Abstract
Most cells in the body have the ability to change their physical locations during physiologic or pathologic events such as inflammation, wound healing, or cancer. When cell migration is directed toward sources of cue chemicals, the process is known as chemotaxis, and it requires linking the sensing of chemicals through receptors on the surfaces of the cells to the directional activation of the motility apparatus inside the cells. This link is supported by complex intracellular signaling pathways, and although details regarding the nature of the molecules involved in the signal transduction are well established, far less is known about how different signaling molecules and processes are dynamically interconnected and how slower and faster signaling events take place simultaneously inside moving cells. In this context, advances in microfluidic technologies are enabling the emergence of new tools that facilitate the development of experimental protocols in which the cellular microenvironment is precisely controlled in time and space and in which signaling-associated changes inside cells can be quantitatively measured and compared. These tools could enable new insights into the intricacies of the biological systems that participate in chemotaxis processes and could have the potential to accelerate the development of novel therapeutic strategies to control cell motility and enhance our abilities for medical intervention during health and disease.
Keywords: microscale, signal transduction processes, signaling networks, neutrophil
Introduction
Of the billions of cells in the human body, almost every cell moves at some point during its lifetime. During development, the migration of embryonic cells helps define the locations and shapes of new organs (1). Carefully choreographed movement of white blood cells through the body is critical in the creation of effective barriers against the spreading of infection (2). Wound healing requires new cells to be brought to the site of damaged tissue and positioned properly for reconstruction processes (3). However, cell movement, as an essential process in all multicellular organisms, is rarely random; most of the time, it is directed by various biochemical or mechanical clues. When soluble chemicals bias cellular motility toward the higher concentration of the chemical stimulus, this process is called chemotaxis.
During chemotaxis, cells use receptors on their surfaces to detect specific chemicals then activate complex intracellular signaling networks and trigger the selective activity of motor proteins. Moving cells continuously reconfigure their actin network and myosin activity to generate the physical forces necessary for displacement and for steering themselves in the direction of the target. This fundamental cell behavior has fascinated biologists and engineers alike by its characteristics of high sensitivity and a large dynamic range, versatility, and conservation of motifs among various cell types. Although tremendous advances have been made in identifying and understanding the molecular components required for cellular chemotaxis in the past 40 years, one problem in particular, namely how all these components assemble into functional systems, remains incompletely solved. Today, we have only fragmentary understanding and are just beginning to gain insights into how known molecules organize to receive signals, amplify them, and convert them into new signals that will activate other molecules, in signaling cascades that diverge and converge repeatedly and involve multiple negative and positive feedback loops. The expertise required for analyzing how information is processed in these complex systems increasingly falls in the engineering realm, and sound engineering principles are increasingly employed toward the understanding of the intracellular chemotaxis circuits for signal amplification, differentiation, or rejection of noise (4–10).
Beyond the scientific thrill of understanding how cells can read and act on soluble extracellular clues, a practical motivation exists for studying chemotaxis. Chemotaxis is important not only in health but also during disease, and the ability to modulate the chemotaxis of target cell populations has therapeutic implications. For example, neutrophils are one of the most effective barriers against microbe invasion and spreading inside the human body, and the failure of leukocytes to promptly arrive at sites of wounds or infection can lead to uncontrollable infections. Even the most potent antibiotics have limited efficiency against infections in individuals with dysfunctional neutrophils and in individuals with impaired neutrophil migration owing to conditions such as diabetes (11) or aging (12), for whom infections that are innocuous to healthy people can quickly evolve into sepsis and death (13). In all these conditions, new therapies that enhance neutrophil activity may be a major benefit. However, the infiltration of normal tissues by overzealous neutrophils and macrophages can produce unnecessary damage and impair organ function (2), e.g., in severe forms of asthma (14), arthritis (15), or ischemia-reperfusion injury (16), whereas the migration of eosinophils and other cells into tissue could exacerbate the symptoms of allergies (17). Preventing these cells from entering tissues may provide temporary relief in these conditions. In cancer, the motility and invasion of malignant cells into local and distant tissues, in the form of metastasis, are responsible for more than 90% of deaths caused by cancer. Blocking the migration of cancer cells may prevent metastasis and extend the lives of patients (18).
To better understand chemotaxis, in terms of biology and the underlying physiology of disease processes, a critical clue may come from a careful, quantitative analysis of the timing of different processes involved. In the past, quantitative measurements and analysis of the temporal dynamics of fundamental biological processes have led to critical advances. Classical examples of how emerging technologies and engineering principles have brought invaluable insights into the fundamental biological processes include the quantitative analysis of the temporal component of electrical pulses in axons (19), mechanical loads in muscle fibers (20), and light pulses for photoreceptors (21). More recently, the combination of new microfluidic devices to modulate the osmolarity of microenvironment and basic engineering principles to analyze the results led to important insights into the mechanism of osmotic adaptation in cells (22, 23). A sustained trend for increasingly more sophisticated experimental tools, which are capable of precise control of cellular microenvironment in time and space, that facilitate quantitative measurements may benefit chemotaxis research and help uncover the intricacies of other complex cellular functions (24–27).
In this review, we evaluate the current state of the art and infer directions for emerging developments of microscale technologies for studying chemotaxis. The emphasis is on the directional migration of bacteria, neutrophils, and Dictyostelium discoideum as model cells in chemotaxis research. We start the discussion with examples from bacteria chemotaxis, which benefited enormously from techniques that allow us to quantify the temporal dimension of responses to changes in attractant or repellant concentrations. These examples show how parallel, overlapping processes that occur at different timescales, some slow and some fast, may be resolved through the analysis of transient responses. We review some early studies toward understanding the eukaryotic response to temporal perturbations. Last, we focus on the benefits that emerging microscale technologies can bring to chemotaxis research and applications. Overall, the conceptual framework for analyzing cellular responses to controlled perturbations may become a useful tool to study chemotaxis and other complex intracellular systems.
Bacteria Chemotaxis and Transient Responses to Perturbations
Today, one of the best understood chemotaxis systems is the one employed by bacteria to find nutrients and avoid dangerous environments. We now know not only the majority of molecules involved but also how they interact and cooperate to achieve the directed motility function (28). Fewer than ten molecular species and a motor are employed inside bacteria no larger than a few micrometers in size to achieve robust gradient sensing and flexible adaptation to environment changes (Figure 1). At the input of the system, molecules of attractant and repellent are recognized by specific receptors on the surface of bacteria. At the output, several motors rotate the flagella (typically four to ten for most of bacteria, such as Escherichia coli) either clockwise (CW) or counterclockwise (CCW) to propel the cell. CCW rotation aligns the flagella in a bundle and propels the cell in a straight line for several times its length, whereas CW rotation breaks the coordination of flagella, which causes bacteria to tumble and change the orientation of subsequent movements.
Figure 1.
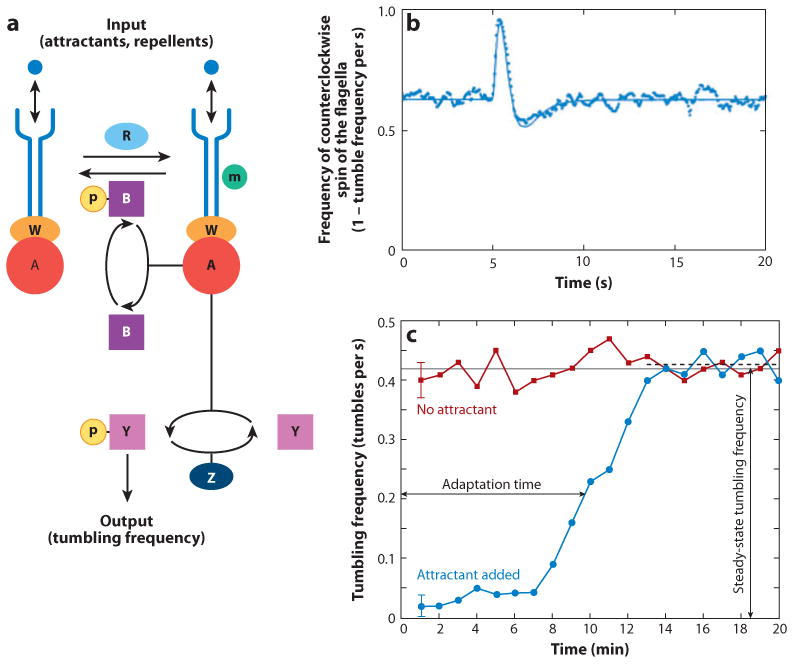
Bacteria chemotaxis. (a) Schematic of signaling in bacteria. Binding of the repellant molecules to the receptors on the surface of the bacteria, through the kinases CheW (W) and CheA (A), induces the phosphorylation of messenger proteins CheY (Y). These will then diffuse to the motors turning the flagella, where they will induce a temporary reversal in the motor's rotation direction. This results in a sudden change in the direction and an overall scattering of bacteria from one location, before the CheY is turned off by removal of the phosphate group (P) through the activity of phosphatase CheZ (Z). Binding of attractant molecules to the receptors inactivates CheA and increases the persistence of migration toward the source of the attractant. An adaptation mechanism, activated by CheA, and actions through the enzymes CheR (R) and CheB (B) can add and remove methyl groups (m) from the receptors, increasing and decreasing the sensitivity of the receptors, respectively. Reproduced from Reference 9. (b) Impulse response of bacteria to one short pulse of attractant (addition followed by quick removal). The probability for counterclockwise (CCW) spin of the flagella (Bias) increases temporarily after a 0.02-s pulse addition of attractant (at time 0+5 s), then decreases below the normal average in the absence of the stimulus, before returning to baseline level. Dots represent experimental data averaged over approximately 300 cells; the line is a curve fit. Adapted from Reference 35. (c) Adaptation after the step addition of attractant (no removal after addition). The frequency of tumbling events (CW) decreases immediately after the addition of attractant (blue line) and then progressively increases as bacteria adapt to the new environment. In the absence of stimulation, the frequency of tumbling events is relatively constant (red line). The frequency of tumbling after adaptation (black dashed line) is almost the same as the steady-state tumbling frequency for unstimulated bacteria (gray line), and the ratio between the two is defined as the precision of adaptation. Adapted from Reference 9.
In between the receptors and motors that turn the flagella, several molecules translate and modulate the signals to assure that bacteria reach their targets in various conditions and avoid dangerous situations. A messenger protein CheY (Y) diffuses between receptors and motors, and in its phosphorylated form, it induces the switch from CWW to CW rotation of the motors and cell tumbling. The CheY protein is phosphorylated by the transfer of phosphoryl groups (P) from the kinase CheA (A) associated with the receptors, and it is dephosphorylated by phosphatase CheZ (Z) in the cytoplasm. CheA can phosphorylate itself when bound to receptors (together with another kinase CheW), and receptors are activated by external repellent molecules. Overall, receptor stimulation by repellents, through CheA, CheY, and CheZ signaling molecules, increases the frequency of tumbling; whereas receptor inactivation by attractant molecules increases the duration of straight runs.
In parallel with the modulation of motor activity, adaptation mechanisms have been devised by bacteria to assure that responses are appropriate at the lowest and the highest concentrations and that bacteria can find nutrients and avoid dangers in the presence of the broadest range of chemoattractant and repellant concentrations. The adaptation mechanism starts from the receptor binding that also activates, through CheA, a pair of enzymes CheR (R) and CheB (B) that add and remove methyl groups (m) from receptors, which modulate the sensitivity of these receptors. CheR adds methyl residues to receptors, which increase their sensitivity, and CheB acts in the opposite way, by removing methyl groups and desensitizing receptors. Whereas CheR is always active, the activity of CheB depends on phosphoryl groups transferred from active CheA. Overall, receptor stimulation through CheA, CheB, and CheR reduces the sensitivity of receptors and results in better adaptation to new conditions. Combined with the modulation of the motility, the adjustment in receptor sensitivity assures that bacteria are more likely to move in the proper direction and remain responsive to small changes in the levels of chemicals in their environment, even under the most extreme conditions.
Experiments that allowed the precise quantification of temporal responses of bacteria to changes in their environment played a critical role in understanding how the chemotaxis system in bacteria works as an integrated network. At the same time, when signaling molecules were identified through specialized molecular biology techniques, the basic circuits for sensing and adaptation were refined through better experimental techniques and biophysical models. Together, these efforts led to the precise identification and quantitative assessment of the roles for each component. One of the first, critical steps toward understanding chemotaxis in bacteria was the distinction between spatial and temporal sensing mechanisms. To differentiate between the spatial sensing mechanism (in which bacteria make comparisons in stimulus concentration along the 2-μm length) and a temporal sensing mechanism (in which bacteria compare concentrations of stimulus over time), an experimental system was designed in which bacteria are initially presented with a uniform attractant concentration and then quickly plunged into a higher uniform concentration of the same attractant (29). Inside this system, a suspension of bacteria is quickly mixed with a solution of attractant twice the final concentration, by quickly passing the two solutions through a rapid mixing device, which is just a tube with twisted wires inside. The residence time of bacteria in the tube was 0.2 s, and observation commenced after an additional 0.5 s. Transitory increases in linear runs, which correspond to CCW rotation of flagella, were present for at least 5 min after attractant concentration increases; and transitory increases in tumbling events, which correspond to CW rotation, were observed for at least 12 s after concentration decreases, which suggest the presence of a temporal sensing mechanism for chemotaxing bacteria. Subsequent experiments using attractants and repellants (30) confirmed temporal sensing mechanisms. They also explained how bacteria can achieve surprising levels of sensitivity by sampling concentrations over a longer distance than their body, which corresponded to the time for displacement over 20–100 body lengths.
A precise experimental system for measurements with single-cell resolution has also been developed and used to monitor changes in the direction of flagella rotation for cells tethered on coverslips (31). This system quantitatively measures the transition times between CW and CCW as functions of attractant or repellant concentrations bound to receptors when bacteria are washed quickly with different solutions. Mathematical models of adaptation and gradient sensing in moving bacteria were then devised on the basis of these quantitative measurements. With the identification of receptor methylation reactions as the basis for receptor adaptation (32, 33) and the role of CheZ (34) in the phosphorylation of CheB and CheR (35) during adaptation, a clearer picture began to emerge on the network organization for temporal sensing in bacteria. Occasional mismatches between models and experimental results led to more discoveries about the biological system itself, e.g., the way that thousands of receptor molecules on the bacterial surface cooperate for the sensitive detection and analysis of different combinations of attractants and repellents (36). Further studies that used genetic manipulations to alter the protein levels of five major signaling molecules in bacteria chemotaxis revealed the robustness of the system, which is capable of precise adaptation after perturbations for wide variations in the levels of biochemical parameters (9).
The steady evolution of our understanding of bacteria's chemotaxis system, enabled by technology and biophysical models, may represent a model to be emulated in the study of eukaryotic cell chemotaxis. Rapidly transitioning the precise measurements of responses to perturbations into mathematical models can accelerate our understanding of chemotaxis mechanisms in eukaryotic cells. Toward the goal of quantitatively defining signaling networks, however, important differences between bacteria and eukaryotic cells must be acknowledged. First, eukaryotic cells are two orders of magnitude larger than bacteria. Although the diffusion of molecular species is fast inside bacteria and can transfer information, the same mechanisms would be too slow and impractical in large eukaryotic cells (37). Also, whereas the concentration differences along bacteria may not be important for all but the steepest gradients (38), the differences in concentration between distinct locations on the surface of eukaryotic cells are significant in the presence of the shallowest physiologic gradients. Second, eukaryotic cells have one order of magnitude more molecular species involved in signaling during chemotaxis than bacteria. The number of interactions, including feedback loops, redundant pathways, and multiplexing circuits for diverse signals, between these molecules is exponentially more complex in eukaryotic cells (39). Finally, the motility apparatus in eukaryotic cells is not only more elaborate but also capable of more various actions than the bacterial flagella. The binary output in bacteria is replaced by motor responses that are more graded, have cumulative temporal and spatial effects on the cell as a whole, and can result in displacement, changes in cell shape, and reorganization of the internal structure of the cell (40). To address these differences, more sophisticated experimental and analysis tools are required to study eukaryotic cell chemotaxis compared with those that were successful in bacteria analysis.
Eukaryotic Chemotaxis and Transitory Gradients in Traditional Assays
Before discussing different assays for the chemotaxis of larger, adhesion-dependent cells that have been developed over time, it is important to briefly review the principles that are incorporated in some of the current models for eukaryotic cell polarization and chemotaxis. The input for the majority of the models is a spatial gradient of chemoattractant that interacts with receptors on the surface of the cell and triggers intracellular pathways that amplify this signal. The output of most of these models is the segregation of distinct regions of actin and acto-myosin activity inside initially homogenous cells. Although actin polymerization defines the fronts, myosin activity defines the backs of these cells (Figure 2). Most current models were developed on the basis of reaction-diffusion principles that were first formulated by Turing in the middle of the last century (41). These models explain the polarization of cells in the direction of gradient with the competition between local stimulator and global inhibitor molecules (42) or the competition between two messenger molecules with distinct dynamics (43). Several other models have been proposed that are centered on distinct principles for the functional polarization of cells in the direction of higher chemoattractant concentrations, and in which reaction-diffusion principles play a minor role or no role. These include differences in the timing of chemoattractant at opposite locations on the cell (44), the overlapping of fast and slow positive feedback loops (5), the noise from activated receptors (45, 46), or the selective stabilization of microtubules at the moving front of the cell (47). Xu et al. (48) proposed one particularly useful model in the context of transitory perturbations. This model considers two divergent signaling pathways that start from stimulated receptors, which stimulate actin polymerization at the fronts and actin-myosin contraction at the backs of cells, and predicts robust polarization of cells in the face of strong perturbations. The source of the robustness is the reciprocal inhibition of the “frontness” and “backness” signals that ultimately can lock the cell in a polarized state (Figure 2). Overall, the purpose of the models is to predict the cellular localization and temporal dynamics of signaling molecules inside cells during polarization. Most of the time, the dynamics of molecules can be revealed through experimental techniques, e.g., by effectively measuring the level of proteins, imaging the activation of molecules tagged with fluorescent markers, and so on. These predictions also play important roles in interpreting the outcomes of cellular chemotaxis experiments using different assays and perturbations.
Figure 2.
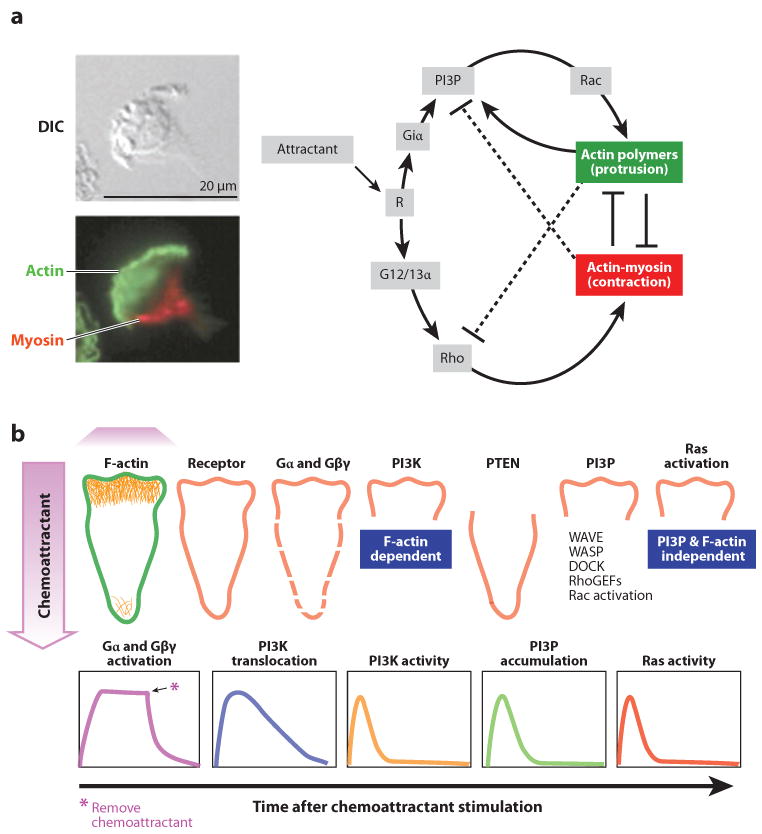
Eukaryotic chemotaxis. (a) Schematic of signaling pathways in a cell that responds to chemokine gradients. (Left) Immediately after stimulation with chemoattractant, the cell starts moving [differential interference contrast (DIC) image]. A moving cell also organizes its cytoskeleton with actin polymerizing at the front of the cell (green fluorescence) and myosin light chain II active at the back of the cell (red fluorescence). Adapted from Reference 139. (Right) Inside the cell, stimulation of specific receptors (R) by the attractant molecules triggers the activation of heteromeric G proteins coupled to these receptors and activates two parallel pathways responsible for self-organizing polarity. Frontness depends upon Giα-mediated production of phosphatidylinositol-3,4,5-triphosphate lipids (PI3P), the activation of Rac GTPase, and the polymerization of actin, ultimately leading to the formation of protrusions at the front of the cell. Backness depends on signals mediated by the G12 and G13 α-subunits of G proteins, including activation of Rho GTPases, Rho-dependent kinase, and myosin II, leading to the acto-myosin contraction at the back of the cell. Adapted from Reference 48. (b) Spatial distribution (first row) and temporal activation dynamics (second row) of molecules that participate in chemotaxis. The activation of F-actin is localized at the front of the cell, toward the source of chemoattractant. Whereas the distribution of the receptors remains uniform on the surface of the cell, the activation of the alpha subunits (Gα) and beta-gamma complex (Gβγ) is higher at the front than at the back. At the front of the cell, the Giα-subunit signals the formation and accumulation of PI3P at the front of the cell, leading to the localized activation of small Ras GTPases and other signaling molecules. Phosphoinositide 3 kinases (PI3K) and phosphoinositide 3 phosphatases (PTEN) play critical roles in the localized generation of PI3P from its precursors. The activation kinetics of most molecules downstream of the G protein coupled to the receptors is now known to be transient in conditions of steady chemoattractant stimulation. Reproduced from Reference 137. Abbreviations: DOCK, dedicator of cytokinesis; RhoGEFs, guanine nucleotide exchange factors for Rho GTPases; WASP, Wiskott-Aldrich syndrome protein; WAVE, WASP family verprolin homologous.
The best known and most commonly used chemotaxis assay is the Boyden chamber (Figure 3). Also known as the transwell assay (49), this assay was developed more than 40years ago and has been extremely useful in identifying various cells that can migrate directionally in response to chemical signals, various chemokines and biochemicals that are responsible for chemotaxis, and the wide range of molecules that are responsible for chemotaxis and directional migration. In this assay, cells are added on one side of a membrane with 3–6-μm diameter pores, chemoattractant is added on the other side, and the number of cells that pass the membrane are counted and compared to the total number of cells added earlier. Although the result of the assay is a number, the assay is far from quantitative, and comparisons between different conditions and between these and controls are usually difficult to interpret. Other assays, in which direct observation of the migrating cells is possible, are better suited for precise measurements and are called Zigmond and Dunn chambers (50, 51). These assays are more difficult to use and thus have a limited range of applications, even in research settings. Finally, chemotaxis assays in which a micropipette filled with chemoattractant is brought close to the target cells require considerable expertise, and because they are analyzed one cell at a time, they have low throughput. When used appropriately, micropipette assays can be informative and can allow precise localizing of the stimulus with respect to the cell surface.
Figure 3.
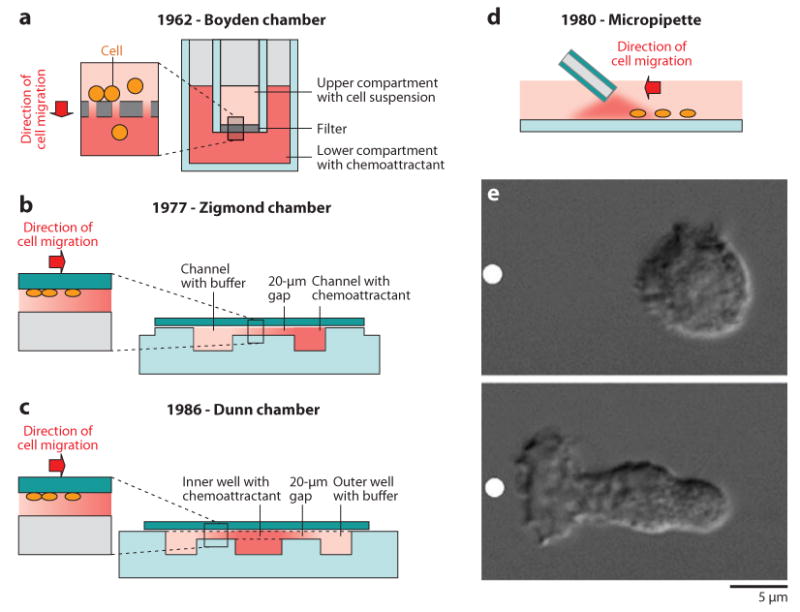
Traditional chemotaxis assays. (a) Schematic of Boyden chamber, first described in 1962. Moving cells follow a gradient that is transiently formed in a membrane with holes separating two compartments, one with cells in buffer and one with the chemoattractant. The number of cells that passes through the membrane is counted and compared with control conditions. (b) Schematic of Zigmond chamber, first described in 1977. Cells are attached to a glass coverslip, positioned over a slide with two larger channels: one filled with chemoattractant solution and one filled with buffer. The channels are connected through a small, 20-μm gap. Diffusion of the chemoattractant between the channels, through the gap, helps establish a gradient in the direction tangent to the coverslip surface. (c) Schematic of Dunn chamber, first described in 1986. This device replaces the two parallel channels in the Zigmond chamber with two concentric wells and maintains the 20-μm gap inside which the gradient is formed. (d) Micropipette assay, first described in the 1980s. Diffusion of the chemoattractant from a micropipette with a micron-size opening. The micropipette is placed at a micron's distance from the coverslip to help establish a chemoattractant gradient in the surrounding buffer solution. Cells will follow the gradient and move radially toward the tip of the micropipette. (e) Polarization of a neutrophil in response to chemoattractant released from a micropipette (white dot). Resting (time 0) and polarized (after 2 min of stimulation) neutrophils are shown. Reproduced from Reference 138.
Most traditional experimental techniques to study cell migration are inadequate for capturing the complexity of the responses of moving cells to spatial and temporal stimuli. The spatial and temporal contexts of stimulation are generally accepted as important, although their relative weight during chemotaxis has been debated for a long time (52). Cells that respond to spatial stimuli must compare the relative concentrations of stimuli at two distinct locations on their surface, whereas cells that respond to temporal stimuli must compare the intensity of stimuli at two different moments in time. In practice, the duality of cellular responses poses a significant challenge for experimental systems that must control the cellular microenvironment in time and space. Although they are most widely used in chemotaxis studies for various cell types and chemoattractants, experimental techniques such as the Boyden chamber can generate spatial chemical gradients that continuously evolve in time. The continuous accumulation of chemoattractant on one side and the alteration of the local concentration of chemokine at the level of pores in the membrane by moving cells cannot be controlled. In other chemotaxis assays, such as the Zigmond chamber (50) or underagarose migration (53, 54), the rapid deterioration of the gradient by the diffusion between small compartments makes these precise conditions during chemotaxis unstable and difficult to estimate. To overcome these limitations, many researchers adapted and customized traditional experimental systems to produce quantitative observations on the temporal responses of cells during chemotaxis.
Transitory Gradients by Diffusion from Source Compartments
The earliest observations of the transitory responses of neutrophils to evolving gradients occurred in the 1970s. In one of the first studies, researchers monitored the migration of single neutrophils toward erythrocytes that were destroyed by laser irradiation (55). They noted a delay between the initiation of the gradient and the response of the neutrophils. However, the delayed response could not be differentiated from the transitory buildup of the chemotactic gradient, which was established through diffusion at a comparable timescale. Precisely defining the time when the chemoattractant reaches cells is a common problem in other experimental models as well. One example is the experimental setup in which solutions in the two compartments of the Zigmond chamber were replaced through Pasteur pipettes placed inside each side chamber (56). Solution changes could be accomplished in 15 s; however, the evolution of the gradient over the cells was much slower. Nonetheless, the responses of neutrophils to increases or decreases in gradient steepness were recorded, and it was noted that during step-up changes, lamellipodia and ruffles formed initially over most of the neutrophil surface. Most ruffles were withdrawn later, and those that remained were limited to a portion of the neutrophil surface, which becomes the front of the moving cell. When the slope of the gradient was decreased, blebs were observed on the surfaces of neutrophils before the cells recommenced locomotion. In a later study, a Zigmond chamber was also employed to observe the polarization of neutrophils after step addition of formyl peptides (fMLP—a common chemoattractant for neutrophils) at different concentrations (57). First, neutrophils were adhered to the surface of a coverslip and then imaged immediately after the coverslip was added to a Zigmond chamber, while the gradient was still transitory. At an optimal concentration, cells assumed irregular morphologies for the first 30 s and showed an increasing proportion of polarized forms over the next 30 min. At higher concentrations, neutrophils assumed multipolar morphologies and polarized poorly, a condition also reported in other studies (58). At lower concentrations, neutrophils polarized immediately on exposure to the attractant. Authors proposed that neutrophil polarization requires the integration of information at high concentrations, whereas cells may polarize toward the site of the first signal from a ligand at smaller concentrations. The duration of adaptive response was directly related to the absolute concentration and the concentration change, which also suggests adaptation and short-term memory.
A device for testing the response of cells to temporal changes of concentration was proposed by Vicker, in which changes of concentration could be accomplished in a controlled fashion (59). The device consisted of a permeable membrane positioned between two compartments, and input and output tubes in each compartment were used to maintain a constant volume of fluid in the device. In this system, gradients developed in approximately 16 s. Although more controlled than other systems, moving cells could only be observed and counted at the end of the experiment. Nonetheless, experiments indicated that the regulation of chemotaxis and chemokinesis in the amoebae Dictyostelium depend on temporal signals and spatial gradients of cyclic AMP. Korohoda et al. (60) designed a more sophisticated setting in which a pocket-like chamber was formed between a glass slide and a coverslip. The coverslip was sealed on three sides and had one side open to a large chamber in which fluid could be replaced quickly with a peristaltic pump. A spatial gradient formed inside the pocket chamber starting approximately 15 min after starting the pump, and the slope of the gradient changed predictably between 30 and 60 min after the addition of chemoattractant. Comparisons between gradients of increasing and decreasing concentrations showed that positive gradients are more effective in the orientation of Dictyostelium cells in response to cAMP (the natural chemoattractant for these cells), folic acid, and calcium, but not in magnesium gradients. Although gradients of magnesium stimulated directional migration of cells when the average concentration was decreasing, stable or decreasing gradients of folic acid, cAMP, and calcium resulted in no orientation, which indicates that distinct pathways for chemotaxis can be activated in the presence of different stimuli.
Transient Concentration Changes in Flow Chambers
To observe the responses of moving cells to faster changes in chemokine concentration (compared to traditional diffusion-based assays), several assays have been developed that rely on convection rather than diffusion to bring the new chemicals close to cells. Most of these are based on a traditional flow chamber, and their major drawback is that no controlled spatial gradient can be established inside, which limits their utility to the study of temporal responses. One early example is a flow chamber used for the recording of Mac-1 integrin (CD11b/CD18) expression after increases in the concentration of fMLP chemoattractant (61). The study showed that Mac-1 expression increases immediately and plateaus within 2–4 min after stimulation and that, in the absence of temporal stimulation, Mac-1 expression levels decayed in approximately 10 min. Repeated small increments in the concentration of the stimulus every 200 s resulted in significantly longer migration paths than single-step increases. To probe the response of neutrophils to even faster stimuli, a new chamber was built between two glass slides, where solutions inside the chamber were changed as fast as 10 s for increasing or decreasing concentrations (62). Chemoattractant was flown in several small tubes, also used as spacers between slides, and connected at the distal end to a 10-way adaptor with nine syringes filled with media of different concentrations. The most interesting observation reported in this study is that neutrophils can reverse their direction of migration by 180° when exposed to temporally decaying gradients of chemoattractant. Most cells reversed their direction by dissolution of the leading edge (lamellipodium) and trailing edge (uropod) and the rebuilding of structures in the new direction. The effect was maximal for fast concentration changes, in 10-s steps, and diminished progressively for shorter and longer steps. Although neutrophils occasionally performed U turns, the authors speculated that a memory mechanism must be reset in cells before they can reorganize and move in an entirely new direction. Notably, chemokinesis (the random migration of cells in the presence of uniform concentrations of chemoattractant) and not chemotaxis (directional migration) was at the origin of neutrophil motility in this study.
To probe the responses of neutrophils to combinations of spatial and temporal gradients, a system for exposing cells to temporal waves of chemoattractant has been reported (63). By alternatively pushing chemoattractant and buffer from two syringes into one observation flow chamber, temporal waves were generated, comparable to those of cAMP experienced by Dictyostelium during spore formation. Whereas a known mechanism assures that cells do not reorient to the back of the wave when exposed to concentration decreases in the presence of spatial gradients in Dictyostelium, a similar behavior has only been speculated in neutrophils. Experiments show that neutrophils are capable of responding to waves with approximately 7-min periodicity, and when treated with a series of four waves with 7-min periodicity, neutrophils exhibited velocity surges in the front of each of the last three waves. During the decaying gradients at the end of the wave, neutrophils did not follow spatial gradients, their pseudopods were small and extended in random directions, and cells did not translocate effectively in any direction. The velocity of neutrophils decreased at the peaks and backs of the last three waves. These results challenge the current assumption that neutrophils migrate toward sites of infection solely in response to standing spatial gradients of soluble chemoattractants. One could speculate that the chemotaxis response to waves of chemoattractant may be more robust to perturbations and noise in the environment and thus better suited to attract neutrophils over large distances to sites of infection in the human body.
The combination of temporal stimuli and molecular probes for intracellular signaling molecules is a powerful strategy to explore the temporal dynamics of cell signaling during chemotaxis (39). To answer the question of how cells respond to stimuli switching their location, and how the tail of the cell is reprogrammed into a front and vice versa, Dictyostelium cells were exposed to increases of cAMP concentration (64) or stimulated by sudden changes in the direction of shear stresses under flow (65). The responses of cells to changes in concentration or direction of flow were analyzed with particular focus on the dynamics of filamentous actin and myosin II. The reports documented the rapid loss of actin markers at the front, which started immediately after the switch, for stimuli and the polymerization of actin starting at the new front within 10–20 s. By comparison, myosin II decreases much more slowly after the switch and takes over 2 min at the tail, then it increases more slowly at the new location of the tail, starting 30 s after the switch. Faster inhibition of the existent front after the switch, even before a new front is formed, indicates the presence of a fast inhibitor mechanism for cell polarization during directional migration. Interestingly, these results contradict many current models of cell polarization derived from the original fast excitation–slow inhibition mechanisms of Turing (42, 66, 67). Reconciling these differences may be important, considering that similar response dynamics were observed earlier in neutrophils, with only differences in the timing of different dynamic processes at the fronts and backs of cells (68).
Micropipettes for Local Gradient Perturbation
Micropipettes are ideally suited to precisely localize stimuli to the surfaces of moving cells because of their small tips. When attached to a micromanipulator, micropipettes can be quickly moved to deliver stimuli in predefined temporal sequences. To probe into the sensitivity of different regions of polarized Dictyostelium, micropipettes were used to locally apply cAMP around moving cells (69). After a brief delay, the local application of cAMP induced the local generation of pseudopods in a region closer to the stimulus. Different refractory periods were noted in different regions of polarized cells. The period was shorter at the leading edge (a few seconds), longer at the tail (an average of 40 s), and intermediate (10 s) on the sides. More elaborate assays, using two micropipettes that can be alternately turned on and off and fluorescently tagged proteins, compared the sensitivity of unpolarized and polarized Dictyostelium cells. Although unpolarized cells are uniformly responsive at their periphery, polarized cells restrict their sensitivity to the leading edge (70, 71). Recently, a biophysical model has been proposed that follows the concept of localized sensitivity and the observation that new pseudopods are formed by splitting existing pseudopods toward increasing concentrations of chemoattractant (72). When neutrophils were probed using chemoattractant released from a micropipette, it was also found that any part from the front to the tail can be stimulated to produce lamellipodia, and different regions of a cell can respond independently to chemotactic gradients (65, 68). Migrating neutrophils reversed their direction of motility immediately after a 0.5-μm micropipette filled with fMLP was placed near the back of the cell (68, 73). The reversal of direction was performed in a sequence that started with the disappearance of the preexisting front, the reversal of cytoplasmic flow, and the formation of a new front instead of the former tail. The formation of a new front at different locations was always preceded by the inactivation and disappearance of the old one.
To probe into the molecular mechanisms of leading and tail formation in more detail, Xu et al. (48) combined the micropipette technique with the use of selective inhibitors of signaling molecules. Using a model for human neutrophils, HL60 cells, which were observed during U-turns upon moving a micropipette from the front to the back of polarized cells, a new model for neutrophil polarization has been suggested (Figure 2). The model proposes that the polarization of cells occurs through the cascade activation of Rho (back), Rac, and ROCK (front) molecules through divergent pathways from G-coupled receptors and the reciprocal inhibition between actin (front) and myosin (back). A critical observation that supports this model is that the trailing edge regained sensitivity to stimulation when cells were treated with drug compounds that selectively inhibit the ROCK kinase. Experiments using chemoattractants flown over the cells from a 5-μm-diameter micropipette (74) revealed even more unexpected aspects of pseudopod formation at the leading edge in moving neutrophils. After the initial pseudopod extension following localized stimulation, a periodic extension and contraction of the pseudopod with an average of 60 s and for more than 12 min was reported. During growth periods, the speed of protrusion was constant and similar for different chemoattractant species. Oscillations were blocked by treating cells with RhoA inhibitors or the activation of ROCK kinase. Interpreting these results in the context of the previously described model of reciprocal inhibition between actin and myosin may suggest a time delay between the two alternative processes that leads to oscillations. Moreover, the existence of distinct pathways of signaling for different chemoattractants was inferred from observations of different effects from the same inhibitor in the presence of different chemoattractants; these pathways were confirmed in recent studies (54).
Micropipette experiments have three important drawbacks. First, the technique requires expertise and is low throughput because only one cell at a time can be studied. Second, chemoattractant delivery to cells is unstable. A careful report from Vicker (75) documents the unpredictable effects of random convection in experimental setups with micropipettes and free solutions. Third, the continuous accumulation of chemoattractant in the solution around cells seriously limits the duration of experiments, which restricts the utility to only the fastest moving cells or short observation windows. Although this may not be a problem for single transient-stimulation experiments, it may become significant if repeated stimulations of the same or different cells are required.
Other Techniques for Temporal Perturbations of Moving Cells
To study the migration of neutrophils in temporally controlled spatial gradients, Ebrahimzadeh et al. (76–79) developed a series of devices that couple chemokine and density gradients. The density gradient is moved physically up or down relative to the filter on which cells reside, by the addition or subtraction of liquid from the lower chamber. Resulting changes of the concentration gradient at the location of the filter and cells appear to modulate cells movement through the filter toward higher concentrations of chemoattractant. Similar to the Boyden chamber assay, one cannot see individual cells during chemotaxis, and cells must be counted on two sides of the filter at the end of the experiment. By comparing the number of neutrophils that pass through the filter in conditions of temporally positive and negative spatial gradients at different velocities, the authors reported substantial migration in positively developing gradients. For decreasing gradients, a critical gradient change velocity of –5 μm min−1, comparable to the average speed of moving neutrophils, blocked chemotaxis. Overall, the results of this study indicate that moving neutrophils are strongly affected by concentration changes over time and require the presence of a positive temporal gradient to maintain chemotaxis.
Another original approach was to sequentially expose neutrophils to low temperature and later to predeveloped, stable spatial gradients (80). Although cold neutrophils are unresponsive, this approach should allow researchers to expose cells directly to well-developed, stabilized gradients upon warming the cells and circumvent transitory events. The lack of neutrophil responses to gradients in this experiment was attributed to the exclusive sensitivity of cells to temporally changing concentrations. A similar approach was used later to synchronize cells during warming and to capture quasi-periodic cycles of chemotactic activity, during which the cells entered phases of undirected motility that lasted for several minutes (81). Importantly, low temperatures may have unexpected effects on cells, similar to the alteration of microtubules that may play important roles during the orientation of neutrophils in response to spatial chemical gradients (47, 82). In the past ten years, microfluidic technologies have enabled the emergence of several new assays that not only overcome the limitations of the modified or the more traditional assays, but also provide new experimental opportunities.
Microfluidic Technologies for the Precise Perturbation of Moving Cells
Among the new technologies with an increasingly broader impact in biology, microfluidic devices are extremely useful for studies of cellular motility and chemotaxis. After the first demonstration of neutrophil chemotaxis in linear gradients of interleukin-8 chemoattractant (83), several devices that generate stable gradients have been reported for studying chemotaxis in bacteria, yeast, various eukaryotic cells (such as neutrophils, lymphocytes, and cancer cell lines), amoeba (Dictyostelium discoideum), and multicellular organisms (e.g., Caenorhabditis elegans). Microfluidic devices have enabled precise studies of concentration-dependent chemotaxis, the effect of steep gradients, combinations of gradients, and responsiveness to inhibitors. Today, three main strategies for making chemical spatial gradients on a scale relevant to the cellular scale have been described in the literature. Devices have been developed that use (a) microfluidic streams of different concentrations that flow in parallel in the same channel, (b) point release of the chemical compounds followed by diffusion in a homogenous space, or (c) diffusion between distinct source and sink reservoirs (Figure 4). Many of these approaches emulate traditional strategies and, at the same time, avoid common shortcomings of these by taking advantage of enabling aspects of microscale technologies. Some microfluidic devices create stable gradients, whereas others enable precise perturbations of the slope, direction, or nature of chemoattractant gradients. Complex devices that integrate semipermeable membranes to decouple convection and diffusion, valves to control flows, or the light-controlled release of caged compounds are also being developed for increased functionality and specific target applications (84).
Figure 4.
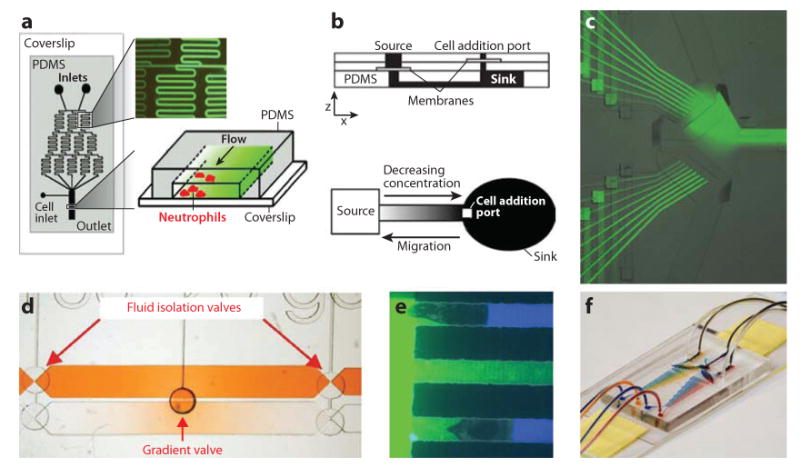
Microfluidic chemotaxis assays. (a) First microfluidic gradient generator for neutrophil chemotaxis. Two solutions, one of chemoattractant and one of buffer, flow continuously through a network of microscale channels to produce streams of different concentrations. Juxtaposition of these streams in the main channel produces a gradient in the direction transversal to the channel. Neutrophils attached to a coverslip inside the main channel are exposed to the chemoattractant gradient and move toward the higher concentrations, in the direction transversal to the channel. PDMS, polydimethylsiloxane. Reproduced from Reference 83. (b) Linear gradients are established by diffusion between a source and a sink. Permeable membranes are employed to ensure a convection-free environment. Cells, introduced to the sink compartment at the beginning of the experiment, migrate toward the chemoattractant source. Reproduced from Reference 124. (c) Rapid switching between independent gradients. Two pairs of nine channels, each carrying different proportions of buffer and fluorescently tagged chemoattractant, are connected through two valves to one main channel where cells are attached to the bottom coverslip. The switch between the two gradients formed by the juxtaposition of streams from each pair of channels can be accomplished in less than 4 s. Reproduced from Reference 101. (d) Evolving radial gradients centered on a microfluidic valve. Two large channels are filled with chemoattractant (top) and cells in buffer (bottom) under the control of microscale valves. Formation of radial gradient is triggered by the opening of a valve between the two channels. Reproduced from Reference 118. (e) Polar stimulation of neutrophils confined in channels. An array of channels with a cross section smaller than the cells is positioned between two compartments, one with chemoattractant and one with buffer, in which solutions are continuously replenished by a self-balanced microfluidic system. Neutrophils that enter these channels and move toward the chemoattractant completely occlude the channels, limiting their chemoattractant exposure to a smaller area at the leading edge. Reproduced from Reference 135. (f) Microfluidic device for switching between two independent gradient generators. Two pairs of solutions (blue and yellow, and blue and red) are mixed inside a network of channels to form two independent gradients, which could be alternatively directed toward a main channel in which cells are attached to the bottom coverslip.
Of all the traditional assays, the Boyden chamber remains the most widely used traditional chemotaxis assay. On one hand, the instability of gradients and poor control over gradient shape in the vicinity of cells are two of its major shortcomings. The local gradient can be well controlled using point release of chemokines from micropipettes; however, these are useful only for short observations of fast moving cells, before the chemokines accumulate in the environment and diffusion degrades the gradient. In sharp contrast, stable and robust gradients can be produced in microfluidic devices at spatial scales relevant for moving cells. Compared to transwell, agarose, or Zigmond chamber assays, microfluidic systems allow not only for better control and linearity of chemokine gradients but also allow designs for various gradient shapes (83) and spatial gradients of different profiles, distinct from those predefined by diffusion principles in traditional assays that can be created at microscale (85–88). On the other hand, traditional assays are still preferred not only for their simplicity but also for the ability to assay almost any cell type. In contrast, in many microfluidic devices, the presence of flow leads to some important limitations, which cannot be used with cells that are sensitive to shear stress. Even for cells that do not react to shear, such as neutrophils and other leukocytes, the presence of flow and the added shear stress from the flow in microfluidic channels has not been demonstrated to alter the migration in the direction of the gradient for a reasonable range of flow rates (89). A recent study demonstrates that the gradient itself can be altered by flow conditions, and the alteration of the gradient profile around one cell in flow could be increased up to 10%, depending on the flow rate (90).
Stable Gradients from Parallel Flow Streams in Microchannels
The basic principle for these microfluidic devices relies on the juxtaposition of streams of different concentrations, flowing in laminar-flow regimes and progressively mixed by diffusion. As long as the flow of solutions is maintained at a constant rate, gradients are extremely well controlled. Streams of different concentration can be produced on the chip starting from just two solutions of different concentrations, and combining and splitting streams in networks of channels. This not only simplifies the logistics of setting up such gradients, but it also enables the assembly of gradients that have different spatial profiles (85–88). A systematic approach for designing devices to generate monotonic gradients has been described by passing two starting solutions through a channel with dividers placed in precise locations to limit diffusion and has been proposed as a universal gradient generator (91). Other devices in which the geometry of the fluidic network is calculated so that different branches have different hydraulic resistances have been described in the literature (92–96). More complex networks capable of handling 16 distinct solutions through 16 independent inlets create complex gradients (97). In addition, the use of valves to control the inlets and outlets in a multiplexed format allows for complex changes between gradients.
Some unexpected insights into cellular chemotaxis have been enabled by quantitative studies on the migration of various cells in stabile gradients in microfluidic devices. In one example, to probe into the role of the temporal history of stimulation on neutrophil chemotaxis, neutrophils that moved toward higher concentrations of interleukin 8 were observed in gradients with different spatial profiles (83). Neutrophils in hill gradient profiles were reported to move toward the higher concentration, overshoot the top, and then return, in a back-and-forth run across the smooth top. In contrast, cells that moved toward higher concentrations in a cliff gradient profile did not overshoot when they reached the highest concentration, which suggests that a sharp downward concentration change has a strong effect on chemotaxis. In another example, steep gradient profiles and high concentrations of compounds that normally function as attractants have been demonstrated to induce directional motility in the opposite direction of the spatial gradient. The discovery of this unexpected behavior, subsequently called fugetaxis, has been enabled by microfluidic devices in which steep, stable gradients at high concentrations can be produced repeatedly and for periods of time long enough to allow systematic studies of the phenomenon (98). In yet another example, the directional motility and velocity of Dictyostelium was measured in response to a broad range of cAMP chemoattractant concentrations and spatial gradients. A basal level of motility was present in Dictyostelium in the absence of the chemoattractant, and directional motility occurred only in the presence of a spatial gradient (99). A new hypothesis suggests that chemotaxis is the result of redirecting stochastic motility in a single direction in stable gradient. This is supported by the observation of maximal migration speed in conditions of the largest difference in number of receptors stimulated at the fronts and backs of cells (at concentrations close to the receptor-binding kinetic constant Kd) and the loss of directionality by the saturation of the same receptors.
An increasing number of these microfluidic devices are emerging that are specifically designed to control the temporal changes of spatial chemokine gradients. The ability to quickly change the direction, spatial slope, or chemical composition of gradients has been a common limitation in traditional systems. Although most microfluidic gradient generators were purposefully designed for stable gradients over time, they had a limited ability to change these gradients in time in a controllable fashion. Approaches that relied on changing the chemokines at the inlet and altering the flow rates were relatively slow (100), on the order of tens of seconds for flow rates compatible with maintaining viable cells inside the channels. However, several microfluidic strategies overcome these limitations. For example, the integration of microfluidic valves on the chip reduced switching times by an order of magnitude compared to the switching of solutions at the inlet at comparable flow rates (101–103). In a different approach, pulses of different temporal duration were encoded in the flow rates at inlets (104) or through the periodic actuation of valves inside the chip (105–107). More sophisticated devices used inflatable structures incorporated into the walls to reverse the positions of adjacent streams by rotation of the fluid (108). Finally, although diffusion occurs in the direction perpendicular to the direction of flow in all these devices, designs in which flow and diffusion are along the same axis and in the opposite direction have also been tested. These approaches require precise control of flow at microscale rates and allow for the formation of sharp gradient profiles (109). However, because of the slow flow rates used to counteract diffusion, any changes of concentration gradient are also slow, and the formation of a new, stable gradient can take more than one minute.
Many of these technologies for switching gradients are slowly making inroads into chemotaxis experiments. Simple microfluidic devices were used to quantify the chemotaxis of the HL60 neutrophil model cell line, in the presence of wortmannin (a common PI3K inhibitor) after reversing the direction of a chemoattractant gradient. Neutrophils displayed normal chemotaxis but had a diminished capability to respond to changes in the direction of the gradient, accomplished by alternately operating two dual-syringe pumps outside the device (103). More complex microfluidic devices, which incorporate different types of valves for fast and precise manipulation of gradients, have also been used to probe the responses of migrating cells to changing conditions. For example, a device that can switch the concentration and direction of chemical gradients in less than 4 s was used to measure the initial directional migration in neutrophils upon stimulation (101). Resting neutrophils may take approximately 5 min to start chemotaxis in this device, a time that is shorter than the 15 min for the neutrophil response in Zigmond chambers (50) and the 20 min in filter assays (77). The difference between these results that explore the same cell behavior can be explained by the faster stabilization of the gradient in the microfluidic device compared to traditional assays.
Recently, several unexpected observations were reported using devices that can quickly switch the direction of gradients. In one example, switching the direction of the gradient results in the immediate and systematic loss of polarization in all observed neutrophils and is followed by a delayed reorganization of the neutrophil polarity for chemotaxis in the new direction. This behavior inside the microfluidic devices was consistent for all observed neutrophils and different from previously reported results in traditional assays. Experiments using the Zigmond chamber or micropipettes reported 40% and 70% U-turns following gradient reversal, respectively (48, 110). In another example, quantitative measurements of neutrophil morphology changes after gradient switches uncovered huge variability in the response times of the same cell to identical perturbations. These observations ultimately led to a new hypothesis regarding the role of cytoskeleton components in signaling for neutrophil orientation in the direction of the gradient (47). Finally, a microfluidic device in which an exponential gradient has been imposed over resting cells in less than 0.5 s has enabled precise control over the concentration and fraction of receptors bound at the fronts and backs of Dictyostelium cells (102). A new model for chemotaxis was developed from these observations, which proposes that differences in the binding of chemoattractant to receptors at the fronts and backs cells, rather than temporal changes in receptor stimulation, are important to induce directional migration in Dictyostelium. In the future, more models can be tested using this experimental system, e.g., models that propose directional polarization based on the time difference in stimulation of the two ends of a cell (44).
In addition to providing new results for new biophysical models of chemotaxis, the use of valves in microfluidic devices can bring several practical advantages. For example, a microfluidic device with valves has been described to accept one droplet of blood at the inlet, from which neutrophils are separated on the chip, and chemotaxis assays are performed within 5 min of blood collection (111). The device circumvents the need for traditional methods to separate neutrophils before the assay, which can introduce artifacts in the chemotaxis assay by artificially activating cells, and may eventually become useful for clinical research. Rapid switches between independent gradients also allow researchers to study the temporal response of moving neutrophils to the sudden addition of active compounds at single-cell resolution (112). In the context of emerging technologies for processing blood samples and the isolation of various types of cells in short times and pure populations, such devices that combine gradient generation and isolation capabilities with complex valve arrangements expand the use of chemotaxis assays beyond research tools into biomedical applications (113).
Point Release of Chemoattractants for Microscale Gradients
Although micropipettes are an all time favorite traditional approach to change concentration profiles around moving cells, only a few examples of microfluidic devices that use comparable approaches have been reported to date. This situation may have less to do with the ability of microfluidic technologies to make precise structures at the same length scale or smaller than the typical micropipette tip but instead with how these point sources can be used. Obviously, the major limitation of these devices is that the opening of the channels has a fixed location on the slide, which takes away much of the flexibility from the traditional micropipette (114). Microfluidic devices that use two pipette-like channels, controlled by valves, or several small openings in the side of a larger channel can generate gradients in small chambers (115, 116). The hybrid implementation of micropipettes in conjunction with microfluidic devices may overcome the positioning limitations, for example, by placing the micropipette close to the slide on which cells are growing. A solution of epithelial growth factor is continuously delivered from the pipette, while the coverslip side acts as a dam. The gradient produced in this system is steeper and better controlled than in the traditional dish (117). Finally, one setup where point release of chemoattractant is used to produce rapidly evolving gradients has been described through the integration of a microfluidic valve at the interface between two compartments. The presence of small, closed compartments increases the precision of control of the radial gradient, especially with respect to the temporal aspects of the gradient (118).
Fast polarization times, between 90 s (56) and 2 min (119, 120), have been reported for neutrophils that respond to chemokines released from micropipettes—faster than any other chemotaxis assay. One emerging approach that circumvents the imprecision of traditional micropipette delivery and the complexity of microfluidic devices with valve actuation takes advantage of the point release of photo-caged compounds in microfluidic streams that flow over target cells. This approach is compatible with a high degree of temporal control and has good temporal resolution. In one recent report, the switch-on time for a caged cyclic AMP (cAMP) was accomplished in 0.8 s, whereas the switch-off by washing the released compound occurred in approximately 0.2 s (121). Moreover, the laser beam could be moved between the front and back of the same cell in 0.25 s. This approach also allows interesting observations, such as the fast translocation of signaling molecules from the cytoplasm to the membrane in Dictyostelium. After a single pulse, the translocation of PH domain-containing proteins peaked at 6 s and then decayed to the initial level in 20 s. Upon continuous stimulation, the translocation peaked with the same dynamics, but decay was slower and reached a normal level in more than 30 s. Although changes in concentration are the fastest of all microfluidic systems (122), the only important limitation is the small number of caged compounds currently available.
Microscale Gradients Produced by Diffusion between Distinct Source and Sink Compartments
To overcome the shortcomings of traditional source and sink assays, several microfluidic devices have been designed in which the moving cells are shielded from flow, and gradients are generated by convection-free diffusion from a source to a sink. One such example is a device fabricated in silicon and glass, in which two compartments are placed in close proximity and connected through a 5-μm-deep microchannel. Cells are aligned at the edge of the channel device passively, by the slow flow induced during cell loading. To prevent a ceaseless flow of contents between adjacent compartments via the communicating channel, a space at the top end of the holder is filled with medium, which bridges the two compartments and equalizes hydrostatic pressures (123). In another variation of sink and source devices, the two compartments were connected through a longer channel that also serves as cell culture chamber. Filters or gels were integrated into devices to minimize convection, and neutrophils settled into the channel through the large pore membrane. A gradient of interleukin 8 was established in a 3-mm-long channel in about 6 h and remained relatively stable for 24 h, which allowed a long-term observation of neutrophil chemotaxis (124, 125). More recently, the ability to converge or disperse cells during chemotaxis through channels of triangular shape, connecting source and sink compartments, has been described (126). In another example, the source, the sink, and the cell compartment were represented by three parallel channels separated by hydrogel blocks, and a gradient was produced by diffusion through the gel into the central channel (127). The migration of dendritic cells toward a gradient of chemoattractant (CCL19) through a collagen matrix (128) and the migration of bacteria toward sources of nutrients (129) have been demonstrated in devices that share this design concept. One advantage of these devices is that solutions in the sink and source compartments can be replaced or renewed at regular time intervals by flowing new solutions into channels (130). Variations of this design have also been described, in which new solutions are continuously flown in, and the migration of cells is observed in several transversal channels that are filled with gel (131). Although the gel is permeable to nutrients and chemical factors that facilitate the formation of linear gradients between the source and sink compartments, its presence also minimizes flow and the shear stress that cells can experience, for example, during vascular invasion assays (132, 133).
A distinct approach to create convection-free and stable gradients in liquid-filled channels, in the absence of dense gels, relies on the balance of pressures at the ends of gradient channels to cancel any convective transport and produce linear, stable gradients. In one approach, symmetric networks of channels matched the higher hydraulic resistance of channels bringing the new solutions in, with the lower hydraulic resistance of transversal channels in which the linear gradients were generated (134). By varying the length of channels, gradients of pheromones of different steepness were created, and cell phenotype and expression of fluorescently tagged genes were monitored at the same time. Distinct phenotypes for the observed yeast cells were noted after a similar history of exposure, which suggests a stochastic decision process with a bimodal outcome. More recently, a different approach took advantage of the fact that, in microfluidic channels, two streams of different compositions can be brought together in one channel and then quickly split again, still keeping their original compositions, and be brought back together again further downstream (135). Excellent symmetry of the channels, easily accomplished by microfluidic technologies, allowed the perfect balance of pressures in the side channels to create almost ideal, zero-flow conditions and linear gradients in the transversal channels. Linear gradients and polar stimulation of the cell that enters the channels have been demonstrated in this device. Unexpected, persistent movement of HL60 cells that enter the channels has been reported, biophysical explanations for which are still to emerge. Switching one of the solutions at the inlets allowed researchers to observe the response of moving cells to the sudden addition of inhibitor drugs to only one side of the cells. One unanticipated result was that the back of the cell appeared to stop first, while the front was still progressing in the direction of the gradient, which suggests a lax coupling between the fronts and backs of cells.
Conclusions
Technologies capable of changing conditions quickly and precisely around moving cells can play critical roles in our understanding of the system-level mechanisms of directional migration in response to chemical gradients. In the past, rapid changes of chemical concentrations around bacteria enabled quantitative insights into the adaptation circuits that allow bacteria to efficiently identify and take advantage of sources of nutrients or avoid dangerous conditions (9, 29, 35). Similar approaches applied to eukaryotic cells facilitated several interesting observations in relation to the initiation and maintenance of polarity in migratory cells. Transient morphological changes (55–58), heterogeneous expression of integrin-binding molecules (61), and oscillations at the leading edge (74) were analyzed in detail immediately after step increases in chemoattractant concentrations and during the initiation phase of migration in cells. More detailed studies of the dynamics of intracellular molecules were also performed, revealing the divergent signaling pathways triggered by stimulation (48) or revealing the differences in the dynamics of actin at the front of the cells and myosin at the back (64, 68) during chemoattractant concentration increases. Several experimental challenges emerged during these studies, in particular related to the limitations of simultaneous temporal and spatial control of the gradients. With the maturing of the microfluidic technologies, most of these challenges can now be effectively addressed, leading to interesting observations of the unexpected capabilities of the moving cells. Simple microfluidic devices that generate stable chemoattractant gradients enabled the discovery of fugetaxis in neutrophils in response to steep chemical gradients (98) as well as observations of intriguing behavior of cells in the presence of sharp or shallow spatial gradient changes (83). Devices capable of abrupt changes in slope or direction of chemoattractant gradients revealed consistent turning or forward-moving behaviors in neutrophils (101), allowed for the systematic study of concentration-dependent responses (99, 102), facilitated the differential analysis of signaling after pulse or step stimulation (122), and enabled the formulation of new hypotheses regarding the role of cytoskeleton in directional sensing (47). Novel uses of similar devices for the screening of fast-acting drugs that modulate chemotaxis (112) and rapid isolation of neutrophils from whole-blood samples (111) are emerging for science and clinical applications.
Keeping up with the accelerating progress in the past few years and moving forward toward a deeper understanding of the complexity of cell chemotaxis requires the expansion of multipronged approaches that combine robust technologies and traditional molecular tools. New devices to generate spatial chemical gradients and precisely change these in time must be engineered, and experimental protocols to take advantage of the new abilities for temporal and spatial control of the environment must be developed. New hypotheses and biophysical models for analyzing the temporal responses from precise perturbations are necessary to integrate experimental results with our vast knowledge of signaling molecules and systems. Further integration with emerging molecular biology techniques for selective perturbation of the intracellular networks can facilitate measurements of elemental responses with high spatial and temporal resolution. Ultimately, even greater challenges lie ahead in translating this new knowledge to clinical problems and studying chemotaxis in the context of disease, from which new opportunities for treatment and cure are poised to emerge.
Acknowledgments
I thank Dr. Jagesh Shah and Dr. Alexander Aranyosi for helpful suggestions and critical reading of this manuscript. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) grant AI076760 and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) grant EB002503.
Footnotes
Disclosure Statement: The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Dormann D, Weijer CJ. Chemotactic cell movement during development. Curr Opin Genet Dev. 2003;13:358–64. doi: 10.1016/s0959-437x(03)00087-x. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–82. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 3.Abe R, Donnelly SC, Peng T, Bucala R, Metz CN. Peripheral blood fibrocytes: differentiation pathway and migration to wound sites. J Immunol. 2001;166:7556–62. doi: 10.4049/jimmunol.166.12.7556. [DOI] [PubMed] [Google Scholar]
- 4.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–95. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–98. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kollmann M, Lovdok L, Bartholome K, Timmer J, Sourjik V. Design principles of a bacterial signaling network. Nature. 2005;438:504–7. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- 7.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–41. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 8.Kitano H. Computational systems biology. Nature. 2002;420:206–10. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 9.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–71. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 10.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–17. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 11.Walrand S, Guillet C, Boirie Y, Vasson MP. In vivo evidences that insulin regulates human polymorphonuclear neutrophil functions. J Leukoc Biol. 2004;76:1104–10. doi: 10.1189/jlb.0104050. [DOI] [PubMed] [Google Scholar]
- 12.Wenisch C, Patruta S, Daxbock F, Krause R, Horl W. Effect of age on human neutrophil function. J Leukoc Biol. 2000;67:40–45. doi: 10.1002/jlb.67.1.40. [DOI] [PubMed] [Google Scholar]
- 13.Kaufmann SHE, Steward MW. Immunology. Washington, DC: Hodder Arnold; 2005. [Google Scholar]
- 14.Beeh KM, Beier J. Handle with care: targeting neutrophils in chronic obstructive pulmonary disease and severe asthma? Clin Exp Allergy. 2006;36:142–57. doi: 10.1111/j.1365-2222.2006.02418.x. [DOI] [PubMed] [Google Scholar]
- 15.Vergunst CE, Gerlag DM, Dinant H, Schulz L, Vinkenoog M, et al. Blocking the receptor for C5a in patients with rheumatoid arthritis does not reduce synovial inflammation. Rheumatology. 2007;46(12):1773–78. doi: 10.1093/rheumatology/kem222. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi T, Hiasa Y, Ohara Y, Miyazaki S, Ogura R, et al. Relation between neutrophil counts on admission, microvascular injury, and left ventricular functional recovery in patients with an anterior wall first acute myocardial infarction treated with primary coronary angioplasty. Am J Cardiol. 2007;100:35–40. doi: 10.1016/j.amjcard.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 17.Turato G, Baraldo S, Zuin R, Saetta M. The laws of attraction: chemokines, neutrophils and eosinophils in severe exacerbations of asthma. Thorax. 2007;62:465–66. doi: 10.1136/thx.2006.070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Condeelis J, Singer RH, Segall JE. The great escape: When cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 19.Huxley AF. Excitation and conduction in nerve: quantitative analysis. Science. 1964;145:1154–59. doi: 10.1126/science.145.3637.1154. [DOI] [PubMed] [Google Scholar]
- 20.Hunter PJ, McCulloch AD, ter Keurs HE. Modelling the mechanical properties of cardiac muscle. Prog Biophys Mol Biol. 1998;69:289–331. doi: 10.1016/s0079-6107(98)00013-3. [DOI] [PubMed] [Google Scholar]
- 21.Burns ME, Baylor DA. Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Annu Rev Neurosci. 2001;24:779–805. doi: 10.1146/annurev.neuro.24.1.779. [DOI] [PubMed] [Google Scholar]
- 22.Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc Natl Acad Sci USA. 2008;105:7165–70. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319:482–84. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toetsch S, Olwell P, Prina-Mello A, Volkov Y. The evolution of chemotaxis assays from static models to physiologically relevant platforms. Integr Biol. 2009;1:170–81. doi: 10.1039/b814567a. [DOI] [PubMed] [Google Scholar]
- 25.Pujic Z, Mortimer D, Feldner J, Goodhill GJ. Assays for eukaryotic cell chemotaxis. Comb Chem High Throughput Screen. 2009;12:580–88. doi: 10.2174/138620709788681952. [DOI] [PubMed] [Google Scholar]
- 26.Bennett MR, Hasty J. Microfluidic devices for measuring gene network dynamics in single cells. Nat Rev Genet. 2009;10:628–38. doi: 10.1038/nrg2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovic A, Howell B, Takayama S. Timing is everything: using fluidics to understand the role of temporal dynamics in cellular systems. Microfluidics Nanofluidics. 2009;6:717–29. [Google Scholar]
- 28.Kirby JR. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu Rev Microbiol. 2009;63:45–59. doi: 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 29.Macnab RM, Koshland DE., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci USA. 1972;69:2509–12. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang N, Macnab R, Koshland DE., Jr Common mechanism for repellents and attractants in bacterial chemotaxis. Science. 1973;181:60–63. doi: 10.1126/science.181.4094.60. [DOI] [PubMed] [Google Scholar]
- 31.Berg HC, Tedesco PM. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:3235–39. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kort EN, Goy MF, Larsen SH, Adler J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc Natl Acad Sci USA. 1975;72:3939–43. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Springer MS, Kort EN, Larsen SH, Ordal GW, Reader RW, Adler J. Role of methionine in bacterial chemotaxis: requirement for tumbling and involvement in information processing. Proc Natl Acad Sci USA. 1975;72:4640–44. doi: 10.1073/pnas.72.11.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Block SM, Segall JE, Berg HC. Impulse responses in bacterial chemotaxis. Cell. 1982;31:215–26. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 35.Segall JE, Block SM, Berg HC. Temporal comparisons in bacterial chemotaxis. Proc Natl Acad Sci USA. 1986;83:8987–91. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duke TA, Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci USA. 1999;96:10104–8. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postma M, Bosgraaf L, Loovers HM, Van Haastert PJ. Chemotaxis: signaling modules join hands at front and tail. EMBO Rep. 2004;5:35–40. doi: 10.1038/sj.embor.7400051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dusenbery DB. Spatial sensing of stimulus gradients can be superior to temporal sensing for free-swimming bacteria. Biophys J. 1998;74:2272–77. doi: 10.1016/S0006-3495(98)77936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–85. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer SJ, Kupfer A. The directed migration of eukaryotic cells. Annu Rev Cell Biol. 1986;2:337–65. doi: 10.1146/annurev.cb.02.110186.002005. [DOI] [PubMed] [Google Scholar]
- 41.Turing AM. The chemical basis of morphogenesis. Phil Trans Royal Soc Lon Series B. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine H, Kessler DA, Rappel WJ. Directional sensing in eukaryotic chemotaxis: a balanced inactivation model. Proc Natl Acad Sci USA. 2006;103:9761–66. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappel WJ, Thomas PJ, Levine H, Loomis WF. Establishing direction during chemotaxis in eukaryotic cells. Biophys J. 2002;83:1361–67. doi: 10.1016/S0006-3495(02)73906-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rappel WJ, Levine H. Receptor noise and directional sensing in eukaryotic chemotaxis. Phys Rev Lett. 2008;100(22):228101. doi: 10.1103/PhysRevLett.100.228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Haastert PJM, Postma M. Biased random walk by stochastic fluctuations of chemoattractant-receptor interactions at the lower limit of detection. Biophys J. 2007;93:1787–96. doi: 10.1529/biophysj.107.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irimia D, Balazsi G, Agrawal N, Toner M. Adaptive-control model for neutrophil orientation in the direction of chemical gradients. Biophys J. 2009;96:3897–916. doi: 10.1016/j.bpj.2008.12.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, et al. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–14. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 49.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–66. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zigmond SH. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977;75:606–16. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zicha D, Dunn GA, Brown AF. A new direct-viewing chemotaxis chamber. J Cell Sci. 1991;99(Pt. 4):769–75. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 52.Zigmond SH. Mechanisms of sensing chemical gradients by polymorphonuclear leukocytes. Nature. 1974;249:450–52. doi: 10.1038/249450a0. [DOI] [PubMed] [Google Scholar]
- 53.Nelson RD, Quie PG, Simmons RL. Chemotaxis under agarose: a new and simple method for measuring chemotaxis and spontaneous migration of human polymorphonuclear leukocytes and monocytes. J Immunol. 1975;115:1650–56. [PubMed] [Google Scholar]
- 54.Heit B, Kubes P. Measuring chemotaxis and chemokinesis: the underagarose cell migration assay. Sci STKE. 2003;2003:PL5. doi: 10.1126/stke.2003.170.pl5. [DOI] [PubMed] [Google Scholar]
- 55.Hu CL, Barnes FS. A theory of necrotaxis. Biophys J. 1970;10:958–69. doi: 10.1016/S0006-3495(70)86345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zigmond SH, Sullivan SJ. Sensory adaptation of leukocytes to chemotactic peptides. J Cell Biol. 1979;82:517–27. doi: 10.1083/jcb.82.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKay DA, Kusel JR, Wilkinson PC. Studies of chemotactic factor-induced polarity in human neutrophils. Lipid mobility, receptor distribution and the time-sequence of polarization. J Cell Sci. 1991;100(Pt. 3):473–79. doi: 10.1242/jcs.100.3.473. [DOI] [PubMed] [Google Scholar]
- 58.Shields JM, Haston WS. Behavior of neutrophil leucocytes in uniform concentrations of chemotactic factors: contraction waves, cell polarity and persistence. J Cell Sci. 1985;74:75–93. doi: 10.1242/jcs.74.1.75. [DOI] [PubMed] [Google Scholar]
- 59.Vicker MG. The regulation of chemotaxis and chemokinesis in Dictyostelium amoebae by temporal signals and spatial gradients of cyclic AMP. J Cell Sci. 1994;107(Pt 2):659–67. doi: 10.1242/jcs.107.2.659. [DOI] [PubMed] [Google Scholar]
- 60.Korohoda W, Madeja Z, Sroka J. Diverse chemotactic responses of Dictyostelium discoideum amoebae in the developing (temporal) and stationary (spatial) concentration gradients of folic acid, cAMP, Ca(2+) and Mg(2+) Cell Motil Cytoskeleton. 2002;53:1–25. doi: 10.1002/cm.10052. [DOI] [PubMed] [Google Scholar]
- 61.Hughes BJ, Hollers JC, Crockett-Torabi E, Smith CW. Recruitment of CD11b/CD18 to the neutrophil surface and adherence-dependent cell locomotion. J Clin Investig. 1992;90:1687–96. doi: 10.1172/JCI116041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albrecht E, Petty HR. Cellular memory: neutrophil orientation reverses during temporally decreasing chemoattractant concentrations. Proc Natl Acad Sci USA. 1998;95:5039–44. doi: 10.1073/pnas.95.9.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geiger J, Wessels D, Soll DR. Human polymorphonuclear leukocytes respond to waves of chemoattractant, like Dictyostelium. Cell Motil Cytoskeleton. 2003;56:27–44. doi: 10.1002/cm.10133. [DOI] [PubMed] [Google Scholar]
- 64.Etzrodt M, Ishikawa HC, Dalous J, Muller-Taubenberger A, Bretschneider T, Gerisch G. Time-resolved responses to chemoattractant, characteristic of the front and tail of Dictyostelium cells. FEBS Lett. 2006;580:6707–13. doi: 10.1016/j.febslet.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 65.Dalous J, Burghardt E, Muller-Taubenberger A, Bruckert F, Gerisch G, Bretschneider T. Reversal of cell polarity and actin-myosin cytoskeleton reorganization under mechanical and chemical stimulation. Biophys J. 2008;94:1063–74. doi: 10.1529/biophysj.107.114702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turing A. The chemical basis of morphogenesis. Philos Trans R Soc London B-Biol Sci. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meinhardt H. Models of biological pattern formation: from elementary steps to the organization of embryonic axes. Curr Top Dev Biol. 2008;81:1–63. doi: 10.1016/S0070-2153(07)81001-5. [DOI] [PubMed] [Google Scholar]
- 68.Gerisch G, Keller HU. Chemotactic reorientation of granulocytes stimulated with micropipettes containing fMet-Leu-Phe. J Cell Sci. 1981;52:1–10. doi: 10.1242/jcs.52.1.1. [DOI] [PubMed] [Google Scholar]
- 69.Swanson JA, Taylor DL. Local and spatially coordinated movements in Dictyostelium discoideum amoebae during chemotaxis. Cell. 1982;28:225–32. doi: 10.1016/0092-8674(82)90340-3. [DOI] [PubMed] [Google Scholar]
- 70.Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765–70. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 71.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–48. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 72.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 73.Keller HU, Naef A, Zimmermann A. Effects of colchicine, vinblastine and nocodazole on polarity, motility, chemotaxis and cAMP levels of human polymorphonuclear leukocytes. Exp Cell Res. 1984;153:173–85. doi: 10.1016/0014-4827(84)90459-2. [DOI] [PubMed] [Google Scholar]
- 74.Zhelev DV, Alteraifi AM, Chodniewicz D. Controlled pseudopod extension of human neutrophils stimulated with different chemoattractants. Biophys J. 2004;87:688–95. doi: 10.1529/biophysj.103.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vicker MG. Ideal and nonideal concentration gradient propagation in chemotaxis studies. Exp Cell Res. 1981;136:91–100. doi: 10.1016/0014-4827(81)90040-9. [DOI] [PubMed] [Google Scholar]
- 76.Ebrahimzadeh PR, Hogfors C, Braide M. Neutrophil chemotaxis in moving gradients of fMLP. J Leukoc Biol. 2000;67:651–61. doi: 10.1002/jlb.67.5.651. [DOI] [PubMed] [Google Scholar]
- 77.Ebrahimzadeh PR, Braide M. The initiation of the neutrophil chemotactic response in filters. J Leukoc Biol. 1999;66:90–94. doi: 10.1002/jlb.66.1.90. [DOI] [PubMed] [Google Scholar]
- 78.Ebrahimzadeh PR, Bazargani F, Afzal F, Hogfors C, Braide M. A subpopulation analysis of f-MLP stimulated granulocytes migrating in filters. Biorheology. 1996;33:231–50. doi: 10.1016/0006-355x(96)00019-4. [DOI] [PubMed] [Google Scholar]
- 79.Braide M, Ebrahimzadeh PR, Strid KG, Bjursten LM. Migration of human granulocytes in filters: effects of gravity and movable gradients of f-MLP. Biorheology. 1994;31:617–30. doi: 10.1016/0006-355X(95)91103-A. [DOI] [PubMed] [Google Scholar]
- 80.Vicker MG, Lackie JM, Schill W. Neutrophil leukocyte chemotaxis is not induced by a spatial gradient of chemoattractant. J Cell Sci. 1986;84:263–80. doi: 10.1242/jcs.84.1.263. [DOI] [PubMed] [Google Scholar]
- 81.Dunn GA, Zicha D. Long-term chemotaxis of neutrophilsin stable gradients: preliminary evidence of periodic behavior. Blood Cells. 1993;19:25–39. discussion p. 41. [PubMed] [Google Scholar]
- 82.Xu J, Wang F, Van Keymeulen A, Rentel M, Bourne HR. Neutrophil microtubules suppress polarity and enhance directional migration. Proc Natl Acad Sci USA. 2005;102:6884–89. doi: 10.1073/pnas.0502106102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeon NL, Baskaran H, Dertinger SKW, Whitesides GM, Van De Water L, Toner M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nat Biotechnol. 2002;20:826–30. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 84.Keenan TM, Folch A. Biomolecular gradients in cell culture systems. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Generation of solution and surface gradients using microfluidic systems. Langmuir. 2000;16:8311–16. [Google Scholar]
- 86.Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Generation of gradients having complex shapes using microfluidic networks. Anal Chem. 2001;73:1240–46. [Google Scholar]
- 87.Rhoads DS, Nadkarni SM, Song L, Voeltz C, Bodenschatz E, Guan JL. Using microfluidic channel networks to generate gradients for studying cell migration. Methods Mol Biol. 2005;294:347–57. doi: 10.1385/1-59259-860-9:347. [DOI] [PubMed] [Google Scholar]
- 88.Neils C, Tyree Z, Finlayson B, Folch A. Combinatorial mixing of microfluidic streams. Lab Chip. 2004;4:342–50. doi: 10.1039/b314962e. [DOI] [PubMed] [Google Scholar]
- 89.Walker GM, Sai J, Richmond A, Stremler M, Chung CY, Wikswo JP. Effects of flow and diffusion on chemotaxis studies in a microfabricated gradient generator. Lab Chip. 2005;5:611–18. doi: 10.1039/b417245k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beta C, Frohlich T, Bodeker HU, Bodenschatz E. Chemotaxis in microfluidic devices–a study of flow effects. Lab Chip. 2008;8:1087–96. doi: 10.1039/b801331d. [DOI] [PubMed] [Google Scholar]
- 91.Irimia D, Geba DA, Toner M. Universal microfluidic gradient generator. Anal Chem. 2006;78:3472–77. doi: 10.1021/ac0518710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Campbell K, Groisman A. Generation of complex concentration profiles in microchannels in a logarithmically small number of steps. Lab Chip. 2007;7:264–72. doi: 10.1039/b610011b. [DOI] [PubMed] [Google Scholar]
- 93.Lee K, Kim C, Ahn B, Panchapakesan R, Full AR, et al. Generalized serial dilution module for monotonic and arbitrary microfluidic gradient generators. Lab Chip. 2009;9:709–17. doi: 10.1039/b813582g. [DOI] [PubMed] [Google Scholar]
- 94.Sun K, Wang ZX, Jiang XY. Modular microfluidies for gradient generation. Lab Chip. 2008;8:1536–43. doi: 10.1039/b806140h. [DOI] [PubMed] [Google Scholar]
- 95.Amarie D, Glazier JA, Jacobson SC. Compact microfluidic structures for generating spatial and temporal gradients. Anal Chem. 2007;79:9471–77. doi: 10.1021/ac0714967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li CW, Chen RS, Yang MS. Generation of linear and nonlinear concentration gradients along microfluidic channel by microtunnel controlled stepwise addition of sample solution. Lab Chip. 2007;7:1371–73. doi: 10.1039/b705525k. [DOI] [PubMed] [Google Scholar]
- 97.Cooksey GA, Sip CG, Folch A. A multi-purpose microfluidic perfusion system with combinatorial choice of inputs, mixtures, gradient patterns, and flow rates. Lab Chip. 2009;9:417–26. doi: 10.1039/b806803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tharp WG, Yadav R, Irimia D, Upadhyaya A, Samadani A, et al. Neutrophil chemorepulsion in defined interleukin-8 gradients in vitro and in vivo. J Leukoc Biol. 2006;79:539–54. doi: 10.1189/jlb.0905516. [DOI] [PubMed] [Google Scholar]
- 99.Song L, Nadkarni SM, Bodeker HU, Beta C, Bae A, et al. Dictyostelium discoideum chemotaxis: threshold for directed motion. Eur J Cell Biol. 2006;85:981–89. doi: 10.1016/j.ejcb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 100.Lin F, Saadi W, Rhee SW, Wang SJ, Mittal S, Jeon NL. Generation of dynamic temporal and spatial concentration gradients using microfluidic devices. Lab Chip. 2004;4:164–67. doi: 10.1039/b313600k. [DOI] [PubMed] [Google Scholar]
- 101.Irimia D, Liu SY, Tharp WG, Samadani A, Toner M, Poznansky MC. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip. 2006;6:191–98. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herzmark P, Campbell K, Wang F, Wong K, El-Samad H, et al. Bound attractant at the leading vs. the trailing edge determines chemotactic prowess. Proc Natl Acad Sci USA. 2007;104:13349–54. doi: 10.1073/pnas.0705889104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu YX, Sai JQ, Richmond A, Wikswo JP. Microfluidic switching system for analyzing chemotaxis responses of wortmannin-inhibited HL-60 cells. Biomed Microdevices. 2008;10:499–507. doi: 10.1007/s10544-007-9158-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.King KR, Wang S, Jayaraman A, Yarmush ML, Toner M. Microfluidic flow-encoded switching for parallel control of dynamic cellular microenvironments. Lab Chip. 2008;8:107–16. doi: 10.1039/b716962k. [DOI] [PubMed] [Google Scholar]
- 105.Azizi F, Mastrangelo CH. Generation of dynamic chemical signals with pulse code modulators. Lab Chip. 2008;8:907–12. doi: 10.1039/b716634f. [DOI] [PubMed] [Google Scholar]
- 106.Xie Y, Wang Y, Chen L, Mastrangelo CH. Fourier microfluidics. Lab Chip. 2008;8:779–85. doi: 10.1039/b718376c. [DOI] [PubMed] [Google Scholar]
- 107.Chen L, Azizi F, Mastrangelo CH. Generation of dynamic chemical signals with microfluidic C-DACs. Lab Chip. 2007;7:850–55. doi: 10.1039/b706304k. [DOI] [PubMed] [Google Scholar]
- 108.Hsu CH, Folch A. Spatio-temporally-complex concentration profiles using a tunable chaotic micromixer. Appl Phys Lett. 2006;89:144102. [Google Scholar]
- 109.Kang T, Han J, Lee KS. Concentration gradient generator using a convective-diffusive balance. Lab Chip. 2008;8:1220–22. doi: 10.1039/b800859k. [DOI] [PubMed] [Google Scholar]
- 110.Zigmond SH, Levitsky HI, Kreel BJ. Cell polarity: an examination of its behavioral expression and its consequences for polymorphonuclear leukocyte chemotaxis. J Cell Biol. 1981;89:585–92. doi: 10.1083/jcb.89.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Agrawal N, Toner M, Irimia D. Neutrophil migration assay from a drop of blood. Lab Chip. 2008;8:2054–61. doi: 10.1039/b813588f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kasuga K, Yang R, Porter TF, Agrawal N, Petasis NA, et al. Rapid appearance of resolvin precursors in inflammatory exudates: novel mechanisms in resolution. J Immunol. 2008;181:8677–87. doi: 10.4049/jimmunol.181.12.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Toner M, Irimia D. Blood-on-a-chip. Annu Rev Biomed Eng. 2005;7:77–103. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fok S, Domachuk P, Rosengarten G, Krause N, Braet F, et al. Planar microfluidic chamber for generation of stable and steep chemoattractant gradients. Biophys J. 2008;95:1523–30. doi: 10.1529/biophysj.107.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chung BG, Lin F, Jeon NL. A microfluidic multi-injector for gradient generation. Lab Chip. 2006;6:764–68. doi: 10.1039/b512667c. [DOI] [PubMed] [Google Scholar]
- 116.Keenan TM, Hsu CH, Folch A. Microfluidic “jets” for generating steady-state gradients of soluble molecules on open surfaces. Appl Phys Lett. 2006;89:114103. [Google Scholar]
- 117.Soon L, Mouneimne G, Segall J, Wyckoff J, Condeelis J. Description and characterization of a chamber for viewing and quantifying cancer cell chemotaxis. Cell Motil Cytoskeleton. 2005;62:27–34. doi: 10.1002/cm.20082. [DOI] [PubMed] [Google Scholar]
- 118.Frevert CW, Boggy G, Keenan TM, Folch A. Measurement of cell migration in response to an evolving radial chemokine gradient triggered by a microvalve. Lab Chip. 2006;6:849–56. doi: 10.1039/b515560f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zadeh AD, Keller H. Chemotactically directed redistribution of alpha-actinin precedes morphological polarization and reversal of polarity in human polymorphonuclear leucocytes (PMNs) Eur J Cell Biol. 2003;82:93–96. doi: 10.1078/0171-9335-00295. [DOI] [PubMed] [Google Scholar]
- 120.Keller HU, Zimmermann A. Shape, movement and function of neutrophil granulocytes. Biomed Pharmacother. 1987;41:285–89. [PubMed] [Google Scholar]
- 121.Beta C, Wyatt D, Rappel WJ, Bodenschatz E. Flow photolysis for spatiotemporal stimulation of single cells. Anal Chem. 2007;79:3940–44. doi: 10.1021/ac070033y. [DOI] [PubMed] [Google Scholar]
- 122.Bae AJ, Beta C, Bodenschatz E. Rapid switching of chemical signals in microfluidic devices. Lab Chip. 2009;9:3059–65. doi: 10.1039/b905521e. [DOI] [PubMed] [Google Scholar]
- 123.Kanegasaki S, Nomura Y, Nitta N, Akiyama S, Tamatani T, et al. A novel optical assay system for the quantitative measurement of chemotaxis. J Immunol Methods. 2003;282:1–11. doi: 10.1016/j.jim.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 124.Abhyankar VV, Lokuta MA, Huttenlocher A, Beebe DJ. Characterization of a membrane-based gradient generator for use in cell-signaling studies. Lab Chip. 2006;6:389–93. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- 125.Abhyankar VV, Toepke MW, Cortesio CL, Lokuta MA, Huttenlocher A, Beebe DJ. A platform for assessing chemotactic migration within a spatiotemporally defined 3D microenvironment. Lab Chip. 2008;8:1507–15. doi: 10.1039/b803533d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim D, Lokuta MA, Huttenlocher A, Beebe DJ. Selective and tunable gradient device for cell culture and chemotaxis study. Lab Chip. 2009;9:1797–800. doi: 10.1039/b901613a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cheng SY, Heilman S, Wasserman M, Archer S, Shuler ML, Wu M. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab Chip. 2007;7:763–69. doi: 10.1039/b618463d. [DOI] [PubMed] [Google Scholar]
- 128.Haessler U, Kalinin Y, Swartz MA, Wu M. An agarose-based microfluidic platform with a gradient buffer for 3D chemotaxis studies. Biomed Microdevices. 2009;11:827–35. doi: 10.1007/s10544-009-9299-3. [DOI] [PubMed] [Google Scholar]
- 129.Diao J, Young L, Kim S, Fogarty EA, Heilman SM, et al. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab Chip. 2006;6:381–88. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- 130.Shamloo A, Ma N, Poo MM, Sohn LL, Heilshorn SC. Endothelial cell polarization and chemotaxis in a microfluidic device. Lab Chip. 2008;8:1292–99. doi: 10.1039/b719788h. [DOI] [PubMed] [Google Scholar]
- 131.Saadi W, Rhee SW, Lin F, Vahidi B, Chung BG, Jeon NL. Generation of stable concentration gradients in 2D and 3D environments using a microfluidic ladder chamber. Biomed Microdevices. 2007;9:627–35. doi: 10.1007/s10544-007-9051-9. [DOI] [PubMed] [Google Scholar]
- 132.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Cell migration into scaffolds under coculture conditions in a microfluidic platform. Lab Chip. 2009;9:269–75. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 133.Vickerman V, Blundo J, Chung S, Kamm R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip. 2008;8:1468–77. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Paliwal S, Iglesias PA, Campbell K, Hilioti Z, Groisman A, Levchenko A. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature. 2007;446:46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- 135.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Polar stimulation and constrained cell migration in microfluidic channels. Lab Chip. 2007;7:1783–90. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang F. The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb Perspect Biol. 2009 doi: 10.1101/cshperspect.a002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sasaki AT, Firtel RA. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur J Cell Biol. 2006;85:873–95. doi: 10.1016/j.ejcb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 138.Weiner OD, Servant G, Welch MD, Mitchison TJ, Sedat JW, Bourne HR. Spatial control of actin polymerization during neutrophil chemotaxis. Nat Cell Biol. 1999;1:75–81. doi: 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wong K, Van Keymeulen A, Bourne HR. PDZRhoGEF and myosin II localize RhoA activity to the back of polarizing neutrophil-like cells. J Cell Biol. 2007;179:1141–48. doi: 10.1083/jcb.200706167. [DOI] [PMC free article] [PubMed] [Google Scholar]


