Abstract
Tocopherols are antioxidants found in chloroplasts of leaves, and it is a matter of current debate whether or not they can affect signaling and gene expression in plant cells. For insight into the possible effects of altered tocopherol composition in chloroplasts on gene expression in the nucleus, the expression of ethylene biosynthesis, perception and signaling genes was investigated in vte1 and vte4 Arabidopsis thaliana mutants, which are impaired in tocopherol (vitamin E) biosynthesis. Changes in gene expression were measured in plants exposed to either salt or water stress, and in young and mature leaves of vte1 and vte4 mutants, which lack tocopherol cyclase and γ-tocopherol methyltransferase, respectively. While transcript levels of ethylene signaling genes in the vte1 mutant and the wild type were similar in all tested conditions, major changes in gene expression occurred in the vte4 mutant, particularly in mature leaves (compared with young leaves) and under salt stress. Accumulation of γ- instead of α-tocopherol in this mutant led to elevated transcript levels of ethylene signaling pathway genes (particularly CTR1, EIN2, EIN3 and ERF1) in mature leaves of control plants. However, with salt treatment, transcript levels of most of these genes remained constant or dropped in the vte4 mutant, while they were dramatically induced in the wild type and the vte1 mutant. Furthermore, under salt stress, leaf age-induced jasmonic acid accumulated in both the vte1 mutant and the wild type, but not in the vte4 mutant. It is concluded that jasmonic acid and ethylene signaling pathways are down-regulated in mature leaves of salt-stressed vte4 plants.
Keywords: Ethylene signaling, Salt stress, Tocochromanols, Vitamin E, Water stress
Introduction
Tocopherols, which are part of the vitamin E group of compounds, are lipid-soluble molecules essential for human nutrition, but they are only synthesized by photosynthetic organisms, including all plants, algae and most cyanobacteria (Horvath et al. 2006, Mène-Saffrané and DellaPenna 2010). Tocopherols play an important role as antioxidants, but there is some controversy concerning whether or not they have additional functions in both animals and plants (Azzi 2007, Maeda and DellaPenna 2007, Traber and Atkinson 2007, Falk and Munné-Bosch 2010). Recent studies on vitamin E-deficient (vte) Arabidopsis thaliana mutants have revealed that one of the most important functions tocopherols play is acting as antioxidants in chloroplasts, thus protecting plants from photoinhibition and photo-oxidative stress (Havaux et al. 2005). Tocopherols, in cooperation with other antioxidants, can scavenge lipid peroxyl radicals, preventing the propagation of lipid peroxidation, and are excellent quenchers and scavengers of singlet oxygen, controlling its levels. Furthermore, studies on vte mutants have revealed an important role for tocopherols in low-temperature adaptation (Maeda et al. 2006, Maeda et al. 2008) and seedling germination and growth (Sattler et al. 2004, Sattler et al. 2006), in both cases influencing gene expression levels though modulation of extraplastidic polyunsaturated fatty acid metabolism. Under water deficit and salt stress, however, it is still unknown to what extent a tocopherol deficiency in these mutants can alter the plant stress response at the gene expression level.
Previous studies have shown a positive correlation between tocopherol accumulation and water stress in several plant species (Munné-Bosch et al. 1999, Munné-Bosch and Alegre 2003), including A. thaliana (Cela et al. 2009). However, nothing is known about how a tocopherol deficiency in A. thaliana affects water stress tolerance. It is noteworthy that increased tocopherol levels in transgenic tobacco plants overexpressing VTE1 from A. thaliana show enhanced tolerance to water stress (Liu et al. 2008). VTE1 encodes a tocopherol cyclase that is essential for the synthesis of all tocopherol forms (Porfirova et al. 2002, Sattler et al. 2003), thus this study indicates that tocopherols, which mainly accumulate in the form of α-tocopherol in leaves (Szymanska and Kruk 2008a, Szymanska and Kruk 2008b), confer resistance to water stress. Protection is thought to be exerted through their antioxidant role in chloroplasts, since reactive oxygen species (ROS) production is known to be exacerbated under drought stress (Smirnoff 1993, Smirnoff 2005), but the exact mechanisms remain unknown. Less information is available on the role of tocopherols in salt stress tolerance, another of the most important stresses limiting crop yields. Salt stress limits growth by different mechanisms, including ion toxicity, causing osmotic unbalance and leading to oxidative stress by increasing ROS production (Miller et al. 2010). However, very little is known about the role of tocopherols in protecting plants from salt stress. Interestingly, it has been shown that decreased total tocopherol contents in HPT:RNAi transgenic tobacco plants, which lack all tocopherol forms, increases the sensitivity to salt stress, while tolerance to osmotic stress and methyl viologen of γ-tocopherol methyltransferase transgenics, which accumulate γ- instead of α-tocopherol, is strongly increased (Abbasi et al. 2007).
Ethylene gas regulates many diverse metabolic and developmental processes in plants, ranging from seed germination to organ senescence, and is considered to play a major role as a signal molecule at low concentrations in the tolerance to environmental stresses, including water and salt stress (Bleecker and Kende 2000, Argueso et al. 2007). Ethylene production is induced by various stresses, as well as by other hormones and developmental cues (Argueso et al. 2007). The rate-limiting step in the two-step biosynthetic pathway is the production of 1-aminocyclopropane-1-carboxylic acid (ACC) by the enzyme ACC synthase (ACS). Conversion of ACC into ethylene gas is then catalyzed by ACC oxidase (ACO). The ACS and ACO enzymes are encoded by multigene families. A. thaliana has nine ACS genes (ACS1, ACS2, ACS4, ACS5, ACS6, ACS7, ACS8, ACS9 and ACS11), which display unique and overlapping expression patterns (Tsuchisaka and Theologis 2004, Argueso et al. 2007). ACS gene expression is differentially regulated by various ethylene-inducing factors. For instance, ACS6 expression is stimulated by ozone, wounding, auxins and ethylene (Vahala et al. 1998, Tian et al. 2002). All but three of the ACS genes are expressed in young rosette leaves; ACS1 is expressed in the vascular tissue, ACS9 appears to be inactive and ACS11 expression is restricted to the trichomes (Tsuchisaka and Theologis 2004).
The ethylene signal transduction pathway, as proposed in A. thaliana, involves five ethylene receptors ETR1, ERS1, ETR2, EIN4 and ERS2, as well as the negative regulator CTR1, the downstream positive regulator EIN2, and the transcription factor EIN3 (among others), which up-regulates yet another transcription factor, ERF1 (Chen et al. 2005, Kendrick and Chang 2008; see also Supplementary Fig. S1). Recent studies have shown that alterations in ethylene signaling affect plant responses to both salt and water stress (Cao et al. 2006, Cao et al. 2008, Cela et al. 2009). For example, ethylene-insensitive mutants are hypersensitive to salt, suggesting that ethylene signaling reduces sensitivity to salt stress (Cao et al. 2008).
In a previous study, it was shown that a defect in ethylene signaling in the ein3-1 mutant can alter tocopherol biosynthesis in A. thaliana (Cela et al. 2009). Since ethylene is a major player in the response of plant to water and salt stress, we aimed here to evaluate not only whether or not a deficiency in specific tocopherol forms in vte1 and vte4 mutants of A. thaliana alters plant response to salt and water stress, but also to what extent a particular differential response in these mutants is associated with a specific response at the expression level of ethylene biosynthesis, perception and/or signaling genes.
Results
Plant response to water and salt stress in vte1 and vte4 mutants
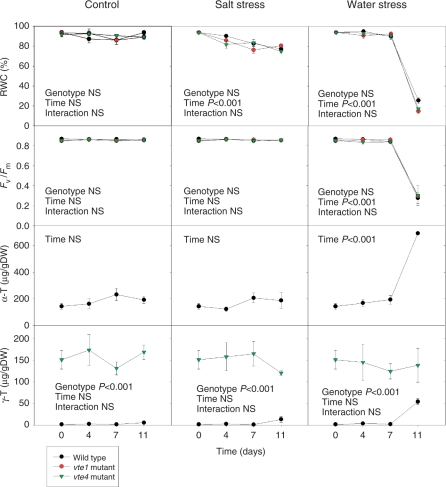
Salt stress (addition of 100 mM NaCl to the nutrient solution) and water stress (by withholding water) for 11 d caused dramatic effects on growth in wild-type A. thaliana plants (Supplementary Fig. S2). Rosette biomass of wild-type plants was reduced by 34 and 57% in response to salt and water stress, respectively. vte1 and vte4 mutants showed a response very similar to that of the wild type, in control and salt- or water-stressed conditions. It is noteworthy that water stress led to a more dramatic phenotype than salt stress; both vte1 and vte4 mutants and the wild type showed a similar response characterized by accelerated senescence in mature leaves, an aspect more evident than that observed under salt stress (Supplementary Fig. S2). Measurements of the relative water content (RWC) and the maximum efficiency of PSII photochemistry (the Fv/Fm ratio), an indicator of photoinhibition to the photosynthetic apparatus, showed that the water stress imposed on plants was much more severe than the salt stress, with RWC decreasing from values >85% in control plants to values of around 20% in water stress and 70% in salt stress (Fig. 1). The Fv/Fm ratio decreased from values around 0.85 in control plants to 0.30 in water-stressed plants, while salt stress did not significantly alter this parameter. Furthermore, in the wild type, the levels of α-tocopherol stayed constant throughout the study in control and salt-stressed plants, while they increased from 190 to 700 μg g−1 DW in water-stressed plants. The levels of γ-tocopherol were much lower than those of α-tocopherol in wild-type plants, but they increased from 4 μg g−1 DW in control to 13 and 54 μg g−1 DW in salt- and water-stressed plants, respectively. As expected, the vte4 mutant showed γ-tocopherol but not α-tocopherol accumulation in leaves, with γ-tocopherol levels around 150 μg g−1 DW under control conditions, similar to α-tocopherol levels in the wild type. However, in contrast to wild-type plants, the vte4 mutant did not increase tocopherol levels under either salt or water stress. As expected, the vte1 mutant did not accumulate either α- or γ-tocopherol in leaves (Fig. 1). Despite significant stress-induced tocopherol increases in wild-type plants, the expression levels of three of the most important genes involved in tocopherol biosynthesis (encoding homogentisate phytyltranferase, VTE2; tocopherol cyclase, VTE1; and γ-tocopherol methyltransferase, VTE4) were not significantly altered (Supplementary Fig. S3).
Fig. 1.
Time-course evolution of relative water content (RWC), maximum efficiency of PSII photochemistry (Fv/Fm ratio) and levels of α- and γ-tocopherol (α-T and γ-T, respectively) in young leaves of the wild type and vte1 and vte4 mutants of A. thaliana exposed to control, salt stress or water stress conditions for 11 d. Data are the mean ± SE of n = 4 experiments. Significant differences between genotypes, time of measurements and their interaction (genotype × time) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant.
Expression of ethylene biosynthesis, perception and signaling genes in response to water and salt stress in vte1 and vte4 mutants
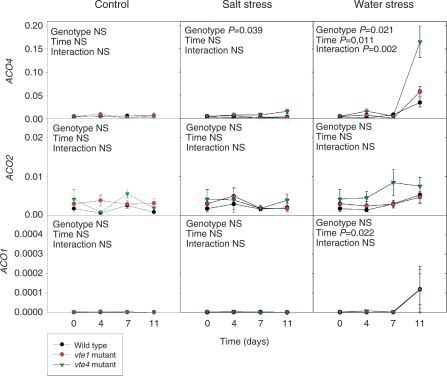
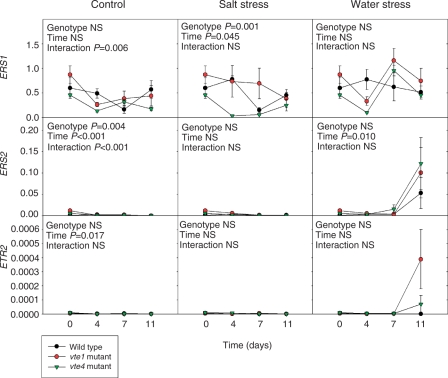
Expression levels were analyzed for the nine genes of the ACS family (ACS1, ACS2, ACS4, ACS5, ACS6, ACS7, ACS8, ACS9 and ACS11), and ACS2, ACS6, ACS8 and ACS11 were shown to have high expression levels in leaves by quantitative PCR (data not shown). Later, it was confirmed that the ACS2 gene showed the highest expression levels by quantitative PCR (Supplementary Fig. S4). Transcripts of both ACS2 and ACS6 increased significantly in response to water deprivation in both mutants and the wild type, while ACS8 also increased in response to water stress but differences were not significant. In contrast, transcript levels of ACS6 increased in the vte4 mutant exposed to salt stress, while levels remained unaltered in the wild type and the vte1 mutant (Supplementary Fig. S4). Transcript levels of ACS11 stayed at values <0.00012 throughout the study (data not shown). Increases in ACS2 and ACS6 transcripts in water-stressed plants were accompanied by significant increases in ACO4 and ACO1 expression levels (Fig. 2). Furthermore, ACO4 expression levels increased more in the vte4 mutant than in the vte1 mutant or the wild type exposed to water stress, a result that was also observed, although to a lesser extent, in plants exposed to salt stress. Expression levels of ethylene perception genes also revealed a differential response in tocopherol mutants (Fig. 3). The vte4 mutant showed lower transcript levels of ERS1 than the vte1 mutant in all tested conditions, although differences were significant under salt stress only. Analysis of the ethylene signaling genes EIN2, EIN3, CTR1 and ERF1 did not reveal any effect of tocopherol deficiency or stress treatments, except for an increase in ERF1 transcript levels under 11 d of water stress in all genotypes (Supplementary Fig. S5).
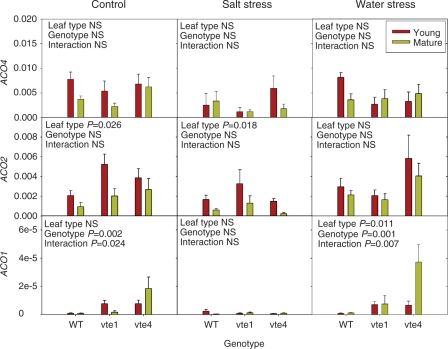
Fig. 2.
Time-course evolution of relative expression of the ethylene biosynthesis genes, ACO4, ACO2 and ACO1 in young leaves of the wild type and vte1 and vte4 mutants of A. thaliana plants exposed to control, salt stress or water stress conditions for 11 d. Data are the mean ± SE of n = 4 experiments. Significant differences between genotypes, time of measurements and their interaction (genotype × time) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant. GAPDH was used as the reference gene to calculate the relative expression of the ethylene biosynthesis genes.
Fig. 3.
Time-course evolution of relative expression of the ethylene perception genes, ERS1, ERS2 and ETR2 in young leaves of the wild type and vte1 and vte4 mutants of A. thaliana plants exposed to control, salt stress or water stress conditions for 11 d. Data are the mean ± SE of n = 4 experiments. Significant differences between genotypes, time of measurements and their interaction (genotype × time) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant. GAPDH was used as the reference gene to calculate the relative expression of the ethylene perception genes.
Influence of leaf age on expression of ethylene biosynthesis, perception and signaling genes and their interaction with salt and water stress
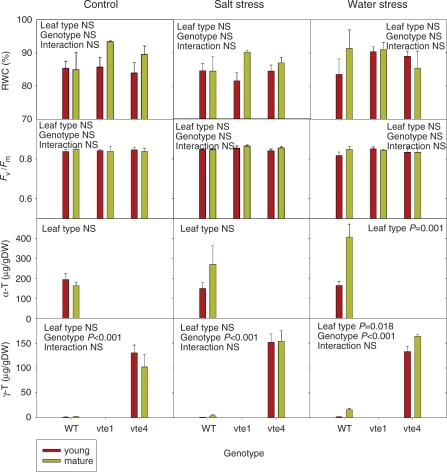
The influence of leaf age on gene expression was evaluated in control conditions and in plants exposed to 7 d of salt or water stress, before stress treatments caused severe cell death in mature leaves. Values of RWC and the Fv/Fm ratio in young and mature leaves did not reveal differences in the degree of stress suffered by both leaf types in all treatments and mutants (Fig. 4). α-Tocopherol increased significantly from 190 to 400 μg g−1 DW in mature leaves compared with young leaves under water stress, but leaf age did not cause any effect under control or salt stress conditions, although levels increased slightly under salt stress. γ-Tocopherol levels changed in parallel to those of α-tocopherol, with a significant increase in wild-type plants under salt stress. Mature leaves of the vte4 mutant also showed significant increases in γ-tocopherol levels in response to water stress (Fig. 4). Again, transcript levels of tocopherol biosynthetic genes did not always correlate with tocopherol levels in leaves. Transcript levels of VTE2 increased in parallel with tocopherol levels in mature leaves of wild-type plants exposed to water stress, but transcript levels of VTE1, VTE2 and VTE4 increased sharply in mature leaves of wild-type plants in response to salt stress with no concomitant increases in tocopherol (Supplementary Fig. S6; and Fig. 4).
Fig. 4.
Relative water content (RWC), maximum efficiency of PSII photochemistry (Fv/Fm ratio) and levels of α- and γ-tocopherol (α-T and γ-T, respectively) in young and mature leaves of the wild type, and vte1 and vte4 mutants of A. thaliana exposed to control, salt stress or water stress conditions for 7 d. Data are the mean ± SE of n = 4 experiements. Significant differences between genotypes, leaf type and their interaction (genotype × leaf type) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant.
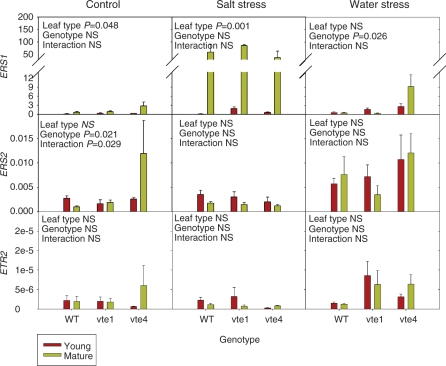
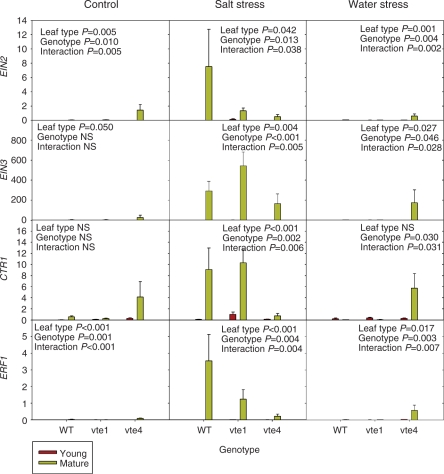
While leaf age did not induce changes in ACS2, ACS6 and ACS8 transcript levels (Supplementary Fig. S7), the ACO1 and ACO2 transcript levels were drastically affected by leaf age, with particularly interesting interactions with salt and water stress (Fig. 5). In control plants, leaf age reduced ACO2 transcript levels, particularly in the wild type and the vte1 mutant, an aspect that was also observed in ACO4 and ACO1 expression, though to a lower extent. These effects were not so evident in the vte4 mutant, to the extreme that ACO1 transcript levels increased in mature leaves of this mutant under control conditions. A similar response was observed under water stress, especially exacerbated in ACO1 transcript levels of the vte4 mutant, which increased particularly in mature leaves of water-stressed plants. In contrast, this response was not observed under salt stress (Fig. 5). Furthermore, transcript levels of ethylene perception genes, namely ERS1, ERS2 and ETR2, increased in mature leaves of control plants of the vte4 mutant only, a response that was also observed only for this mutant for the ERS1 gene under water stress (Fig. 6). Major changes in gene expression of ethylene signaling genes also occurred in mature leaves of the vte4 mutant under both control and water-stressed conditions (Fig. 7). Elevated levels of all the ethylene signaling transcripts, particularly the downstream components CTR1, EIN2, EIN3 and ERF1, were observed in mature leaves of the control and water-stressed vte4. However, with salt treatment these transcript levels remained constant or dropped in the vte4 mutant, while they were dramatically induced in the wild type (particularly CTR1, EIN2 and ERF1). Furthermore, the vte1 mutant and wild-type plants behaved similarly in all tested conditions, particularly for CTR1, EIN3 and ERF1 expression (Fig. 7).
Fig. 5.
Relative expression of the ethylene biosynthesis genes, ACO4, ACO2 and ACO1 in young and mature leaves of the wild type and vte1 and vte4 mutants of A. thaliana plants exposed to control, salt stress or water stress conditions for 7 d. Data are the mean ± SE of n = 4 experiments. Significant differences between genotypes, leaf type and their interaction (genotype × leaf type) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant. GAPDH was used as the reference gene to calculate relative expression of the ethylene biosynthesis genes.
Fig. 6.
Relative expression of the ethylene perception genes, ERS1, ERS2 and ETR2 in young and mature leaves of the wild type and vte1 and vte4 mutants of A. thaliana plants exposed to control, salt stress or water stress conditions for 7 d. Data are the mean ± SE of n = 4 experiments. Significant differences between genotypes, leaf type and their interaction (genotype × leaf type) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant. GAPDH was used as the reference gene to calculate relative expression of the ethylene perception genes.
Fig. 7.
Relative expression of the ethylene signaling genes, EIN2, EIN3, CTR1 and ERF1 in young and mature leaves of the wild type and vte1 and vte4 mutants of A. thaliana plants exposed to control, salt stress or water stress conditions for 7 d. Data are the mean ± SE of n = 4 experiments. Significant differences between genotypes, leaf type and their interaction (genotype × leaf type) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant. GAPDH was used as the reference gene to calculate relative expression of the ethylene signaling genes.
Changes in jasmonic acid levels
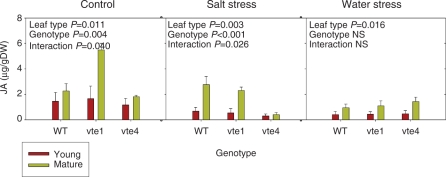
To unravel whether or not those changes in gene expression could be associated with a modulation of oxylipin levels, a time course of the evolution of endogenous jasmonic acid levels was evaluated under control, salt and water stress conditions (Supplementary Fig. S8), and in young and mature leaves at 7 d of stress (Fig. 8). Jasmonic acid levels did not significantly differ between vte1 or vte4 mutants and the wild type in young leaves during the 11 d of treatment (Supplementary Fig. S8). However, jasmonic acid levels increased in mature leaves of the vte1 mutant under control conditions, but not in those of the vte4 mutant or the wild type. Under salt stress, leaf age-induced jasmonic acid accumulated in both the vte1 mutant and the wild type, but not in the vte4 mutants (Fig. 8). No differences between mutants and the wild type were observed in jasmonic acid levels under water stress (Supplementary Fig. S8).
Fig. 8.
Jasmonic acid (JA) levels in young and mature leaves of the wild type and vte1 and vte4 mutants of A. thaliana plants exposed to control, salt stress or water stress conditions for 7 d. Data are the mean ± SE of n = 4 experiments. Significant differences between genotypes, leaf type and their interaction (genotype × leaf type) are given in the panels (ANOVA, P ≤ 0.05). NS, not significant.
Discussion
A number of environmental stress factors have been associated with an increase in photosynthesis-derived ROS, and the levels of tocopherols and other antioxidants have been related to stress tolerance. Whereas stress-tolerant plants normally display increased tocopherol levels, the most sensitive ones show net tocopherol loss under stress, which is thought to lead to oxidative damage and cell destruction (reviewed in Munné-Bosch and Alegre 2002). However, the function of tocopherols might be more complex, especially considering their role in cellular signaling. Tocopherols are part of an intricate signaling network controlled by ROS, antioxidants and hormones, and are therefore good candidates to influence signaling in plant cells. Tocopherols, ascorbate and glutathione are interdependent in the control of ROS levels (Foyer and Noctor 2005, Kanwischer et al. 2005, Munné-Bosch 2005, Asada 2006), and ROS are crucial modulators of gene expression in chloroplasts (Apel and Hirt 2004) and the nucleus (Vandenabeele et al. 2003). Furthermore, ROS and hormones interact in the regulation of signal transduction and gene expression in plants (Pastori and Foyer 2002). By controlling ROS levels and the extent of lipid peroxidation in chloroplasts, tocopherols may indirectly regulate the amounts of extraplastidic lipid peroxidation products and affect gene expression in the nucleus. Hofius et al. (2004) showed that tocopherols may affect source–sink transitions and may alter gene expression in plants, thus providing the first steps towards understanding the role of tocopherols in signaling in plant cells. Later, it was found that tocopherols play an essential role in low-temperature adaptation (Maeda et al. 2006, Maeda et al. 2008, Song et al. 2010) and seedling germination and growth (Sattler et al. 2004, Sattler et al. 2006), in both cases influencing gene expression levels though modulation of extraplastidic polyunsaturated fatty acid metabolism. Under water deficit and salt stress, however, it was still unknown to what extent a tocopherol deficiency can alter the plant stress response at the gene expression level.
The results clearly indicate that major changes in transcript levels of ethylene biosynthesis, perception and signaling genes occur in the vte4 mutant, which lacks α-tocopherol, but accumulates γ-tocopherol in leaves. Transcript levels of ACS6 increased in the vte4 mutant exposed to 11 d of salt stress, while levels were unaltered in the wild type and the vte1 mutant. ACO4 expression levels also increased more in the vte4 mutant than in the vte1 mutant or the wild type exposed to severe water stress, a result that was also observed, although to a lesser extent, in plants exposed to salt stress, probably partly because the salt stress was not as severe as the water stress. Another important factor controlling the extent of tocopherol effects on the expression of ethylene biosynthesis, perception and signaling genes is leaf age. In control plants, reduced ACO2 transcript levels were observed in mature leaves of the wild type and the vte1 mutant, while ACO1 transcript levels increased in those of the vte4 mutant. Furthermore, transcript levels of ERS1, ERS2 and ETR2 increased in mature leaves of the vte4 mutant only, and elevated levels of CTR1, EIN2, EIN3 and ERF1 were observed in mature leaves of the control and water-stressed vte4 mutant. However, with salt treatment, these transcript levels remained constant or dramatically dropped in the vte4 mutant. In addition, leaf age-induced jasmonic acid accumulated in both the vte1 mutant and the wild type, but not in the vte4 mutant under salt stress. It has been shown that the transcription factor ERF1 acts downstream of the interaction between the ethylene and jasmonic acid pathways and is a key element in the integration of both signals for the regulation of defense response genes (Lorenzo et al. 2003), thereby suggesting that jasmonic acid and ethylene signaling transcript levels are down-regulated in mature leaves of the salt-stressed vte4 mutant.
Interestingly, this differential response at the gene expression level in the vte4 mutant results in a similar resistance to salt and water stress, as indicated by the Fv/Fm ratio and plant biomass, which were similar in the vte4 mutant and the wild type. Similar stress sensitivity in the vte4 mutant and the wild type was also observed when plants were subjected to light, heat and cold stress (Bergmüller et al. 2003). Indeed, neither the vte1 nor the vte4 mutants were more sensitive than the wild type to any of the stress conditions applied in the present study, thus suggesting that γ-tocopherol is at least as efficient as its α homolog in the protection of plants against salt and water stress. It appears, however, that the mechanisms to achieve this similar response differ between the mutants, as indicated by changes in transcript levels of ethylene biosynthesis genes under both salt and water stress. Obviously, this does not exclude many other mechanisms that may operate in these mutants, and their response to salt and water stress may not necessarily be the same as their response to other types or magnitudes of stresses. Indeed, transgenic tobacco accumulating γ- instead of α-tocopherol was more sensitive to high doses of salt stress (≥200 mM NaCl) than the wild-type strain, which accumulates α-tocopherol (Abbasi et al. 2007). Taken together, this suggests that plants constitutively accumulating γ- instead of α-tocopherol show an altered response to salt stress, since sustained accumulation of γ-tocopherol represses jasmonic acid and ethylene signaling response pathways, which leads to reduced expression of transcription factor ERF1. One may speculate that this will predispose plants to suffer the negative effects of salt stress when the duration and/or magnitude of stress is increased (Abbasi et al. 2007), effects that were not observed in our study due to exposure of plants to low doses of salinity. It is interesting to note that such effects were specific to the plant response to salinity and were not observed under water deficit.
Furthermore, the apparent lack of differential response in the vte1 mutant might be associated with the antioxidant potential of the tocopherol precursor, 2,3-dimethyl-5-phytylhydroquinol, which accumulates in this mutant (Liebler and Burr 2000). Although the vte1 mutant lacks both tocopherols and plastochromanol, which account for 5–10% of total tocochromanols in A. thaliana grown under low light conditions (Zbierzak et al. 2010), this mutant accumulates plastoquinol, an important lipid-soluble antioxidant which in addition to tocopherols protects chloroplasts from photo-oxidative stress (Szymanska and Kruk 2010). It is therefore very likely that the accumulation of plastoquinol and 2,3-dimethyl-5-phytylhydroquinol can substitute for the lack of tocochromanols in the vte1 mutant and therefore no apparent phenotype in plant response to water deficit and salt stress in the low light conditions used in the present study was observed.
It is concluded that jasmonic acid and ethylene signaling pathways are down-regulated in mature leaves of the salt-stressed vte4 mutant, thus indicating that an altered tocopherol composition in chloroplasts of this mutant alters the physiological response of mature leaves to salt stress. It is still to be determined, however, how enhanced γ-tocopherol levels lead to reduced jasmonic acid and ethylene signaling gene expression levels in mature leaves, and why this occurs under salt stress conditions only.
Materials and Methods
Plant material, growth conditions and treatments
Seeds of A. thaliana Columbia ecotype (Col 0) and vte1 and vte4 mutants, which were provided by Kathleen Brückner (University of Kiel, Germany), were used in this study. vte1 and vte4 mutants have T-DNA insertions in the VTE1 and VTE4 genes, which encode tocopherol cyclase and γ-tocopherol methyltransferase, respectively, so that the vte1 mutant lacks both α- and γ-tocopherol, and the vte4 mutant lacks α-tocopherol but accumulates γ-tocopherol (Porfirova et al. 2002, Bergmüller et al. 2003). Plants were grown in pots containing a mixture of peat : perlite : vermiculite (1 : 1 : 1, v/v/v) in a constant environment chamber (8 h photoperiod, 90–110 μmol quanta m−2 s−1, air temperature 21–23°C) and were watered with Hoagland's solution every 3 d during 8 weeks until the experiment started. Then, plants were exposed for 11 d to either (i) water deficit (water-stressed plants) by withholding water; (ii) salt stress by watering with Hoagland's solution but with an additional 100 mM of NaCl; or (iii) watering with Hoagland's solution (controls).
For analysis of tocopherols, jasmonic acid and the genes involved in the biosynthesis, perception and signaling of ethylene, young leaves were collected in the middle of the photoperiod at 0, 4, 7 and 11 d of treatment, immediately frozen in liquid nitrogen and stored at −80°C until analyses. Young and mature leaves were additionally collected at day 7 of the experiment (before severe necrosis occurred in mature leaves of water-stressed plants) to examine the influence of leaf age.
Biomass, leaf water content and the Fv/Fm ratio
Biomass was determined after 11 d of treatment by measuring the rosette weight after drying the leaves in an oven at 80°C for 24 h. RWC was determined as 100 (FW − DW)/(TW − DW), where FW is the fresh weight, TW is the turgid weight after re-hydrating the leaves at 4°C in darkness for 24 h, and DW is the dry weight after oven-drying the leaves at 80°C to constant weight. The Fv/Fm was determined by using a pulse-modulated fluorimeter mini-PAM (Walz) after 1 h of dark adaptation, as described (Genty et al. 1989).
Analyses of tocopherols
For the extraction of α- and γ-tocopherol, leaf samples were ground in liquid nitrogen and extracted with ice-cold methanol using a Branson 2510 ultrasonic cleaner (Bransonic) for 45 min. The samples were then centrifuged for 15 min at 4°C and transferred to vials for analysis. The HPLC analysis was carried out as described (Amaral et al. 2005). In brief, the HPLC equipment consisted of an integrated system with a Waters 600 controller pump, a Waters 714 plus auto-sampler and an FP-1520 fluorescence detector (Jasco). Tocopherols were separated on an Inertsil 100A (5 μm, 30 × 250 mm, GL Sciences Inc.) normal-phase column operating at room temperature. The mobile phase used was a mixture of n-hexane and 1,4-dioxane (95.5 : 4.5, v/v) at a flow rate of 0.7 ml min−1, and the injection volume was 10 μl. Detection was carried out for excitation at 295 nm and emission at 330 nm. Quantification was based on the fluorescence signal response compared with authentic standards of each compound (Sigma-Aldrich).
Gene expression analyses
RNA was isolated from leaf material using the RNEasy Plant Mini Kit (Qiagen) following the kit instructions, but incubated with DNase [RNase-Free DNase Set (Qiagen)] to eliminate the genomic DNA. The RNA was used for cDNA synthesis using GeneAmp RNA PCR (Applied Biosystems).
The most important genes related to ethylene biosynthesis, perception and signaling were analyzed by real-time PCR for each sample. At the same time, genes of tocopherol biosynthesis were analyzed only in wild-type plants. cDNAs were amplified using LightCycler 480 SYBR Green 1 Master (Roche), and the specific primers for each gene (Supplementary Table S1) were purchased from Eurofins MWG Operon. For the great majority of primers, the conditions were: 45 cycles of 95°C (10 s); 63°C (20 s); 72°C (20 s). For quantification, the ΔCT method was used with the equation: ratio (reference gene, target gene) = 2CT (reference gene)−CT (target gene), where CT is the cycle number at which enough amplified product accumulates to yield a detectable fluorescent signal, the reference gene was GAPDH (which encodes glyceraldehyde-3-phosphate dehydrogenase) and the target gene was the gene of interest.
Jasmonic acid analyses
Levels of jasmonic acid were analyzed by UPLC-MS/MS (ultra-performance liquid chromatography–tandem mass spectrometry) with a modification of the method described by Abreu and Munné-Bosch (2009). In brief, leaf samples were ground in liquid nitrogen and, after addition of deuterated jasmonic acid as an internal standard, extracted with methanol using sonication. The supernatant was dried completely under a nitrogen stream and the extract reconstituted in methanol. The extracts were then filtered through a 0.22 μm PTFE filter (Waters), and injected into the UPLC-MS/MS system. The UPLC system consisted of an Acquity Waters binary pump equipped with an autosampler and a UV detector. For analysis of the extracts, a HALO C18 column (2.1 × 75 mm, 2.7 μm) was used with a binary solvent system comprising acetonitrile (A) and deionized water (B), both containing 0.005% (v/v) glacial acetic acid. Separations were performed using a gradient of increasing acetonitrile content, at a constant flow rate of 0.4 ml min−1. The gradient used was: 0 min, 1% A; 0–4.5 min, increasing to 45% A; 4.5–5 min, increasing to 99% A; 5–6 min, 99% A; 6–6.2 min, decreasing to 1% A; 6.2–7 min, 1% A. MS/MS analyses were performed on an API 3000 triple quadrupole mass spectrometer (PE Sciex). All the analyses were performed using the Turbo Ion spray source in negative ion mode. MRM acquisition was carried out by monitoring the following transition: JA, 209/59; and for the deuterated internal standard: JA* (d5), 214/62. Quantification by MS/MS using the MRM method was performed as described (Abreu and Munné-Bosch 2009). Results were corrected taking into account the specific recovery rates using the internal deuterated standard.
Statistical analyses
Statistical differences between measurements of the different treatments and ages were analyzed with analysis of variance (ANOVA) using SPSS software. Differences were considered significant at a probability level of P < 0.05.
Supplementary data
Supplementary data are available at PCP online.
Funding
This research was supported by the Generalitat de Catalunya [ICREA Academia prize awarded to S.M.-B.]; the National Institutes of Health [(1R01GM071855) to C.C.]; the Comissionat per a Universitats i Recerca del Departament d'Innovació, Universitats i Empresa of the Generalitat de Catalunya [fellowship awarded J.C.].
Supplementary Material
Acknowledgments
We are very grateful to the Serveis Científico-tècnics (University of Barcelona) for technical assistance and Kathleen Brückner (University of Kiel, Germany) for providing seeds of vte1 and vte4 mutants.
Glossary
Abbreviations
- ACC
1-aminocyclopropane-1-carboxylic acid
- ACO
ACC oxidase
- ACS
ACC synthase
- ANOVA
analysis of variance
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- ROS
reactive oxygen species
- RWC
relative water content
- UPLC-MS/MS
ultra-high performance liquid chromatography–tandem mass spectrometry
References
- Abbasi A, Hajirezaei M, Hofius D, Sonnewald U, Voll LM. Specific roles of α- and γ-tocopherol in abiotic stress responses of transgenic tobacco (Nicotiana tabacum L.) Plant Physiol. 2007;143:1720–1738. doi: 10.1104/pp.106.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu ME, Munné-Bosch S. Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J. Exp. Bot. 2009;60:1261–1271. doi: 10.1093/jxb/ern363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral JS, Casal S, Torres D, Seabra RM, Oliveira BPP. Simultaneous determination of tocopherols and tocotrienols in hazelnuts by a normal phase liquid chromatographic method. Anal. Sci. 2005;21:1545–1548. doi: 10.2116/analsci.21.1545. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Argueso CT, Hansen M, Kieber JJ. Regulation of ethylene biosynthesis. J. Plant Growth Regul. 2007;26:92–105. [Google Scholar]
- Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A. Molecular mechanism of α-tocopherol action. Free Radic. Biol. Med. 2007;43:16–21. doi: 10.1016/j.freeradbiomed.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Bergmüller E, Porfirova S, Dörmann P. Characterization of an Arabidopsis mutant deficient in gamma-tocopherol methyltransferase. Plant Mol. Biol. 2003;52:1181–1190. doi: 10.1023/b:plan.0000004307.62398.91. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H. Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, et al. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2006;143:707–719. doi: 10.1104/pp.106.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Chen SY, Zhang JS. Ethylene signaling regulates salt stress response. Plant Signal. Behav. 2008;3:761–763. doi: 10.4161/psb.3.10.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cela J, Falk J, Munné-Bosch S. Ethylene signaling may be involved in the regulation of tocopherol biosynthesis in Arabidopsis thaliana. FEBS Lett. 2009;583:992–996. doi: 10.1016/j.febslet.2009.02.036. [DOI] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller E. Ethylene signal transduction. Ann. Bot. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J, Munné-Bosch S. Tocochromanol funtions in plants: antioxidation and beyond. J. Exp. Bot. 2010;61:1549–1566. doi: 10.1093/jxb/erq030. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B, Briantais J, Baker NR. The relationship between quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta. 1989;990:87–92. [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dörmann P. Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell. 2005;17:3451–3469. doi: 10.1105/tpc.105.037036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D, Hajirezaei M, Geiger M, Tschiersch H, Melzer M, Sonnewald U. RNAi-mediated tocopherol deficiency impairs photoassimilate export in transgenic potato plants. Plant Physiol. 2004;135:1256–1268. doi: 10.1104/pp.104.043927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath G, Wessjohann L, Bigirimana J, Monica H, Jansen M, Guisez Y, et al. Differential distribution of tocopherols and tocotrienols in photosynthetic and non-photosynthetic tissues. Phytochemistry. 2006;67:1185–1195. doi: 10.1016/j.phytochem.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Kanwischer M, Porfirova S, Bergmüller E, Dörmann P. Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol. 2005;137:713–723. doi: 10.1104/pp.104.054908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick MD, Chang C. Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 2008;11:479–485. doi: 10.1016/j.pbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebler DC, Burr JA. Antioxidant reactions of α-tocopherolhydroquinone. Lipids. 2000;35:1045–1047. doi: 10.1007/s11745-000-0617-8. [DOI] [PubMed] [Google Scholar]
- Liu X, Hua X, Guo J, Qi D, Wang L, Liu Z, et al. Enhanced tolerance to drought stress in transgenic tobacco plants overexpressing VTE1 for increased tocopherol production from Arabidopsis thaliana. Biotechnol. Lett. 2008;30:1275–1280. doi: 10.1007/s10529-008-9672-y. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras O, Sánchez-Serraro JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, DellaPenna D. Tocopherol functions in photosynthetic organisms. Curr. Opin. Plant Biol. 2007;10:260–265. doi: 10.1016/j.pbi.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Maeda H, Sage TL, Isaac G, Welti R, DellaPenna D. Tocopherols modulate extraplastidic polyunsaturated fatty acid metabolism in Arabidopsis at low temperature. Plant Cell. 2008;20:452–470. doi: 10.1105/tpc.107.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Song W, Sage TL, DellaPenna D. Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell. 2006;18:2710–2732. doi: 10.1105/tpc.105.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mène-Saffrané L, DellaPenna D. Biosynthesis, regulation and functions of tocochromanols in plants. Plant Physiol. Biochem. 2010;48:301–309. doi: 10.1016/j.plaphy.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S. The role of α-tocopherol in plant stress tolerance. J. Plant Physiol. 2005;162:743–748. doi: 10.1016/j.jplph.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S, Alegre L. The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 2002;21:31–57. [Google Scholar]
- Munné-Bosch S, Alegre L. Drought-induced changes in the redox state of α-tocopherol, ascorbate, and the diterpene carnosic acid in chloroplasts of Labiatae species differing in carnosic acid contents. Plant Physiol. 2003;131:1816–1825. doi: 10.1104/pp.102.019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munné-Bosch S, Schwarz K, Alegre L. Enhanced formation of α-tocopherol and highly oxidized abietane diterpenes in water-stressed rosemary plants. Plant Physiol. 1999;121:1047–1052. doi: 10.1104/pp.121.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori G, Foyer CH. Common components, networks, and pathways of cross-tolerance to stress: the central role of ‘redox’ and abscisic acid-mediated controls. Plant Physiol. 2002;129:460–468. doi: 10.1104/pp.011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfirova S, Bergmüller E, Tropf S, Lemke R, Dörmann P. Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl Acad. Sci. USA. 2002;99:12495–12500. doi: 10.1073/pnas.182330899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Cahoon EB, Coughlan SJ, DellaPenna D. Characterization of tocopherol cyclases from higher plants and cyanobacteria. Evolutionary implications for tocopherol synthesis and function. Plant Physiol. 2003;132:2184–2195. doi: 10.1104/pp.103.024257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell. 2004;16:1419–1432. doi: 10.1105/tpc.021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Mène-Saffrané L, Farmer EE, Krischke M, Müller MJ, DellaPenna D. Non-enzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell. 2006;18:3706–3720. doi: 10.1105/tpc.106.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. Antioxidants and Reactive Oxygen Species in Plants. Oxford: Blackwell Publishing; 2005. [Google Scholar]
- Song W, Maeda H, DellaPenna D. Mutations of the ER to plastid lipid transporters TGD1, 2, 3 and 4 and the ER oleate desaturase FAD2 suppress the low temperature-induced phenotype of Arabidopsis tocopherol-deficient mutant vte2. Plant J. 2010;62:4–18. doi: 10.1111/j.1365-313X.2010.04212.x. [DOI] [PubMed] [Google Scholar]
- Szymanska R, Kruk J. Tocopherol content and isomers’ composition in selected plant species. Plant Physiol. Biochem. 2008a;46:29–33. doi: 10.1016/j.plaphy.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Szymanska R, Kruk J. Gamma-tocopherol dominates in young leaves of runner bean (Phaseolus coccineus) under a variety of growing conditons: the possible functions of gamma-tocopherol. Phytochemistry. 2008b;69:2142–2148. doi: 10.1016/j.phytochem.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Szymanska R, Kruk J. Plastoquinol is the main prenyllipid synthesized during acclimation to high light conditions in Arabidopsis and is converted to plastochromanol by tocopherol cyclase. Plant Cell Physiol. 2010;51:537–545. doi: 10.1093/pcp/pcq017. [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traber MG, Atkinson J. Vitamin E, antioxidant and nothing more. Free Radic. Biol. Med. 2007;43:4–15. doi: 10.1016/j.freeradbiomed.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylase synthase gene family members. Plant Physiol. 2004;136:2982–3000. doi: 10.1104/pp.104.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahala J, Schlagnhaufer CD, Pell EJ. Induction of an ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol. Plant. 1998;103:45–50. [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, et al. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl Acad. Sci. USA. 2003;100:16113–16118. doi: 10.1073/pnas.2136610100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbierzak AM, Kanwischer M, Wille C, Vidi P, Giavalisco P, Lohmann A, et al. Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem. J. 2010;425:389–399. doi: 10.1042/BJ20090704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.