of spheroplasts. Each curve is the average of 10–20 swelling kinetics, fitted with two-exponential equations. Control spheroplasts were isolated from yeast which had been transformed with the empty vector used for transformation (pYeDP60u, negative control) or with an Arabidopsis MIP (AtTIP1;2) known to show water channel activity (positive control). (B) Rate constants of water flow in spheroplasts. Results are averages ±SD (error bars) of the analysis of (n) kinetics as shown in (A): HvPIP2;5, (4); HvTIP1;1, (8); HvTIP2;3, (8), HvPIP2;2, (8); HvPIP2;7, (2, no error bar shown, values of 4.5 and 3.6 s−1); HvPIP1;2, (3); HvTIP1;2 (4); negative control, (3); and positive control (4). Rate constants of swelling of barley-MIP expressing spheroplasts were significantly higher (P <0.001) than that of (negative) control spheroplasts containing the empty vector used for transformation.
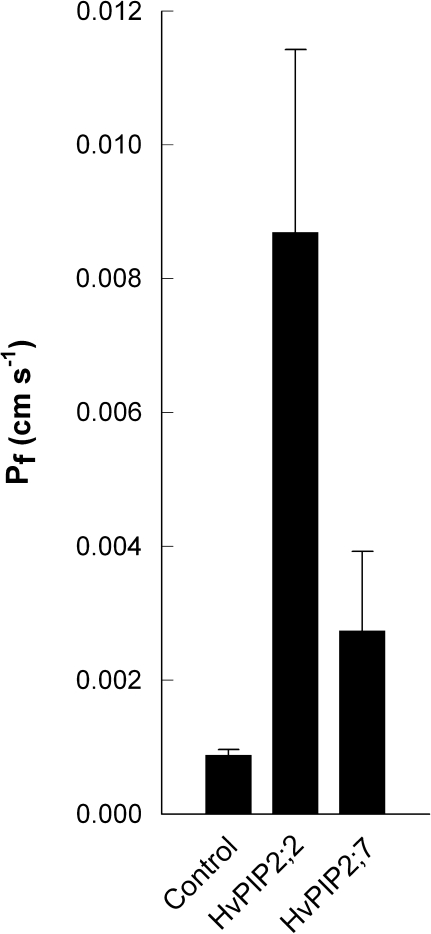
To test the validity of spheroplast data, water channel function of two barley MIPs was tested through an independent approach by conducting swelling assays of Xenopus leavis oocytes which had expressed these MIPs. In agreement with spheroplast data, expression of HvPIP2;2 and HvPIP2;7 increased significantly the osmotic water permeability of oocytes (Fig. 6). Preincubation with 50 μM HgCl2 of oocytes expressing HvPIP2;2 reduced osmotic water permeability by 50–70% (not shown); subsequent recovery of osmotic water permeability following the addition of reducing agents (e.g. dithiothreitol) was not tested. The opposite permeability difference between HvPIP2;2 and HvPIP2;7 as observed in spheroplasts versus oocytes might have been due to different amounts of protein being expressed. This was not tested.
Fig. 6.
Test of water channel function of HvPIP2;2 and HvPIP2;7 through transient (3 d) expression in Xenopus laevis oocytes. Results are means ±SD (error bars) of six (control) and eight (HvPIP2;2, HvPIP2;7) oocytes, analysed from one representative batch. Osmotic water permeability of oocytes expressing HvPIP2;2 or HvPIP2;7 was significantly higher (P <0.001) than that of water-injected (control) oocytes, as was the difference in water permeability between HvPIP2;2 and HvPIP2;7-expressing oocytes (P <0.001).
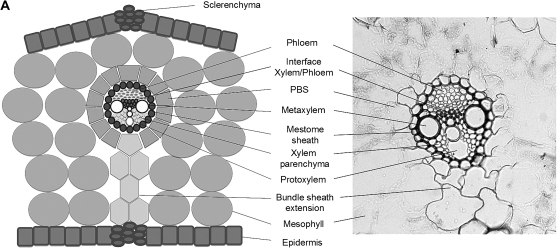
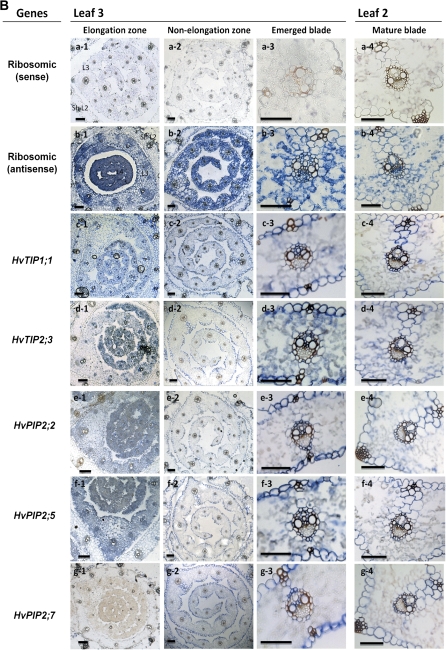
Tissue localization of expression
In situ hybridization was used to determine the tissue distribution of expression of candidate aquaporins (Fig. 7). HvTIP1;1 was expressed in most leaf tissues, including mesophyll, particularly during the earlier stages of leaf development. Expression was particularly pronounced in the epidermis, in the part facing mesophyll, and in the xylem. Expression in phloem was hardly detectable. Expression of HvTIP2;3 was evident in the epidermis and bundle sheath extensions of large lateral veins; it was also detectable in the mestome bundle sheath and to a considerable extent in the mesophyll of the emerged and mature blade. HvPIP2;2 was expressed mostly in the epidermis and in vascular bundles, particularly the mestome sheath and xylem parenchyma, and including bundle sheath extension. There was little expression detectable in mesophyll cells during the later leaf developmental stages. The pattern of expression of HvPIP2;5 was similar to that of HvPIP2;2, with mesophyll expression being detectable particularly during the earlier developmental stages Expression of HvPIP2;7 was not detectable in the elongation zone and was largely confined to the epidermis, vascular bundles (mestome sheath, xylem parenchyma) and bundle sheath extensions of mature leaf tissue.
Fig. 7.
Tissue-specific expression of barley MIPs in leaf regions. (A) Scheme and micrograph of a cross-section of a mature blade, identifying tissues. (B) In situ hybridization data. Expression is shown as blue colour, and was detected using antisense probes of genes of interest. An antisense designed against 18S ribosomal RNA was used as positive control. A sense probe was tested for all genes as negative control and is shown representatively for ribosomal RNA. PBS, parenchymatous bundle sheath; L3, leaf three; Sh L2, sheath of leaf two; scale bar=50 μm.
Subcellular localization of gene product
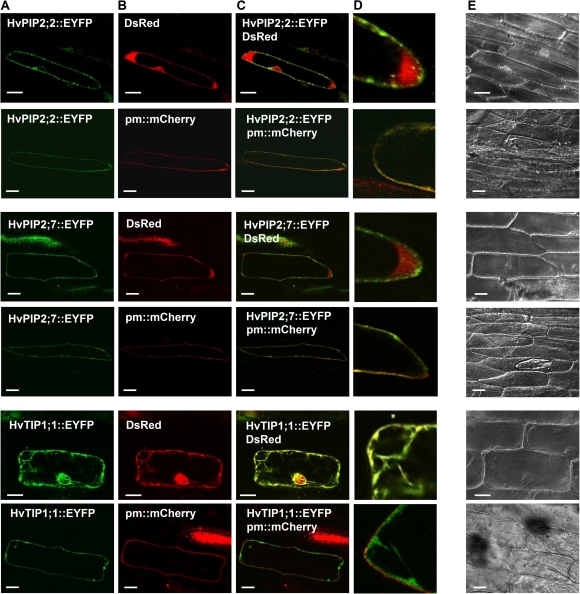
It was beyond the scope of this study to test the subcellular localization of all candidate MIPs. Therefore, a selection of candidate MIPs was tested. Constructs which contained the cDNA encoding the yellow fluorescent proteins at the 3'-end of the aquaporin genes were transiently co-expressed with cytoplasmic and plasma membrane markers in onion epidermis. HvPIP2;2 was mainly localized to the plasma membrane, as was HvPIP2;7 (Fig. 8). Some of HvPIP2;2 appeared to be localized around the nucleus, most likely in the endoplasmic reticulum, reflecting protein trafficking through the secretory pathway. HvTIP1;1 would have been expected to localize to the tonoplast but failed to show a clear-cut localization in this membrane. Instead, HvTIP1;1 appeared to localize to cytoplasmic strands. Since HvTIP1;1 did not co-localize with a cytoplasmic (DsRed) or plasma membrane marker (pm::mCherry, bottom two rows in Fig. 8), localization in strands of cytoplasm could have reflected the secretory pathway. However, HvTIP1;1 might also have been localized to the tonoplast. What appeared like cytoplasmic strands might have been invaginations of the tonoplast. Also, the strands may be transvacular, consisting of a tonoplast ‘sheath’ and inner cytoplasmic ‘core’ (Reisen et al., 2005). It is also possible that the HvTIP1;1 fusion protein was cleaved and that the yellow fluorescent protein was cytosolic. However, in this case, the nucleus should have been marked entirely, which was not the case.
Fig. 8.
Subcellular localization of barley MIPs as studied through transient (2 d) expression in onion epidermis. Expression was viewed in epidermal peels using an Olympus FV1000 confocal laser scanning microscope. (A) Candidates genes fused to the enhanced yellow fluorescent protein (EYFP); (B) Cytoplasm marker pSAT6-DsRed2-N1 (DsRED, red fluorescence) and pBIN20-plasma membrane-mCherry marker (pm mCherry, red fluorescence); (C) overlay of (A) and (B); (D) magnified parts of cells shown in (C); (E) transmitted light micrographs of cells shown in (A) to (D). Scale bar (A, B, C, E)=50 μm, Scale bar (D)=25 μm.
Casparian bands in leaves
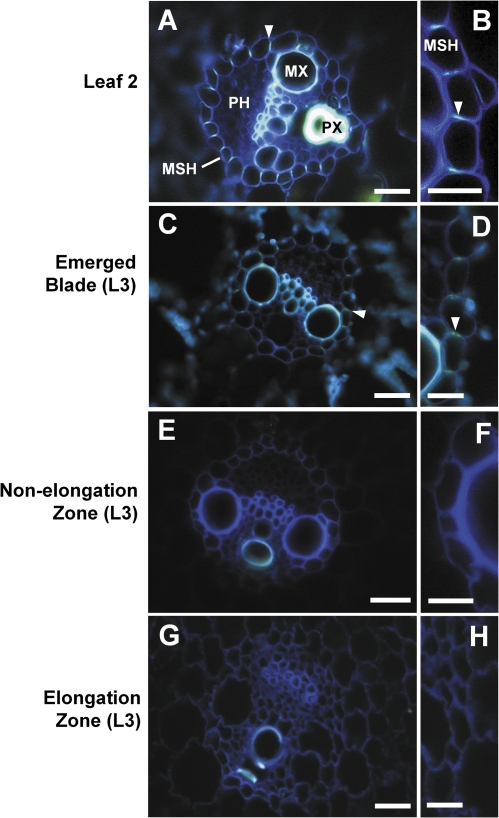
The occurrence of Casparian bands in the mestome sheath of vascular bundles was studied by staining hand-cut cross-sections of leaf regions with berberin-hemisulphate/toluidine blue and viewing the sections under UV-light (Brundrett et al., 1988). Casparian bands showed as yellow fluorescence, as indicated by arrows in Fig. 9. There were no Casparian bands visible in the elongation and non-elongation zone of the growing leaf three (Fig. 9E–H). By contrast, Casparian bands could be detected in the emerged portion of leaf three and were most pronounced in the mature blade of the older leaf two (Fig. 9A–D).
Fig. 9.
Casparian bands in the mestome sheath in different developmental regions of barley leaves. Cross-sections were stained with berberin-hemisulphate/toluidine blue and viewed under UV-light (Brundrett et al., 1988). Occurrence of Casparian bands (yellow fluorescence) is indicated by arrows. Xylem vessels showed autofluorescence. MX, metaxylem; PX, protoxylem; MSH, mestome sheath; PH, phloem. Scale bar (A, C, E, G)=50 μm, scale bar (B, D, F, H)=25 μm.
Discussion
The present study provides the most comprehensive functional, transcriptional, and phylogenetic analysis of barley aquaporins so far. The strict transcriptional control of barley aquaporins reported here during leaf development provides a platform for future studies into the physiological function of these aquaporins, now that certain aquaporin family members can be associated with or excluded from features intrinsic to these leaf developmental stages (e.g. growth, photosynthesis, transpiration). Not only are MIP genes differentially expressed between leaf regions, but they belong to the most differentially expressed genes of more than 21 000 sequences on the barley Affimetrix chip. In particular, the water channel HvPIP2;7 is the third most differentially expressed gene between the emerged blade and elongation zone (lower expression). Finally, we are not aware of any other study which has reported, for a family as large as MIPs, that most family members are under a transcriptional control during leaf development that resembles more an ‘On’ and ‘Off’ type than a gradual regulation. The results provide an opportunity to study underlying mechanisms involving (common) promoter control, not just of certain aquaporins but of other genes which are expressed development-specific such as cuticle wax genes (Richardson et al., 2007a) or putative potassium transporters (Boscari et al., 2009). The common functional links between these groups of genes in the leaf elongation zone are water movement and cell expansion.
Developmental pattern of aquaporin expression in leaves
Five genes, including one PIP1 (HvPIP1;2), were expressed at such low levels in leaf regions that it was difficult to conclude on their pattern of expression. A sixth gene (HvSIP2;1) was expressed so uniformly between leaf tissues that it turned out to be a suitable reference gene of expression. The 17 remaining genes analysed, which included all other known barley PIPs, were expressed particularly in either growing (seven genes) or emerged, mature leaf tissue (ten genes). It can be concluded from these data that differential expression during leaf development is the rule rather than the exception for barley MIPs and that all MIPs which facilitate the diffusion of water across the plasma membrane are under developmental or environmental control. There is no obvious reason why the control of water channel activity of one particular aquaporin through post-translational regulation and trafficking (Johansson et al., 2000; Tournaire-Roux et al., 2003; Maurel, 2007; Zelazny et al., 2007; Boursiac et al., 2008) should not provide sufficient means to meet the requirements specific to growing and mature leaf tissue. The present finding that many different aquaporins show development-specific expression points to these aquaporins fulfilling tissue-specific functions. Individual aquaporins with localization to specific tissues may play important roles but their relative abundance in whole tissue is lower because of their specific localization.
None of the MIPs tested was expressed highest or lowest in the non-elongation zone. Instead, the start (elongation zone) and end point (emerged, mature blade) of a cell's ontogeny was accompanied by maximum or minimum MIP expression. This contrasts with the only comparable study, on maize (Hachez et al., 2008), where expression of some PIPs was highest in the non-elongation zone. A continuous increase in expression during leaf development was only observed for ZmPIP1;5. The studies on maize and barley are the only ones of their kind. It cannot be said which study presents the rule and which the exception with respect to aquaporin expression. The most notable difference between barley and maize is that barley is a C3 and maize a C4 plant. How that could explain the differences in the expression pattern of aquaporins is not clear.
The sheath of leaf two transpires and photosynthesizes, similar to the mature blade. The sheath is also located close to the apical meristem, where the intensity of incident light is lower, as in the elongation zone. Together, this may explain why the expression of some genes, notably HvTIP1;1, in sheath tissue was more comparable with that in the leaf elongation zone, while the expression of other genes, notably HvPIP2;4, was more comparable with that in the mature blade.
It is not known what regulates the differential expression of barley MIPs during leaf development. The most pronounced difference in microenvironment between the elongation zone and the other leaf regions analysed is the intensity and quality of light which reaches the cells. The elongation zone of leaf three is enclosed by the sheaths of leaves one and two, whereas the non-elongation zone is only enclosed by the sheath of leaf two. The sheaths are green and photosynthetic. As a result, the light which reaches the non-elongation and, particularly, the elongation zone will have a higher ratio of far-red to red light than the light which reaches the emerged and mature blade. This could enable regulation of MIP expression through the phytochrome system.
Similar to the present study on barley, Alexandersson et al. (2005) observed for Arabidopsis that no MIP family member was expressed exclusively in leaf tissue, and that expression of MIPs, in particular PIPs, was generally higher in root tissue (see also Jang et al., 2004). This generally observed pattern of expression may reflect the function of the root as being the major site for uptake of water and mineral nutrients.
Roles of particular MIPs during barley leaf growth
HvPIP2;5, HvTIP1;1, and HvTIP2;3 were expressed abundantly and highest in growing tissue of roots and leaves (Fig. 3; Table 1). The same was observed for HvPIP1;1 (HvPIP1;6) in a previous study (Wei et al., 2007). These four MIPs, all of which show water channel activity (Figs 5, 6; Wei et al., 2007), seem to have a role which is specific to growth, regardless of the organ. By contrast, the water channel HvPIP2;2 was expressed particularly in growing tissue of leaves and may have a growth-related function which is leaf-specific.
High expression of TIP1;1 isoforms in meristematic and elongating shoot tissue has been reported for maize (Chaumont et al., 1998), tulip (Tulipa gesneriana, Balk and de Boer, 1999), cauliflower (Brassica oleracea, Barrieu et al., 1998), and oilseed rape (Brassica napus, Frangne et al., 2001) and appears to be a common characteristic associated with growth. Arabidopsis plants which lack AtTIP1;1 (and AtTIP1;2) protein do not show any phenotype or change in growth rate under normal growth conditions (Schüssler et al., 2008). This does not preclude a role of AtTIP1;1 or the barley homologue HvTIP1;1 in facilitating water uptake and vacuole enlargement during leaf cell expansion. For example, the high expression of the water channel HvTIP2;3 points to some redundancy in function among TIPs.
In the elongation zone of barley, HvPIP1;1 and HvPIP2;5 accounted for 90% or more of the expression of PIP1s and PIP2s, respectively. Their closest maize homologues (based on sequence identity), ZmPIP1;1, and ZmPIP2;1 together with ZmPIP2;2, also accounted for the bulk of expression of PIP1s and PIP2s in the elongation zone (Hachez et al., 2008). It appears from these two studies on grasses, that dominant PIP isoforms are conserved in elongating leaf tissue.
The barley water channel HvPIP2;5 was expressed abundantly in the leaf elongation zone and this included the mesophyll in this leaf region. Mesophyll constitutes most of the tissue volume of leaves and this could explain why HvPIP2;5 accounted for more than 90% of PIP2 expression in the elongation zone. Together, this renders HvPIP2;5 a prime candidate to mediate plasma membrane water flow in growing mesophyll cells. In growing epidermal cells, this function appears to be carried out by HvPIP1;1 and HvPIP2;2, both of which are expressed highest in the epidermis (Fig. 7; Wei et al., 2007). Trans-tonoplast movement of water in growing leaf tissues seems to be facilitated by the abundantly expressed HvTIP1;1 and HvTIP2;3.
It is not known whether water reaches epidermal cells in the elongation zone directly through the mesophyll or through bundle sheath extensions, from where it diffuses radially within the epidermis. In the latter case, many membranes and hydraulic resistances have to be overcome. A potential hydraulic limitation of cell expansion growth could be avoided by high expression of aquaporins such as HvPIP1;1 in the epidermis, leading to a higher cell hydraulic conductivity in the epidermis of elongating compared with mature leaf tissue (Volkov et al., 2007). The comparatively low water transport activity of HvPIP1;1 (Wei et al., 2007) may be partially compensated for by high expression levels and heteromerization (Fetter et al., 2004; Zelazny et al., 2007) with the concurrently expressed HvPIP2;2 and HvPIP2;5.
The mestome sheath of grass leaves can be suberized (O'Brien and Carr, 1970; O'Brien and Kuo, 1975; for a review, see Fricke, 2002). The present data show that, in barley, Casparian-band like structures increase during leaf development (Fig. 9). The mestome sheath may fulfil a role in leaves that is comparable with that of the endodermis in roots (Wu et al., 2005; Heinen et al., 2009). As Casparian bands, and possibly suberization, increase during leaf development, so does the hydraulic resistance to water movement along the apoplast. Increase in expression of HvPIP2;7 during leaf development in vascular bundles could compensate for the formation of this apoplastic barrier by facilitating the movement of transpiration water through the membranes along a cell-to-cell pathway. Such a pathway has recently been supported by a study on Tradescantia (Ye et al., 2008). In rice, OsPIP2;7 is expressed in leaves predominantly in the mesophyll, and over-expression results in increased transpirational water loss (Li et al., 2008). It is possible that PIP2;7 isoforms play a key role in facilitating transpirational water flow through functioning in different tissues in different plant species. The considerable expression of HvPIP2;7 in the non-transpiring non-elongation zone might be in preparation for the displacement of cells into the open atmosphere (past the point of emergence from the sheath of leaf two). This displacement can occur in as little as 10 h (Richardson et al., 2007b).
Between 98–99 of every 100 water molecules which enter the leaf elongation zone along the xylem are lost through the emerged blade; only 1–2 molecules are used for expansion growth (Fricke, 2002). Therefore, it is surprising that water channels such as HvPIP2;5 and HvTIP1;1 are expressed at so much lower levels in mature leaf tissue. Could it be that their water channel activity is an experimental disguise of their true function in planta or that the need rapidly to equilibrate osmotically across membranes is much higher in growing than in mature plant tissue?
Supplementary data
Supplementary data can be found at JXB online.
Supplementary File S1. Details of primers used.
Supplementary File S2. Annotation and protein sequences of barley MIPs and of MIPs of Arabidopsis, maize and rice used for construction of phylogenetic trees.
Supplementary File S3. Differential expression of major intrinsic proteins (MIPs) between developmental regions of leaf three of barley as revealed by microarray analysis.
Supplementary File S4. qPCR data of the expression of candidate barley MIPs in different leaf regions, together with their statistical analysis.
Supplementary File S5. qPCR data for genes tested or used as reference of expression.
Acknowledgments
We thank Brendan and Eugene for provision of additional laboratory space. This project was funded through a research grant from SFI (Science Foundation Ireland, grant no 07/RFP/EEEOBF277, to WF) and a PhD fellowship from IRCSET (Irish Research Council for Science, Engineering and Technology, to TK). Rothamsted Research is grant-aided by the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK. Thanks also to Patrick Bienert (Université catholique Louvain-la-Neuve, Belgium), Susan Smith (Rothamsted Research), and Geneviève Conejero (PHIV-CIRAD Montpellier) for help with analyses of spheroplasts, oocytes, and in situ hybridization, respectively. We would like to thank two anonymous referees for their helpful comments on an earlier version of the manuscript.
References
- Alexandersson E, Fraysse L, Sjovall-Larsen S, Gustavsson S, Fellert M, Karlsson M, Johanson U, Kjellbom P. Whole gene family expression and drought stress regulation of aquaporins. Plant Molecular Biology. 2005;59:469–484. doi: 10.1007/s11103-005-0352-1. [DOI] [PubMed] [Google Scholar]
- Balk PA, de Boer AD. Rapid stalk elongation in tulip (Tulipa gesneriana L. cv. Apeldoorn) and the combined action of cold-induced invertase and the water-channel protein γTIP. Planta. 1999;209:346–354. doi: 10.1007/s004250050642. [DOI] [PubMed] [Google Scholar]
- Barrieu F, Thomas D, Marty-Mazars D, Charbonnier M, Marty F. Tonoplast intrinsic proteins from cauliflower (Brassica oleracea L. var. botrytis): immunological analysis, cDNA cloning and evidence for expression in meristematic tissues. Planta. 1998;204:335–344. doi: 10.1007/s004250050264. [DOI] [PubMed] [Google Scholar]
- Bienert GP, Moller AL, Kristiansen KA, Schulz A, Moller IM, Schjoerring JK, Jahn TP. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. Journal of Biological Chemistry. 2007;282:1183–1192. doi: 10.1074/jbc.M603761200. [DOI] [PubMed] [Google Scholar]
- Boscari A, Clement M, Volkov V, Golldack D, Hybiak J, Miller JA, Amtmann A, Fricke W. Potassium channels in barley: cloning, functional characterisation and expression analyses in relation to leaf growth and development. Plant, Cell and Environment. 2009;32:1761–1777. doi: 10.1111/j.1365-3040.2009.02033.x. [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Boudet J, Postaire O, Luu DT, Tournaire-Roux C, Maurel C. Stimulus-induced down-regulation of root water transport involves reactive oxygen species-activated cell signaling and plasma membrane intrinsic protein internalization. The Plant Journal. 2008;56:207–218. doi: 10.1111/j.1365-313X.2008.03594.x. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Breitling R, Herzyk P. Rank-based methods as a non-parametric alternative of the t-statistic for the analysis of biological microarray data. Journal of Bioinformatics and Computational Biology. 2005;3:1171–1189. doi: 10.1142/s0219720005001442. [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Enstone DE, Peterson CA. A berberine–aniline blue staining procedure for suberin, lignin, and callose in plant tissue. Protoplasma. 1988;146:133–142. [Google Scholar]
- Calamita G, Gena P, Meleleo D, Ferri D, Svelto M. Water permeability of rat liver mitochondria: a biophysical study. Biochimica et Biophysica Acta. 2006;1758:1018–1024. doi: 10.1016/j.bbamem.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Herman EM, Chrispeels MJ. Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiology. 1998;117:1143–1152. doi: 10.1104/pp.117.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Mitani N, Yamaji N, Ma JF. HvLsi1 is a silicon influx transporter in barley. Plant Journal. 2009;57:810–818. doi: 10.1111/j.1365-313X.2008.03728.x. [DOI] [PubMed] [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise R. A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiology. 2004;134:960–968. doi: 10.1104/pp.103.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson J, Johanson U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biology. 2008;8:45. doi: 10.1186/1471-2229-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetter K, Van Wilder V, Moshelion M, Chaumont F. Interactions between plasma membrane aquaporins modulate their water channel activity. The Plant Cell. 2004;16:215–228. doi: 10.1105/tpc.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexas J, Ribas-Carboö M, Hanson DT, Bota J, Otto B, Cifre J, McDowell N, Medrano H, Kaldenhoff R. Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. The Plant Journal. 2006;48:427–439. doi: 10.1111/j.1365-313X.2006.02879.x. [DOI] [PubMed] [Google Scholar]
- Frangne N, Maeshima M, Schäffner AR, Mandel T, Martinoia E, Bonnemain JL. Expression and distribution of a vacuolar aquaporin in young and mature leaf tissues of Brassica napus in relation to water fluxes. Planta. 2001;212:270–278. doi: 10.1007/s004250000390. [DOI] [PubMed] [Google Scholar]
- Fricke W. Biophysical limitation of cell elongation in cereal leaves. Annals of Botany. 2002;90:157–167. doi: 10.1093/aob/mcf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C, Heinen RB, Draye X, Chaumont F. The expression pattern of plasma membrane aquaporins in maize leaf highlights their role in hydraulic regulation. Plant Molecular Biology. 2008;68:337–353. doi: 10.1007/s11103-008-9373-x. [DOI] [PubMed] [Google Scholar]
- Hachez C, Moshelion M, Zelazny E, Cavez D, Chaumont F. a. Localization and quantification of plasma membrane aquaporin expression in maize primary root: a clue to understanding their role as cellular plumbers. Plant Molecular Biology. 2006;62:305–323. doi: 10.1007/s11103-006-9022-1. [DOI] [PubMed] [Google Scholar]
- Hachez C, Zelazny E, Chaumont F. Modulating the expression of aquaporin genes in planta: a key to understand their physiological functions? Biochimica et Biophysica Acta. 2006b;1758:1142–1156. doi: 10.1016/j.bbamem.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Hamann T, Møller BL. Improved cloning and expression of cytochrome P450s and cytochrome P450 reductase in yeast. Protein Expression and Purification. 2007;56:121–127. doi: 10.1016/j.pep.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Hanba YT, Shibasaka M, Hayakawa T, Kasamo K, Terashima I, Katsuhara M. Overexpression of the barley aquaporin HvPIP2;1 increases internal CO2 conductance and CO2 assimilation in the leaves of transgenic rice plants. Plant and Cell Physiology. 2004;45:521–529. doi: 10.1093/pcp/pch070. [DOI] [PubMed] [Google Scholar]
- Heinen RB, Ye Q, Chaumont F. Role of aquaporins in leaf physiology. Journal of Experimental Botany. 2009;60:2971–2985. doi: 10.1093/jxb/erp171. [DOI] [PubMed] [Google Scholar]
- Hukin D, Doering-Saad C, Thomas CR, Pritchard J. Sensitivity of cell hydraulic conductivity to mercury is coincident with symplasmic isolation and expression of plasmalemma aquaporin genes in growing maize roots. Planta. 2002;215:1047–1056. doi: 10.1007/s00425-002-0841-2. [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conejero G, Rodriguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiology. 2009;150:1955–1971. doi: 10.1104/pp.109.138008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JY, Kim DG, Kim YO, Kim JS, Kang H. An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Molecular Biology. 2004;54:713–725. doi: 10.1023/B:PLAN.0000040900.61345.a6. [DOI] [PubMed] [Google Scholar]
- Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjövall S, Fraysse L, Weig AR, Kjellbom P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiology. 2001;126:1358–1369. doi: 10.1104/pp.126.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P. The role of aquaporins in cellular and whole plant water balance. Biochimica et Biophysica Acta. 2000;1465:324–342. doi: 10.1016/s0005-2736(00)00147-4. [DOI] [PubMed] [Google Scholar]
- Kaldenhoff R, Fischer M. Functional aquaporin diversity in plants. Biochimica et Biophysica Acta. 2006;175:1134–1141. doi: 10.1016/j.bbamem.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Katsuhara M. Molecular mechanisms of water uptake and transport in plant roots: research progress with water channel aquaporins. Plant Root. 2007;1:22–26. [Google Scholar]
- Katsuhara M, Akiyama Y, Koshio K, Shibasaka M, Kasamo K. Functional analysis of water channels in barley roots. Plant and Cell Physiology. 2002;43:885–893. doi: 10.1093/pcp/pcf102. [DOI] [PubMed] [Google Scholar]
- Katsuhara M, Hanba YT. Barley plasma membrane intrinsic proteins (PIP aquaporins) as water and CO2 transporters. Pflügers Archive–European Journal of Physiology. 2008;456:687–691. doi: 10.1007/s00424-007-0434-9. [DOI] [PubMed] [Google Scholar]
- Katsuhara M, Hanba Y, Shiratake K, Maeshima M. Expanding roles of plant aquaporins in plasma membranes and cell organelles. Functional Plant Biology. 2008;35:1–14. doi: 10.1071/FP07130. [DOI] [PubMed] [Google Scholar]
- Katsuhara M, Koshio K, Shibasaka M, Hayashi Y, Hayakawa T, Kasamo K. Over-expression of a barley aquaporin increased the shoot/root ratio and raised salt sensitivity in transgenic rice plants. Plant and Cell Physiology. 2003a;44:1378–1383. doi: 10.1093/pcp/pcg167. [DOI] [PubMed] [Google Scholar]
- Katsuhara M, Koshio K, Shibasaka M, Kasamo K. Expression of an aquaporin at night in relation to the growth and root water permeability in barley seedlings. Soil Science and Plant Nutrition. 2003b;49:883–888. [Google Scholar]
- Knipfer T, Besse M, Verdeil JL, Fricke W. Aquaporin-facilitated water uptake in barley (Hordeum vulgare L.) roots. Journal of Experimental Botany. 2011 doi: 10.1093/jxb/err075. doi:10.1093/jxb/err075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipfer T, Fricke W. Root pressure and a solute reflection coefficient close to unity exclude a purely apoplastic pathway of radial water transport in barley (Hordeum vulgare) New Phytologist. 2010;187:159–170. doi: 10.1111/j.1469-8137.2010.03240.x. [DOI] [PubMed] [Google Scholar]
- Li GW, Zhang MH, Cai WM, Sun WN, Su WA. Characterization of OsPIP2;7, a water channel protein in rice. Plant and Cell Physiology. 2008;49:1851–1858. doi: 10.1093/pcp/pcn166. [DOI] [PubMed] [Google Scholar]
- Liu K, Nagase H, Huang CG, Calamita G, Agre P. Purification and functional characterization of aquaporin-8. Biology of the Cell. 2006;98:153–161. doi: 10.1042/BC20050026. [DOI] [PubMed] [Google Scholar]
- Ma LF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M. A silicon transporter in rice. Nature. 2006;440:688–691. doi: 10.1038/nature04590. [DOI] [PubMed] [Google Scholar]
- Marini AM, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. Molecular Cell Biology. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C. Plant aquaporins: novel functions and regulation properties. FEBS Letters. 2007;581:2227–2236. doi: 10.1016/j.febslet.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ. The vacuolar membrane protein gamma-TIP creates water specific channels in Xenopus oocytes. EMBO Journal. 1993;12:2241–2247. doi: 10.1002/j.1460-2075.1993.tb05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Verdoucq L, Luu DT, Santoni V. Plant aquaporins: membrane channels with multiple integrated functions. Annual Review of Plant Biology. 2008;59:595–624. doi: 10.1146/annurev.arplant.59.032607.092734. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. A multi-color set of in vivo organelle markers for colocalization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Norholm MH, Jensen JK, Halkier BA. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Research. 2006;34:e122. doi: 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TP, Carr DJ. A suberized layer in the cell walls of the bundle sheath of grasses. Australian Journal of Biological Sciences. 1970;23:275–287. [Google Scholar]
- O'Brien TP, Kuo J. Development of the suberized lamella in the mestome sheath of wheat leaves. Australian Journal of Botany. 1975;23:783–794. [Google Scholar]
- Obroucheva NV, Sin'kevich IA. Aquaporins and cell growth. Russian Journal of Plant Physiology. 2010;57:153–165. [Google Scholar]
- Pata MO, Wu BX, Bielawski J, Xiong TC, Hannun YA, Ng CK. Molecular cloning and characterization of OsCDase, a ceramidase enzyme from rice. The Plant Journal. 2008;55:1000–1009. doi: 10.1111/j.1365-313X.2008.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen D, Marty F, Leborgne-Castel N. New insights into the tonoplast architecture of plant vacuoles and vacuolar dynamics during osmotic stress. BMC Plant Biology. 2005;5:13. doi: 10.1186/1471-2229-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A, Boscari A, Schreiber L, Kerstiens G, Jarvis M, Herzyk P, Fricke W. Cloning and expression analysis of candidate genes involved in wax deposition along the growing barley (Hordeum vulgare) leaf. Planta. 2007a;226:1459–1473. doi: 10.1007/s00425-007-0585-0. [DOI] [PubMed] [Google Scholar]
- Richardson A, Wojciechowski T, Franke R, Schreiber L, Kerstiens G, Jarvis M, Fricke W. Cuticular permeance in relation to wax and cutin development along the growing barley (Hordeum vulgare) leaf. Planta. 2007b;225:1471–1481. doi: 10.1007/s00425-006-0456-0. [DOI] [PubMed] [Google Scholar]
- Sakurai J, Ishikawa F, Yamaguchi T, Uemura M, Maeshima M. Identification of 33 aquaporin genes and analysis of their expression and function. Plant and Cell Physiology. 2005;46:1568–1577. doi: 10.1093/pcp/pci172. [DOI] [PubMed] [Google Scholar]
- Schünmann PHD, Ougham HJ, Turk KS. Leaf extension in the slender barley mutant: delineation of the zone of cell expansion and changes in translatable mRNA during leaf development. Plant, Cell and Environment. 1994;17:1315–1322. [Google Scholar]
- Schnurbusch T, Hayes J, Hrmova M, Baumann U, Ramesh SA, Tyerman SD, Langridge P, Sutton T. Boron toxicity tolerance in barley through reduced expression of the multifunctional aquaporin HvNIP2;1. Plant Physiology. 2010;153:1706–1715. doi: 10.1104/pp.110.158832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüssler MD, Alexandersson E, Bienert GP, Kichey T, Laursen KH, Johanson U, Kjellbom P, Schjoerring JK, Jahn TP. The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. The Plant Journal. 2008;56:756–767. doi: 10.1111/j.1365-313X.2008.03632.x. [DOI] [PubMed] [Google Scholar]
- Shen L, Gong J, Caldo RA, Nettleton D, Cook D, Wise RP, Dickerson JA. BarleyBase: an expression profiling database for plant genomics. Nucleic Acids Research. 2005;33:D614–D618. doi: 10.1093/nar/gki123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Wada M, Ludewig U, Schaaf G, von Wiren N, Fujiwara T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. The Plant Cell. 2006;18:1498–1598. doi: 10.1105/tpc.106.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournaire-Roux C, Sutka M, Javot H, Gout E, Gerbeau P, Luu DT, Bligny R, Maurel C. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature. 2003;425:393–397. doi: 10.1038/nature01853. [DOI] [PubMed] [Google Scholar]
- Volkov V, Hachez C, Moshelion M, Draye X, Chaumont F, Fricke W. Water permeability differs between growing and non-growing barley leaf tissues. Journal of Experimental Botany. 2007;58:377–390. doi: 10.1093/jxb/erl203. [DOI] [PubMed] [Google Scholar]
- Wei W, Alexandersson E, Golldack D, Miller AJ, Kjellbom PO, Fricke W. HvPIP1;6, a barley (Hordeum vulgare L.) plasma membrane water channel particularly expressed in growing compared with non-growing leaf tissues. Plant and Cell Physiology. 2007;48:1132–1147. doi: 10.1093/pcp/pcm083. [DOI] [PubMed] [Google Scholar]
- Wu X, Lin J, Lin Q, Wang J, Schreiber L. Casparian strips in needles are more solute-permeable than endodermal transport barriers in roots of Pinus bungeana. Plant and Cell Physiology. 2005;46:1799–1808. doi: 10.1093/pcp/pci194. [DOI] [PubMed] [Google Scholar]
- Ye Q, Holbrook NM, Zwieniecki MA. Cell-to-cell pathway dominates xylem-epidermis hydraulic connection in Tradescantia fluminensis (Vell. Conc.) leaves. Planta. 2008;227:1311–1317. doi: 10.1007/s00425-008-0703-7. [DOI] [PubMed] [Google Scholar]
- Zelazny E, Borst JW, Muylaert M, Batoko H, Hemminga MA, Chaumont F. FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proceedings of the National Academy of Sciences, USA. 2007;104:12359–12364. doi: 10.1073/pnas.0701180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.