Abstract
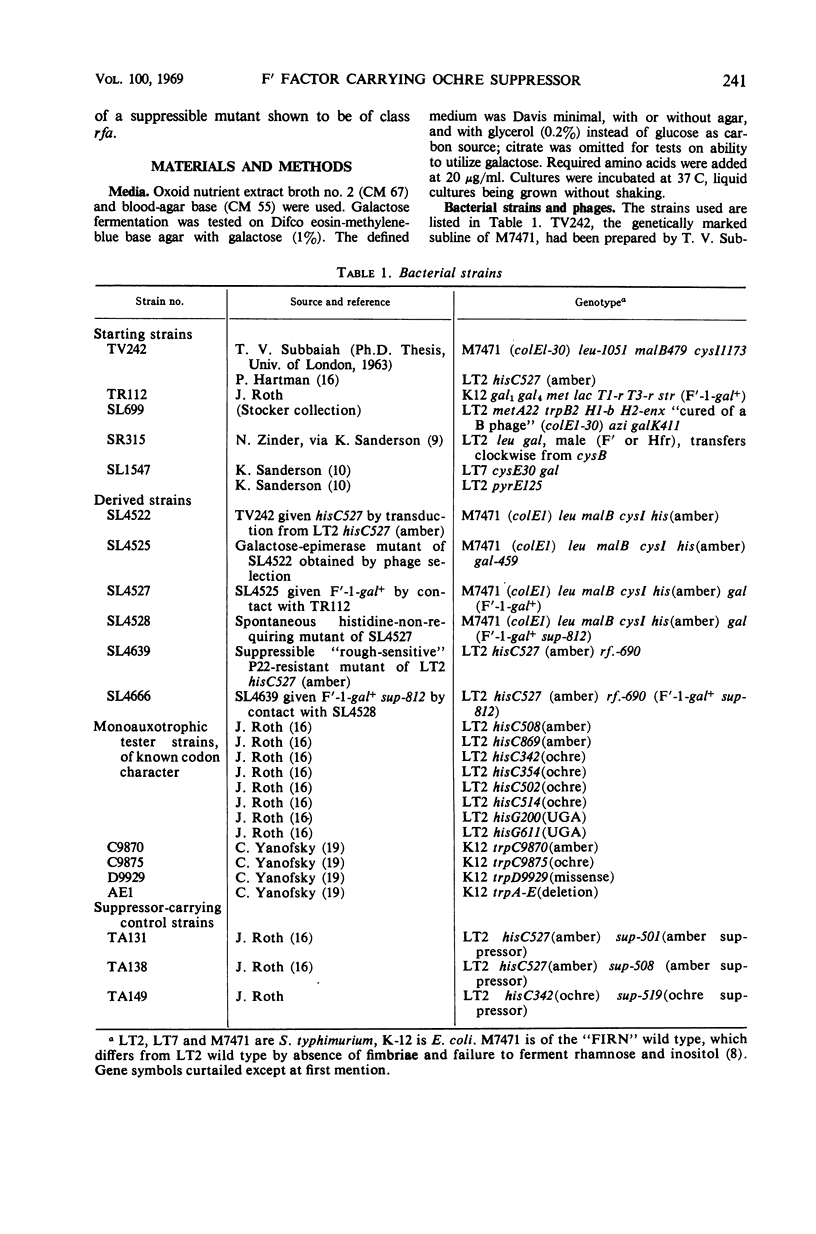
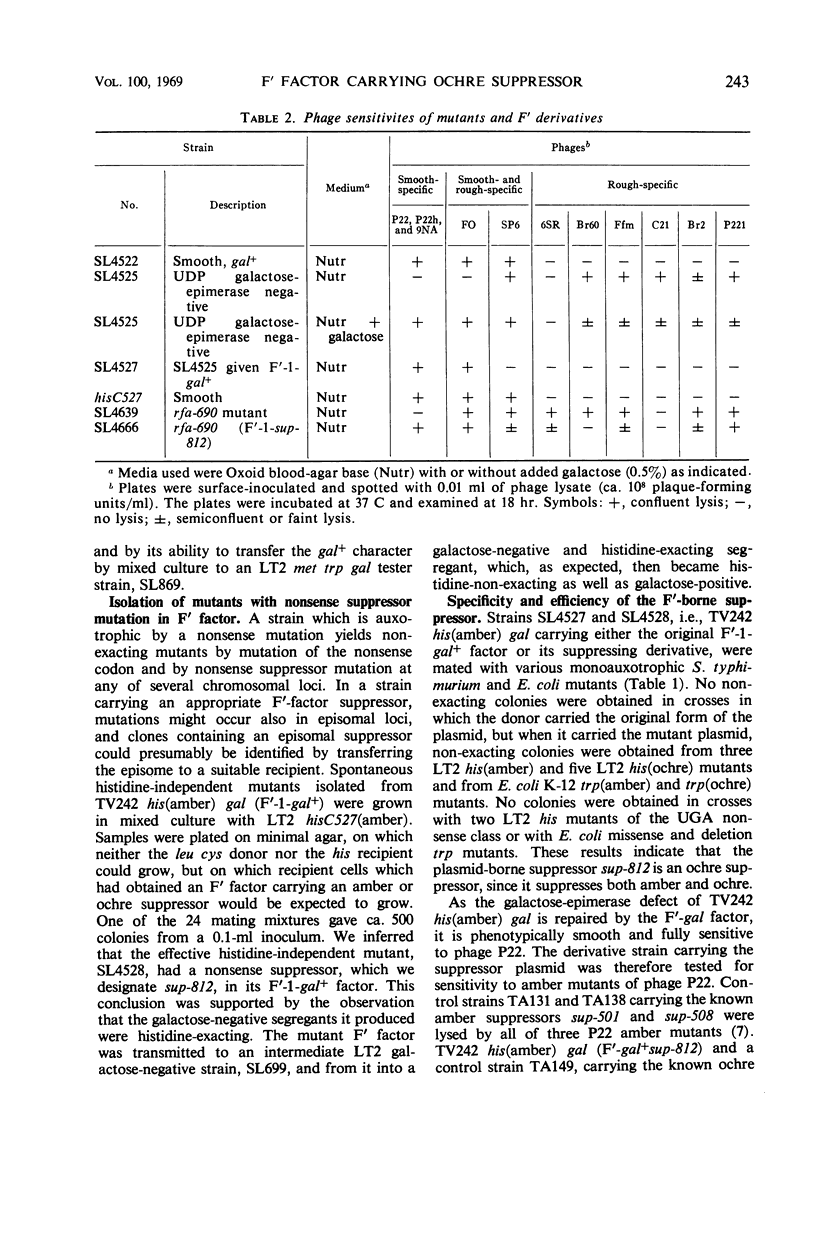
A Salmonella typhimurium strain was given the amber mutation hisC527 by transduction, made galactose-negative by mutation, then infected with the F′-1-gal factor. Of 107 spontaneous and mutagen-induced histidine-independent mutants tested, 3 proved to result from suppressor mutations within the F′ factor. The mutant F′ factors, when transferred to S. typhimurium and E. coli auxotrophs, suppressed amber and ochre but not UGA or missense mutants, and are inferred to carry ochre suppressor genes. Attempts to isolate an F′ amber suppressor mutant were unsuccessful. A suppressor F′ factor was transferred to 14 rough mutants which had been isolated from LT2 hisC527 (amber) by selection for resistance to phage P22.c2. One rough mutant was partly suppressed, as shown by its acquisition of O agglutinability and by alterations in its phage resistance pattern. Phage P22h grown on the suppressed mutant contransduced its rf. gene with cysE+ and with pyrE+, and the affected locus is inferred to be rfaL. Both the original and the mutant F′ factors conferred resistance to the rough-specific phage Br60, which is therefore “female-specific.”
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkowitz D., Hushon J. M., Whitfield H. J., Jr, Roth J., Ames B. N. Procedure for identifying nonsense mutations. J Bacteriol. 1968 Jul;96(1):215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson G., Adelberg E. A. Map positions and specificities of suppressor mutations in Escherichia coli K-12. Genetics. 1965 Aug;52(2):319–340. doi: 10.1093/genetics/52.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggertsson G. Mapping of ochre suppressors in Escherichia coli. Genet Res. 1968 Feb;11(1):15–20. doi: 10.1017/s0016672300011150. [DOI] [PubMed] [Google Scholar]
- GORINI L., KAUFMAN H. Selecting bacterial mutants by the penicillin method. Science. 1960 Feb 26;131(3400):604–605. doi: 10.1126/science.131.3400.604. [DOI] [PubMed] [Google Scholar]
- Garen A. Sense and nonsense in the genetic code. Three exceptional triplets can serve as both chain-terminating signals and amino acid codons. Science. 1968 Apr 12;160(3824):149–159. doi: 10.1126/science.160.3824.149. [DOI] [PubMed] [Google Scholar]
- Gemski P., Jr, Stocker B. A. Transduction by bacteriophage P22 in nonsmooth mutants of Salmonella typhimurium. J Bacteriol. 1967 May;93(5):1588–1597. doi: 10.1128/jb.93.5.1588-1597.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolstad R. A., Prell H. H. Amber-Mutanten des Salmonella-Bakteriophagen P22. Mol Gen Genet. 1968;101(2):189–190. doi: 10.1007/BF00336585. [DOI] [PubMed] [Google Scholar]
- Morgenroth A., Duguid J. P. Demonstration of different mutational sites controlling rhamnose fermentation in FIRN and non-FIRN rha-strains of Salmonella typhimurium: an essay in bacterial archaeology. Genet Res. 1968 Apr;11(2):151–169. doi: 10.1017/s0016672300011320. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E. Information transfer in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1965 Jun;53(6):1335–1340. doi: 10.1073/pnas.53.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E. Revised linkage map of Salmonella typhimurium. Bacteriol Rev. 1967 Dec;31(4):354–372. doi: 10.1128/br.31.4.354-372.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer E. R., Beckwith J. R., Brenner S. Mapping of suppressor loci in Escherichia coli. J Mol Biol. 1965 Nov;14(1):153–166. doi: 10.1016/s0022-2836(65)80237-6. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Levine M. A phage P22 gene controlling integration of prophage. Virology. 1967 Feb;31(2):207–216. doi: 10.1016/0042-6822(67)90164-x. [DOI] [PubMed] [Google Scholar]
- Smith J. D., Abelson J. N., Clark B. F., Goodman H. M., Brenner S. Studies on amber suppressor tRNA. Cold Spring Harb Symp Quant Biol. 1966;31:479–485. doi: 10.1101/sqb.1966.031.01.062. [DOI] [PubMed] [Google Scholar]
- Stretton A. O., Kaplan S., Brenner S. Nonsense codons. Cold Spring Harb Symp Quant Biol. 1966;31:173–179. doi: 10.1101/sqb.1966.031.01.025. [DOI] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Revised linkage map of Escherichia coli. Bacteriol Rev. 1967 Dec;31(4):332–353. doi: 10.1128/br.31.4.332-353.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield H. J., Jr, Martin R. G., Ames B. N. Classification of aminotransferase (C gene) mutants in the histidine operon. J Mol Biol. 1966 Nov 14;21(2):335–355. doi: 10.1016/0022-2836(66)90103-3. [DOI] [PubMed] [Google Scholar]
- Wilkinson R. G., Stocker B. A. Genetics and cultural properties of mutants of Salmonella typhimurium lacking glucosyl or galactosyl lipopolysaccharide transferases. Nature. 1968 Mar 9;217(5132):955–957. doi: 10.1038/217955a0. [DOI] [PubMed] [Google Scholar]
- YAMAMOTO N., ANDERSON T. F. Genomic masking and recombination between serologically unrelated phages P22 and P221. Virology. 1961 Aug;14:430–439. doi: 10.1016/0042-6822(61)90334-8. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Ito J. Nonsense codons and polarity in the tryptophan operon. J Mol Biol. 1966 Nov 14;21(2):313–334. doi: 10.1016/0022-2836(66)90102-1. [DOI] [PubMed] [Google Scholar]



