Abstract
Investigation of gene function in diverse organisms relies on knowledge of how the gene products interact with each other in their normal cellular environment. The Bimolecular Fluorescence Complementation (BiFC) Assay1 allows researchers to visualize protein-protein interactions in living cells and has become an essential research tool. This assay is based on the facilitated association of two fragments of a fluorescent protein (GFP) that are each fused to a potential interacting protein partner. The interaction of the two protein partners would facilitate the association of the N-terminal and C-terminal fragment of GFP, leading to fluorescence. For plant researchers, onion epidermal cells are an ideal experimental system for conducting the BiFC assay because of the ease in obtaining and preparing onion tissues and the direct visualization of fluorescence with minimal background fluorescence. The Helios Gene Gun (BioRad) is commonly used for bombarding plasmid DNA into onion cells. We demonstrate the use of Helios Gene Gun to introduce plasmid constructs for two interacting Arabidopsis thaliana transcription factors, SEUSS (SEU) and LEUNIG HOMOLOG (LUH)2 and the visualization of their interactions mediated by BiFC in onion epidermal cells.
Protocol
I. Plasmid preparation
The plasmid DNA used for bombardment must be of high purity and at a concentration of 500ng/μl or greater. For BiFC involving two plasmid constructs, 25 μg of each plasmid is needed. In other words, 1:1 molar ratio of the two plasmids should be mixed to give rise to 50 μg total plasmid DNA for the preparation of cartridges. Be mindful that adding more DNA than 50 μg may cause agglomeration of the gold particles and should be avoided. Commercially available plasmid DNA extraction kits are recommended for isolating plasmid DNAs.
Plasmid vectors should be relatively small in size such as pUC19. We cloned SEU and LUH cDNA into pUC19-SPYNE and pUC19-SPYCE, respectively3. Binary vectors for Agrobacterium-mediated transformation are too large and inappropriate for the bombardment.
II. Cartridge preparation
Cartridge preparation involves coating gold particles with plasmid DNA and loading them into plastic tubing that is subsequently cut into half inch long pieces, which can be loaded into the gene gun cartridge holders. Cartridge preparation is described in great details in a separate Jove article4 and is thus not described here. For bombardment of onion cells, we recommend mixing 50 μg of plasmid DNA with 12.5 mg of gold particles per preparation, which yields about up to 50 cartridges at 1 μg DNA/0.25 mg gold/cartridge. Each cartridge is for one shot. The gold particles can be either 0.6 μm or 1.0 μm in diameter (Cat #1652262 or Cat #1652263; Biorad). We also recommend using 0.5 mg/ml PVP solution for the cartridge preparation. For BiFC, combine the two plasmid DNAs of each putative interacting partner in the same cartridge preparation.
After cartridge preparation, it is important to test the cartridges by firing the newly made cartridges at a helium pressure between 150 to 200 psi into a square Parafilm that is wrapped around a Petri dish. This will test if the gold particles can be propelled efficiently from the cartridges; you should see faint gold particles on the Parafilm. This gives you an idea of the area that each shot will cover. You may notice different amounts of gold particles propelled onto the Parafilm at different helium pressures (at 150-200 psi range). If there is no significant difference, choose the lower pressure for bombardment.
III. Preparing onion tissues
On the day of shooting, peel off outer skin from a large yellow onion. Using a clean and sharp razor blade, cut 4-5 layers deep a rectangular area of about 2 X 8 cm. Take out the rectangular onion tissue, discard the outer two layers, cut the remaining layers of onion into about 2X1.5 cm pieces. Place the onion pieces in a Petri dish containing filter paper (Whatman 3MM paper cut into circles) moistened with sterile water. Cover the Petri dish and they are ready to go.
IV. Bombardment
Load the cartridges into the white round cartridge holders. Cartridges containing different plasmid constructs should be loaded into different cartridge holders. Each cartridge holder can hold up to 12 cartridges for 12 shots.
Insert the battery and the fresh barrel liner into the Helios Gene Gun. Make sure that both the barrel liner and the gun have respective o-rings attached.
First, insert an empty cartridge holder into the gene gun, with mark #12 on the cartridge holder facing the top of the gun. Advance cartridge holder once or twice to make sure it is inserted correctly.
Attach gun to the helium tank and adjust pressure between 150 to 200 psi.
Wear ear protection and fire gun a few times with the empty cartridge holder to clean out the chamber of the gun and to make sure that the pressure is stable.
Remove the empty cartridge holder and insert a loaded cartridge holder. Advance the cartridge holder so that the cartridge you want to fire is positioned at bottom (6 o'clock) of the cartridge holder. Shooting a piece of onion here provides a form of negative control for background fluorescence.
Place onion pieces in the middle of an empty Petri dish with the inner most layer of the onion facing up. Rest the gene gun's barrel liner on Petri dish and fire the gun by holding down the side button while pulling the trigger. Transfer the bombarded onion to a Petri dish containing moist 3MM filter paper.
Advance to the next cartridge and fire a different onion piece. Repeat this step until four to five pieces of onions are shot. Each onion piece is shot once.
Change to a different cartridge holder loaded with cartridges containing a different plasmid or different plasmid combination. Remember to change the barrel liner as well to prevent contamination. Shoot four to five onion pieces.
After bombardment, return all onions to perti dishes. Cover the Petri dishes with lids. Wrap them with Parafilm to prevent drying. Incubate the onion pieces at room temperature in the dark for 16-48 hours.
Turn off gas and release pressure before detaching the gene gun from the helium tank. Unscrew the barrel liner, and remove the cartridge holder and the used (or unused) cartridges.
Cleaning cartridge holders and barrel liners by submerging them in a beaker with soapy water. Place the beaker in a bath sonicator and sonicate for 20 minutes. Afterwards, rinse them with water to remove all soap residues. Soak them in 70% ethanol for an hour and then dry them on paper towels.
V. Observation
After incubating bombarded onion pieces for 16-20 hours at room temperature, we can observe GFP or YFP fluorescence using a fluorescence microscope such as the Zeiss Axio Observer.z1 used in this study.
Use forceps with flat ends to slowly peel a single cell layer off the inner epidermis of the onion (the layer that directly faces the gun during the bombardment). Place the peel onto a drop of water on a slide. Add coverslip but be careful to minimize bubbles.
Under bright field of the microscope, first check for cytoplasmic streaming to ensure that the cells are alive. To do so, expose the slide to the bright field light for a few minutes to warm up the onion tissue and then look for moving plastids. Cytoplasmic streaming is not always easy to see, so don't get discouraged if you can't see any.
For fluorescence detection, use appropriate excitation and detection filters for GFP or YFP. It is important to include negative controls such as onion pieces shot with a non-fluorescent plasmid. Faint yellow signals may appear from wounded cells, air bubbles, or debris. It is also important to include positive controls. A plasmid containing constitutively expressed fluorescent reporter such as 35S::GFP is an excellent positive control. Visible GFP signals from onion epidermal cells (Figure 1A, D) indicated that the cartridges, the onion tissues, and the gene gun firing are properly prepared and executed. Subsequently, we observed interactions between SEU-pSPYNE and LUH-pSPYCE in onion pieces bombarded with cartridges containing a mixture of 35S::SEU-pSPYNE and 35S::LUH-pSPYCE DNAs. Faint YFP fluorescence was observed in nucleus only (Figure 1 B, E), indicating the direct physical interaction between SEU and LUH that brought the split YFP fragments together in the nucleus. Note that the YFP signal from BiFC is significantly weaker than the GFP signal from 35S::GFP.
Representative Results
In our study reported here, the 35S::GFP positive control plasmid gives off strong fluorescence that fills the entire cell, including both the nucleus and cytoplasm (Figure 1A, D). GFP protein is small enough to diffuse into nucleus without a nuclear localization signal. In contrast, SEU and LUH are two Arabidopsis transcriptional factors previously shown to interact in a yeast two-hybrid assay2. SEU-pSPYNE (SEU fused to the N-terminal fragment of YFP) and LUH-pSPYCE (SEU fused to the N-terminal fragment of YFP), both too large to diffuse into the nucleus, contain nuclear localization signals2,5. The YFP fluorescent signal detected in the onion cell nucleus (Figure 1 B, E) indicates that SEU-pSPYNE and LUH-pSPYCE can interact in onion nuclei. No fluorescent signal is detected in onions bombarded with plasmid mix of vector plasmid pSPYNE containing the N-terminal fragment of YFP and LUH-pSPYCE (Figure 1C, F).
The BiFC-mediated YFP fluorescence is significantly weaker than the GFP-mediated fluorescence from the 35S::GFP plasmid. In our study, they also showed different subcellular localization (compare Figure 1A with B). Additionally, the number of fluorescent cells is significantly lower for BiFC, as BiFC only works when both partner plasmids successfully enter and are expressed in the same onion cells.
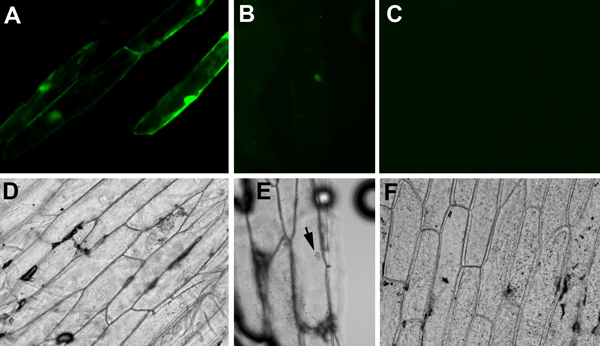 Figure 1. Fluorescent (A, B, C) and bright field (D, E, F) images of onion cells bombarded with different plasmid constructs. (A) and (D), Onion cells bombarded with 35S::GFP. GFP diffuses through out the cytoplasm and nucleus. (B) and (E), Onion cells bombarded with equal molar mixes of 35S::SEU-pSPYNE (SEU fused to the N-terminal fragment of YFP) and 35S::LUH-pSPYCE (LUH fused to the C-terminal fragment) plasmid DNA. An interaction between SEU-pSPYNE and LUH-pSPYCE is shown by a fluorescent nucleus. (C) and (F), A negative control showing onion cells bombarded with mixes of vector pUC19-SPYNE and LUH-pSPYCE plasmids. No fluorescent signal is detected.
Figure 1. Fluorescent (A, B, C) and bright field (D, E, F) images of onion cells bombarded with different plasmid constructs. (A) and (D), Onion cells bombarded with 35S::GFP. GFP diffuses through out the cytoplasm and nucleus. (B) and (E), Onion cells bombarded with equal molar mixes of 35S::SEU-pSPYNE (SEU fused to the N-terminal fragment of YFP) and 35S::LUH-pSPYCE (LUH fused to the C-terminal fragment) plasmid DNA. An interaction between SEU-pSPYNE and LUH-pSPYCE is shown by a fluorescent nucleus. (C) and (F), A negative control showing onion cells bombarded with mixes of vector pUC19-SPYNE and LUH-pSPYCE plasmids. No fluorescent signal is detected.
Discussion
For first time users, we recommend practicing the entire procedure, from cartridge preparation to observation, with a single plasmid constitutive reporter such as 35S::GFP or 35S::GUS. We used a 35S::GFP plasmid obtained from Drs. Steve Mount and Xiaoning Zhang6 (Figure 1A, D). The gene shot control cartridges sold at RioRad (Cat 165-2244) are not suitable for plants.
For BiFC, a second positive control is necessary, which should consist of two known interacting partners fused to the C-terminal and N-terminal fragments of YFP, respectively.
For BiFC, it is necessary to include several negative controls: (1) protein X fused to the N-terminal fragment of YFP plus a vector coding for the C-terminal fragment; (2) protein Y fused to the C-terminal fragment of YFP plus a vector coding for the N-terminal fragment; (3) vector only.
A diffusion screen can be used on the base of the Helios Gene Gun barrel when firing the shot, which serves to reduce tissue damage. In our study, we did not use the diffusion screen.
During microscopic viewing of YFP fluorescence mediated by BiFC, exposure time to the excitation light should be minimized to reduce photo bleaching.
Cartridges stored in the scintillation vials containing desiccants are good for several months.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank Drs. Steve Mount and Xiaoning Zhang for the 35S::GFP pGlowbug construct and the Hokensen Doctoral Fellowship to C. H. Research in Z. L's laboratory is supported by the US National Science Foundation (IOB0616096 and MCB0744752). Z.L. is partially supported by the University of Maryland Agricultural Experiment Station.
References
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9(4):789–789. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Sitaraman J, Bui M, Liu Z. LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 2008;147(2):672–672. doi: 10.1104/pp.108.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40(3):428–428. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Woods G, Zito K. Preparation of gene gun bullets and biolistic transfection of neurons in slice culture. J Vis Exp. 2008 doi: 10.3791/675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhakanandam S, Nole-Wilson S, Bao F, Franks RG. SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol. 2008;146(3):1165–1165. doi: 10.1104/pp.107.114751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XN, Mount SM. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009;150(3):1450–1450. doi: 10.1104/pp.109.138180. [DOI] [PMC free article] [PubMed] [Google Scholar]


