Abstract
Xvent homeobox genes encode transcription factors that repress organizer genes and are essential for dorsoventral specification during early embryogenesis in Xenopus. In contrast to the Xvent-2 gene subfamily, Xvent-1 subfamily members, including PV.1A, have been proposed as indirect targets of Bone Morphogenetic Protein-4 (BMP-4) signaling. Because PV.1A is a critical downstream mediator of, and tightly regulated by, BMP-4 signaling, we hypothesized that its promoter contains a direct BMP-4 response element to effect this transcriptional regulation. We demonstrate that direct regulation by BMP-4 is necessary for transcription of PV.1A: its proximal promoter contains cis-acting binding elements for Smads and Oaz crucial to induction in response to BMP-4 signaling. In addition to these direct cis-acting BMP-4 responsive elements, an indirect Xvent-2 response element and several repressive elements exist in the PV.1A promoter to regulate its transcription. In summary, PV.1A undergoes combinatorial regulation during early Xenopus development as both the direct target of BMP-4 signaling and as the direct and indirect target of positive and negative regulatory factors.
Introduction
Dorsoventral patterning in developing Xenopus embryos is established in part by a gradient of Bone Morphogenetic Protein (BMP) signaling generated by the BMP antagonists Chordin, Noggin, and Follistatin in the extracellular space. Binding of BMPs, which effect ventralization, to antagonists prevents their interaction with their cognate receptors, leading to embryo dorsalization in overexpression studies [1], [2], [3].
In vertebrates, BMPs play critical roles in dorsoventral patterning of the early embryonic mesoderm and specification of the epidermis. In Xenopus, BMP-2, BMP-4, and BMP-7 ventralize the early mesoderm and negatively regulate neurogenesis [4], [5]; BMP-4 in particular is instrumental in tissue patterning and fate determination during embryonic development [6]. Intracellular BMP-4 signaling is mediated through Smad proteins, which translocate into the nucleus to activate the transcription of target genes [7], and ectopic expression of these targets, including the homeobox-containing genes Xvent-1 [8], PV.1 [9], Xvent-2 [10], and Xmsx-1 [11], recapitulates the effects of BMP-4 signals. Inhibition of BMP-4 signaling represses transcription of Xvent homeobox proteins [12], while BMP-4 overexpression induces ectopic expression of Xvents during early embryonic development [12], [13], [14].
Xvent family members can suppress dorsal fates, and promote ventral, when ectopically expressed [15]. BMP-4 signaling induces the expression of Xvent members that repress transcription of dorsal-specific genes [13], [14], [16] and rescue the dorsalized phenotype affected by dominant-negative Type I BMP receptor (DN-BR) [17]. The Xvent family comprises two subfamilies subdivided on the basis of their amino acid sequence: Xvent-2 (Xvent-2 [10], Xbr-1b [18], Xom [19], Vox [20] and Xvent-2B [21]) and Xvent-1 (Xvent-1 [8], Xvent-1B [21], PV.1 [9], and PV.1A).
Xvent-2 transcription is regulated by Smad1/4 and its co-activator Oaz in response to BMP-4 signaling [22]. However, the transcriptional regulation of these Xvent families differs. According to previous reports, the Xvent-2, but not the Xvent-1 [15], family is a direct target of BMP-4 signaling [16], [22], [23]. Nevertheless, expression of Xvent-1B, a highly conserved Xvent-1 paralogue, is induced indirectly by BMP-4 signaling [9].
In order to understand how BMP-4 signaling regulates the transcription of PV.1A, we isolated genomic DNA (gDNA) encompassing PV.1A and its 5′-flanking region (−2525 bp) and analyzed the regulatory elements in its promoter with regard to BMP-4 signaling during early embryonic development. The region 180-bp upstream of the major transcriptional initiation site promoted full reporter-gene activity and contained cis-acting elements responding directly to BMP-4 signaling. The proximal region of the promoter possessed putative binding sites for Smads and Oaz, both of which were necessary for response to BMP-4 signaling. Additionally, we determined that the promoter contains a BMP-4 response element (BRE) and an Xvent-2 response element (XRE). Furthermore, we identified negative cis-acting elements that responded to the dorsal-specific transcription factors AP-1 and Goosecoid in the proximal PV.1A promoter. These results suggest that the proximal PV.1A promoter contains multiple cis-acting elements, including direct, indirect, and negative response elements, as well as those binding transcriptional co-activators, allowing BMP-4 signals to converge and maintain tight transcriptional regulation during early embryonic development in Xenopus.
Materials and Methods
Ethics Statement
Approval from the Institutional Animal Care and Use Committee (IACUC) is not required for the experimental use of amphibians or reptiles in Korea. All members of our research group attended educational and training courses on appropriate care and usage of experimental animals. Adult X. laevis were entrained in 12 hr light/dark (LD 12∶12 h) cycles at 18°C in containers from the Institutes of Laboratory Animal Resources built to specifications for laboratory animal maintenance.
DNA and RNA preparation
cDNAs encoding BMP-4, DN-BR, Smad1(3.4SA), Smad1(3SA), Oaz, dominant-negative Xvent-2 (DN-Xvent-2) [21], Goosecoid, c-Jun, and c-Fos were all subcloned into the pSP64T expression vector. Smad1 wild-type sequence and that of its 3SA and 4SA mutants was subcloned into pSP64TEN. Smad1(3.4SA) was generated from Smad1(3SA) by replacement of the linker region of Smad1(4SA). Smad2 and Smad2(EPSM) were subcloned into the pCS2+ vector [24]. Each vector was linearized with the appropriate restriction enzyme and used for in vitro transcription using a MEGAscript kit according to manufacturer's instructions (Ambion, Austin, TX). Synthetic RNAs were quantitated via ethidium bromide staining by comparison to a standard RNA control (Invitrogen, Carlsbad, CA).
Cloning of PV.1A genomic DNA
PV.1A genomic DNA (gDNA) was isolated by screening a Xenopus muscle gDNA library (Clontech) with a PCR-amplified cDNA probe corresponding to the segment from +48 to +215 bp of Xenopus PV.1A cDNA. PCR primers were (upstream) 5′-CCTTCAGCATGGTTCAACAG-3′ and (downstream) 5′-CATCCTTTCTCCTTGGCATCTCCT-3′. Approximately 7.0×106 plaque forming units (pfu) were screened using an ECL system in accordance with manufacturer's instructions (GE Healthcare Biosciences, Pittsburgh, PA) to identify putative positive clones, which were subsequently isolated and analyzed by restriction mapping and Southern blotting. A 3.8 kb DNA fragment in a positive clone was subcloned into the pBluescript SK(-) plasmid (Stratagene, Cedar Creek, TX). Both strands of DNA were sequenced by the Sanger method to confirm the identity of the clone.
PV.1A promoter constructs
A 3.8 kb fragment from a positive clone containing 2.5 kb of 5′-flanking region was subcloned into the pGL-2 basic plasmid (Promega, Madison, WI); the clone was designated the -2525 construct. Serially deleted PV.1A promoter mutants were made from this -2525 construct by PCR amplification (Table 1). PCR conditions were as follows: 1 minute at 94°C, 1 minute at 54°C, and 1 minute at 72°C for 30 cycles. PCR amplification products were digested with XhoI/HindIII and inserted into similarly digested pGL-2 basic plasmid. A triple-repeat BMP-4-response element (BRE) was generated by annealing two complementary oligos:
Table 1. Primers used for serially deleted reporter gene constructs.
| Primer name | Sequences (5′ → 3′) | |
| Upstream primers | -2525 | AGTCCTCGAGTACCTGCAACTTACTCGC |
| -399 | AGTCCTCGAGCCAGTCTCCTGGTGTGACTT | |
| -374 | AGTCCTCGAGCCAACATAAAAGGATAAAGG | |
| -351 | AGTCCTCGAGAGAGGTTGTTCTTATTGGTG | |
| -330 | AGTCCTCGAGGCTCAATAACAACATCAAGG | |
| -300 | AGTCCTCGAGAACCTACATTATCTCTTTCC | |
| -262 | AGTCCTCGAGTCTCTGCTGTCTGTCCATGGGA | |
| -240 | AGTCCTCGAGTTCTGTGCCGGCCAATGCTAAT | |
| -204 | AGTCCTCGAGCCTCCAATATCACAAGGTGAA | |
| -180 | AGTCCTCGAGACTAACCTGACAGACTCACTGG | |
| -162 | AGTCCTCGAGACTGGAGCCAGGACCAGG | |
| -136 | AGTCCTCGAGCTACAAGTGAGAACATAA | |
| -103 | AGTCCTCGAGTAGCCCATTCTGATAGCC | |
| -180 MT | AGTCCTCGAGACTAACCTGACCAACTCACTGG | |
| ORE(M) -180 | AGTCCTCGAGTAACCTGACAGACTCACTAAAGCCAGGAC | |
| Downstream primer | AGTCAAGCTTGATGGAGCCGCTGGAGTTGTG |
5′-ACTAACCTGACAGACTCACTAACCTGACAGACTCACTAACCTGACAGACTC-3′ and 5′-GAGTCTGTCAGGTTAGTGAGTCTGTCAGGTTAGTGAGTCTGTCAGGTTAGT-3′ before subcloning into the pGL-2 basic plasmid.
Embryo injection and explant culture
Xenopus embryos were obtained by in vitro fertilization after induction of female frogs with 500 units of human chorionic gonadotropin (Sigma, St. Louis, MO). RNAs were injected into the animal pole of 2-cell stage embryos; animal caps were dissected from injected embryos at stage 8.5 and incubated to stage 11 in 0.5X modified Barth's saline [0.5X MBS: 44 mM NaCl, 0.5 mM KCl, 0.35 mM CaCl2, 0.5 mM MgCl2, 2.5 mM HEPES (pH 7.8), 1.25 mM NaHCO3] for RT-PCR, measurement of luciferase activity, and electrophoretic mobility shift assays (EMSAs).
RNA isolation and RT-PCR
Bmp-4 mRNA (0.5 ng) was injected into the animal pole of 2-cell stage Xenopus embryos. At stage 7.5, embryos were treated with 25 µg/mL cycloheximide (CHX) in 0.5X MBS and maintained until control untreated embryos reached stage 8.5. Animal caps were then dissected from the injected embryos and incubated until stage 11 in 0.5X MBS containing 25 µg/mL CHX. Total RNA was isolated from whole embryos or animal caps using TRIzol reagent following the manufacturer's instructions (Invitrogen, Carlsbad, CA) and treated with DNase I to remove gDNA contamination. RT-PCR was performed with Superscript II (Invitrogen, Carlsbad, CA), as described by the manufacturer, with 2 µg total RNA per reaction. PCR was performed according to the following conditions: 1 minute at 94°C, 1 minute at each annealing temperature, 1 minute at 72°C; 20-28 cycles of amplification (Table 2).
Table 2. Primers used for PCR amplification.
| Gene Name | Sequences (5′ → 3′) | Annealing Temp. (°C) | Cycles |
| EF-1α | U: CAGATTGGTGCTGGATATGCD: ACTGCCTTGATGACTCCTAG | 56 | 20 |
| Xvent-2 | U: AACGGGAAATCCAAGATGGCD: TTTTGTTTGTCCTGCGGGAG | 57 | 23 |
| Xvent-1 | U: TTCCCTTCAGCATGGTTCAACD: GCATCTCCTTGGCATATTTGG | 57 | 25 |
| PV.1A | U: CCTTCAGCATGGTTCAACAGD: CATCCTTCTTCCTTGGCATCTCCT | 60 | 27 |
| Goosecoid | U: ACAACTGGAAGCACTGGAD: TCTTATTCCAGAGGAACC | 57 | 28 |
Luciferase assays
Levels of luciferase reporter activity were measured by the luciferase assay system according to manufacturer's instructions (Promega, Madison, WI). Five different groups of animal caps (3 to 5 animal caps per group) were harvested and homogenized in 10 µL lysis buffer per animal cap. 10 µL animal cap homogenate were assayed with 50 µL luciferase substrate and activity determined by luminometer (EG & G Berthold, Bad Wildbad, Germany). All experiments were repeated at least three times using independently derived sample sets.
In vitro translation and electrophoretic mobility shift assays
cDNAs encoding Smads 1, 3, and 4 were PCR-amplified and used as templates for in vitro translation. In vitro translated proteins were prepared using the TNT Quick Coupled Transcription/translation System according to manufacturer's instructions (Promega, Madison, WI). Promoter fragments or 3x-repeat oligonucleotides were amplified by annealing two complementary oligos, respectively, and labeled with α-32P-ATP using T4 polynucleotide kinase (Promega, Madison, WI). Labeled DNA probes were incubated with 10 µg animal cap protein extract or 1 µg in vitro translated protein at room temperature for 20 minutes in 10 µL binding buffer (2.5% glycerol, 5 mM MgCl2, 50 ng/µL Poly [dI·dC], and 0.05% NP-40). DNA/protein complexes were separated on 4% polyacrylamide gels. Competition experiments were performed using a 100-fold excess of unlabeled DNA probe.
Site-directed mutagenesis
Mutagenesis was performed by an overlap extension PCR method using the several oligonucleotides in accordance with instructions (Table 3).
Table 3. Primers used for mutant gene constructs.
| Mutated site | Name | Primer name | Sequences (5′ → 3′) |
| XRE | XRE(M) | M-1 | ATGCTTACGTTCCTTAGCCCATTC |
| M-2 | GAATGGGCTAAGGAACGTAAGCAT | ||
| -180 | AGTCCTCGAGACTAACCTGACAGACTCACTGG | ||
| Rev. | ACATCACTGTTCCAGGAAGGCAGG |
Nucleotide sequence accession number
The PV.1A (accession number; AF133122) cDNA sequence has been submitted to GenBank.
Statistical analysis
The data are presented as mean and standard deviation of measurements from at least three separate and independent experiments. Differences were considered significant at P<0.05.
Results
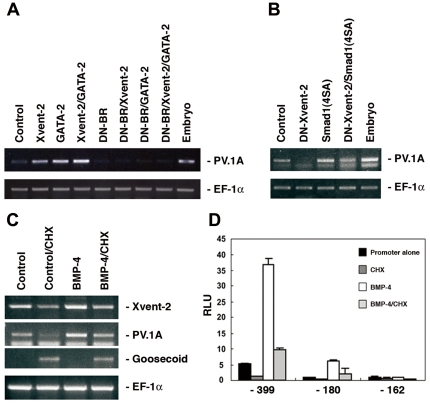
Concomitant overexpression of Xvent-2 and GATA-2 is not sufficient for complete activation of PV.1A transcription in the absence of direct BMP-4 signaling
Previous work posits that, unlike Xvent-2, Xvent-1 subfamily members are not under the direct transcriptional control of BMP-4 [21]. Moreover, expression of Xvent-2 and GATA-2 is sufficient for transcription, in the presence of cycloheximide (CHX), of Xvent-1 on the ventral side of the embryo [15]. However, in the absence of BMP-4, we observed abolition of PV.1A transcription despite continuing transcription of Xvent-2 and GATA-2, indicating that transcription of Xvent-1 subfamily members relies to a greater degree on BMP-4 signaling than that of its direct targets Xvent-2 and GATA-2. Furthermore, the increases in Xvent-2 and GATA-2 transcription were less sensitive to BMP-4 overexpression than that of PV.1A, in contrast to our expectation (data not shown).
We examined the necessity of BMP-4 signaling in the regulation of PV.1A transcription, finding that BMP-4 contributes both directly and indirectly to the process (Fig. 1A). We first assessed the capacity of Xvent-2 and GATA-2 to rescue the dramatic repression of PV.1A affected by dominant-negative Type I BMP receptor (DN-BR) in animal cap explants (Fig. 1A); co-injection of mRNAs encoding these factors could not reverse this repression. This suggests that although PV.1A transcription is augmented by Xvent-2 and GATA-2, its transcriptional activation is in fact dependent on functional BMP signaling. Conversely, the reduction in PV.1A transcription occurring in the presence of dominant-negative Xvent-2 (DN-Xvent-2) was mitigated by co-injection of a constitutively active Smad1 mutant (Smad1(4SA), with four serine residues altered to alanine at the inhibitory phosphorylation sites in its linker region; Fig. 1B).
Figure 1. PV.1A is a direct target of BMP-4 signaling.
(A, B) Animal caps injected with the indicated RNAs (0.5 ng/embryo) were dissected from stage 8.5 embryos and incubated until stage 11. Total RNA was isolated for RT-PCR and assayed to evaluate PV.1A expression. (C, D) Xenopus embryo animal poles were injected at the 2-cell stage with the specified promoter constructs (20 pg/embryo) in the presence or absence of BMP-4 (0.5 ng/embryo). At stage 7.5, embryos were treated with 25 µg/mL CHX in 0.5X MBS until control embryos reached stage 8.5. Animal caps were then dissected from the injected embryos and incubated to stage 11 in 0.5X MBS for RT-PCR analysis (C) or measurement of luciferase activity (D). Luciferase activity was measured as described in Materials and Methods. Gene expression was normalized to EF-1α transcription. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
We next wished to examine the effects of BMP-4 signaling on PV.1A expression in animal cap explants under CHX treatment. As shown in Figure 1C, prior to CHX addition, the synthesis of BMP-4 led to activation of Xvent-2 and PV.1A transcription (Fig. 1C). As a control, transcriptional induction of Goosecoid was examined to confirm that CHX could block protein synthesis. Notably, Goosecoid was expressed even in the presence of BMP-4 in CHX-treated animal cap explants; its expression is known to be negatively regulated by repressor protein(s) induced by BMP signaling [25]. Further, in reporter assays using various deletion constructs of the PV.1A promoter, 2 of 3 reporter activities were positively affected by BMP-4 in CHX-treated animal cap explants (Fig. 1D). The results indicate that PV.1A is in part a direct target of BMP-4 signaling.
Isolation of the PV.1A 5′-flanking region
The injection of Bmp-4 mRNAs enhances embryonic expression of Xvent family members [8], [9], [10]. To analyze the mechanics of PV.1A transcription in regard to BMP-4 regulation, we cloned and characterized its genomic locus. A 3.8-kb fragment containing the PV.1A 5′-flanking region and a part of its coding region was cloned from a gDNA library (Genbank accession number; AF133122). The fragment contained the promoter and sequence upstream to -2525 bp, including a TATA-like element proximal to the transcriptional start-site (TATAA). The proximal sequence of our isolated PV.1A promoter was nearly identical to that of Xvent-1B, and differed from those of Xbr-1a/Xvent-2 and Xvent-2B; additionally, it contained putative BMP-4 and Oaz-response elements (BREs and OREs, respectively) at positions similar to the sites found in the Xbr-1a/Xvent-2 and Xvent-2B promoters (Fig. S1A).
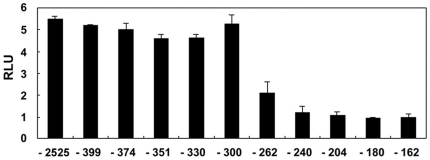
Identification of a positive-regulatory element in the 5′-flanking region of PV.1A
Our detection of putative BREs and OREs, believed to be under direct regulation by the BMP-4 signaling pathway, in the PV.1A 5′ genomic flanking region led us to probe the nature of the cis-acting elements that can evoke PV.1A transcription. We created serial deletions of 5′-flanking sequences (Fig. S1B), and compared promoter activities among these mutants after the injection of equimolar concentrations of each reporter construct. Promoter activities were measured via luciferase assay in stage 11 animal caps. Deletion of PV.1A from 2525 bp to 300 bp (-300 construct) upstream of the major transcriptional initiation site did not alter promoter activity. However, the -262 construct had only half the capacity for transcriptional induction of the -300 construct (Fig. 2), indicating that the region between −300 and −262 bp contains an element that positively influences PV.1A promoter activity.
Figure 2. Identification of a positive-regulatory element in the 5′-flanking region of PV.1A.
A putative element positively regulating PV.1A transcription was identified between −300 and −262 bp from the transcriptional start site. 2-cell stage embryos were injected at their animal poles with 20 pg of serially deleted PV.1A promoter constructs driving luciferase reporter transcription. Animal caps were dissected at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity as described in Materials and Methods. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
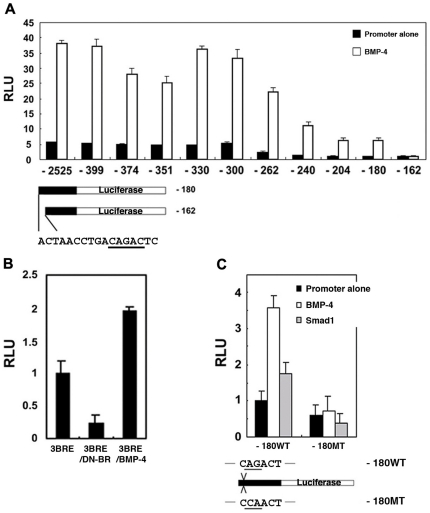
Identification of a BMP-4 response element in the 5′-flanking region of PV.1A
To identify and analyze putative BREs in the 5′-flanking region of PV.1A, we first quantitated the promoter activities of our serial deletion mutants in the presence of BMP-4, finding that ten deletion constructs retaining at least 180 bp of sequence 5′ of the major transcription initiation site responded to BMP-4 signaling, while the activity of the -162 construct was completely abolished (Fig. 3A).
Figure 3. Identification of a BMP-4-response element in the 5′-flanking region of PV.1A.
(A) Serially deleted promoter constructs were injected (20 pg/embryo) with or without BMP-4 (0.5 ng/embryo) into 2-cell stage embryos. Animal caps were dissected from injected embryos at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity. A putative BRE was detected between −180 and −162 bp from the transcription start site based on reporter gene expression. The underlined sequence (CAGA) is a consensus binding-site for Smad proteins. Luciferase activity was measured as described in Materials and Methods. (B) 3BRE is a luciferase fusion construct with a triple BRE repeat. The 3BRE construct (20 pg/embryo) was co-injected with DN-BR or BMP-4 (each 0.5 ng/embryo) into 2-cell stage embryos. Animal caps were dissected at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity as described in Materials and Methods. (C) The -180 WT and -180 MT constructs (20 pg/embryo) were injected with or without BMP-4 or Smad1 (each 0.5 ng/embryo) into 2-cell stage embryos. -180 MT indicates a construct with mutated BRE. Luciferase activity was measured as described in Materials and Methods. The sequences underlined indicate alterations in the original sequences. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
Interestingly, CAGA sequences, which comprise a portion of the BRE, can be found within the region between 180 and 162 bp upstream of the PV.1A transcription start site. We therefore further analyzed this putative BRE by site-directed mutagenesis of our -180 construct, as well as through a synthetic threefold BRE tandem repeat construct (3BRE). The 3BRE construct responded positively to BMP-4 and negatively to DN-BR (Fig. 3B); when CAGA was mutated to CCAA in the core BRE region of the -180 construct, response to BMP-4 and Smad1 was abolished as well (Fig. 3C). Furthermore, in electrophoretic mobility shift assays (EMSAs) we found a direct interaction of the 3BRE construct with Smad1 and Smad4 (Fig. S2A). Interestingly, Smad3 also bound to the BRE (Fig. S2B), indicating that this response element alone is insufficient to precisely specify Smad binding, which instead appears to necessitate other cis-acting elements.
We examined promoter fragments of differing length in extracts of uninjected animal cap explants by competition assay, using competitor probes to determine whether endogenous proteins exhibit specific interactions with core Smad-binding sequences. The appearance of bands in all labeled probes was dramatically diminished by competition with unlabeled wild-type -180 probe, but not wild-type -160 or -180 probe harboring a mutation in the CAGA region (Fig. S2C). These data suggest that PV.1A is directly regulated by BMP-4 signaling through binding of Smad1 and Smad4 to the BRE within its promoter.
Binding of Smad mutants to the 5′-flanking region of PV.1A
We examined the association of the BMP-4 signaling molecules Smad1 and Smad4 with the minimal PV.1A promoter (180 bp upstream of the transcriptional start site) in response in BMP-4 stimulation, using the wild-type (-180 WT) and mutant (-180 MT) constructs. Expression of wild-type Smad1, Smad4, and Smad1(4SA) significantly increased PV.1A promoter activity in the -180 WT construct. In addition, co-injection of Smad1 and Smad4 further increased promoter activity of the -180 WT construct compared with either mRNA alone (Fig. S3A). Dominant-negative Smad1 (Smad1(3SA), with the three serine residues in its C-terminal SSVS mutated to alanines); Smad1(3.4SA), generated from Smad1(3SA) by replacing the linker region with that of Smad1(4SA); and mutant Smad2 isoforms did not alter transcriptional activation of this minimal promoter (Fig. S3A, B). Furthermore, expression of the -180 MT construct, in which the CAGA sequences in BRE were altered to CCAA, was unaffected by all Smads (Fig. S3A, B). These results demonstrate that the CAGA sequences of the BRE in the minimal PV.1A promoter are critical and essential for the response to BMP-4 and that the -180 region contains the specific response region for BMP signaling mediated through Smad effectors.
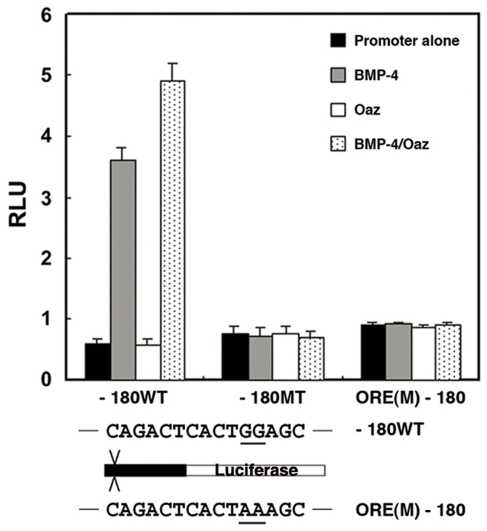
Oaz cooperates with BMP-4 signaling in the 5′-flanking region of PV.1A
A putative ORE lies in similar regions in the PV.1A and Xvent-2 promoters (Fig. S1A). We therefore examined possible cooperativity of Oaz, a known transcriptional co-activator of Xvent-2 family members [16], [22], with BMP-4. As expected, we observed additive reporter activity in animal cap explants on co-injecting Oaz and Bmp-4 with the -180 promoter construct (Fig. 4). We then examined whether the putative ORE element could mediate this additive transcriptional induction. When the putative ORE sequence, TGGAGC, was mutated to TAAAGC in the -180 promoter construct, BMP-4 and Oaz cooperativity were completely abolished in luciferase reporter assays (Fig. 4). Notably, the response to BMP-4 and Oaz was also diminished when the BRE or ORE in the -180 promoter construct was mutated (Fig. 4). These data indicate that the BRE and ORE in the PV.1A promoter are both necessary for the appropriate transcriptional response to BMP-4 signaling.
Figure 4. Oaz and BMP-4 cooperate to regulate PV.1A transcription.
-180, -180 MT, and ORE(M)-180 constructs (20 pg/embryo) were injected with or without BMP-4 (0.5 ng/embryo) and Oaz (0.1 ng/embryo) into the animal poles of 2-cell stage embryos. Animal caps were dissected from injected embryos at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity as described in Materials and Methods. The sequences underlined depict alterations to the original sequences. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
Xvent-2 enhances PV.1A promoter activity
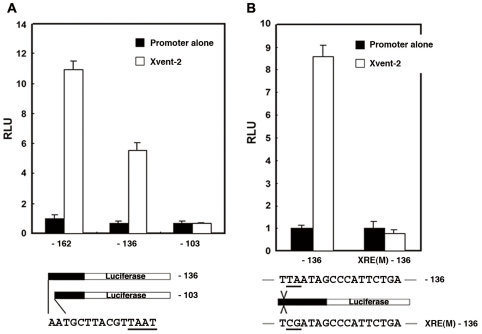
Xvent-1, which, unlike Xvent-2, is not under direct control of BMP-4 signaling, can be transcriptionally up-regulated by Xvent-2 [21]. To identify the Xvent-2 response element (XRE) in the PV.1A promoter, we co-injected constructs containing serial deletion mutants of the PV.1A promoter driving reporter-gene expression into 2-cell stage embryos with Xvent-2 mRNA. We found that the Xvent-2 response region in the PV.1A promoter differed from the BRE (Fig. 5A): Xvent-2 mediated transcriptional induction of the -136 construct was abrogated in the -103 construct. Notably, the region between -136 and -103 bp contained a TAAT homeobox-protein binding motif, and reporter activity of the -136 construct increased approximately 10-fold in the presence of Xvent-2 (Fig. 5A).
Figure 5. Positive regulation by Xvent-2 of the 5′-flanking region of PV.1A.
(A) Serial-deletion mutant constructs (−162, −136, and −103; 20 pg/embryo) of the PV.1A promoter were co-injected with Xvent-2 (0.5 ng/embryo) into the animal poles of 2-cell stage embryos. Animal caps were dissected from injected embryos at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity (as described in Materials and Methods). A putative Xvent-2 binding site was detected between −136 and −103 bp from the major transcription initiation site. The underlined sequence, TAAT, is a consensus binding-site for homeobox proteins. (B) The −136 and XRE(M)-136 constructs were injected (20 pg/embryo) with or without Xvent-2 (0.5 ng/embryo) at the animal poles of 2-cell stage embryos. Luciferase activity was measured as described in Materials and Methods. The sequences underlined indicate alterations to original sequences. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
To determine whether this TAAT motif constituted the core XRE critical for Xvent-2 mediated PV.1A expression, we co-injected point mutant constructs [XRE(M) -136] into 2-cell stage embryos with Xvent-2 mRNA. In the XRE(M) -136 construct-injected embryos, promoter activity was not induced by Xvent-2, while the WT -136 construct retained activity (Fig. 5B). These results indicate that the XRE lies between -136 and -103 bp of the PV.1A promoter and that the TAAT sequence is the core cis-element for Xvent-2 binding and transcriptional transctivation.
Dorsal-specific transcription factors decrease PV.1A promoter activity
To identify negative trans-acting elements within the PV.1A promoter, mRNAs encoding the dorsal- and neural-specific transcription factors Goosecoid and AP-1 were co-injected with the -2525 PV.1A promoter construct into 2-cell stage embryos. Both Goosecoid and AP-1 significantly down-regulated PV.1A promoter activity (Fig. S4). These results suggest that dorsal- and neural-specific transcription factors (particularly Goosecoid) are involved in repression of the PV.1A promoter in the dorsal mesoderm and neural ectoderm. In addition, the downregulation of PV.1A promoter activity by AP-1 implies the existence of uncharacterized cross-talk among several signaling pathways, including BMP-4, FGF, and activin.
Discussion
Here, we examined the role of BMP-4 signaling on transcriptional regulation of PV.1A, a BMP-4 target gene encoding a transcription factor instrumental in the formation of ventral mesoderm during early Xenopus development. While BMP-4 was not previously thought to directly impact the expression of Xvent-1 family members, we find that BMP-4 signaling plays a substantial role in modulation of PV.1A expression and that Xvent-2, in contrast, occupies a subsidiary role.
We first investigated whether PV.1A was under direct regulation by BMP-4. Our findings indicated PV.1A transcription in the presence of CHX and BMP-4, suggestive of a direct contribution of BMP-4 signaling to gene induction. Additionally, co-injection of Xvent-2 into embryos with DN-BR mediated abrogation of BMP-4 signaling could not rescue reduced PV.1A expression, suggesting that Xvent-2 alone is insufficient for full PV.1A expression. Further, the decrease in PV.1A expression caused by DN Xvent-2 was mitigated by constitutively active Smad1, suggesting that basal PV.1A expression does not require Xvent-2, and PV.1A expression indeed appeared much more sensitive to BMP-4 signaling than to Xvent-2. In a previous study, we established slight induction of Xvent-2 expression (∼2-fold) by BMP-4 signaling [16], in contrast to our current demonstration of BMP-4's dramatic effect on PV.1A expression (≥7-fold induction).
We reasoned that analysis of the PV.1A promoter would elucidate the mechanics of its combinatorial regulation. We isolated the PV.1A genomic locus, including the 5′-flanking region (−2525 bp), and generated serial promoter deletions to investigate their several contributions to gene induction. The region 180 bp upstream of the major transcriptional initiation site had full reporter activity in response to BMP-4 signals; and within this region, putative binding sites for Smad family members and Oaz were necessary and sufficient to mediate the effects of BMP-4 signaling. Additionally, we detected a cooperative interaction between these factors in promoting PV.1A transcription. Notably, the BRE in the PV.1A promoter was insufficient for Smad specificity, which was achieved only in the context of an ORE proximal to a BRE.
Using serial-deletion and point-mutation containing promoter fragments to drive reporter-gene expression, we determined that the PV.1A promoter contained a positive-regulatory cis-element and an XRE within 399 bp upstream of the major transcriptional initiation site: this latter element merely potentiated BMP-4 signaling for full transcriptional activation of PV.1A. We also investigated the negative regulation of the PV.1A promoter via co-injection of the dorsal-specific transcription factors AP-1 and Goosecoid, determining that each can repress PV.1A transcription. Our results suggest that Smads, Oaz, and Xvent-2 stimulate PV.1A expression and that the PV.1A promoter harbors the unique characteristic of possessing multiple cis-acting response elements for its regulation, including direct, indirect, co-activator, and negative response elements, to regulate the early embryonic development of Xenopus laevis.
A comparison of PV.1A 5′ sequence to promoters of other Xvent family members, including Xbr-1a/Xvent-2 and Xvent-2B, found minor conservation of regulatory motifs, principally in the BRE and ORE, while PV.1A and Xvent-1 share considerable sequence identity upstream of their transcriptional start sites (Fig. 2A). Xvent-1, however, is an indirect target of BMP-4 signaling [21]. Xvent-2 is not competent to rescue PV.1A expression in the absence of BMP-4 signal transduction; and the induction of each depends on BMP-4, suggesting that although the PV.1A and Xvent-1B promoters are nearly identical and even they belong to same family, their expression is differentially regulated. For instance, other regulatory elements, such as introns, may influence their transcription.
The zinc-finger protein Oaz is a DNA-binding co-factor that associates directly with the MH2 domain of Smad1 in vitro [22]. A complex of Smad1, Smad4, and Oaz binds to a BRE in the Xvent-2 promoter, to enhance Xvent-2 transcription upon BMP-4 stimulation of cultured cells [22]. We likewise find this requirement for Oaz in induction of the PV.1A promoter. Our data demonstrate a critical role for the BRE in the PV.1A BMP-4 response, as transcriptional activation is quelled by its mutation; the ORE imparts Smad specificity. 3BRE sequence bound Smad1, Smad4, and Smad3, thereby implying that the BRE is insufficient to confer Smad specificity, which might require other cis-acting elements. Although we did not examine their interaction, Oaz and Smads interact directly [22]. The ORE might mediate PV.1A transcription through interactions between Oaz, Smad1, and Smad4 at the BRE.
As shown in Fig. 1A, Xvent-2 failed to rescue the DN-BR induced reduction in PV.1A transcripts, demonstrating that PV.1A is a direct target of BMP-4 signaling and that its expression requires BMP-4 signaling. Xvent-2 has dual activities during early embryonic development. In the dorsal embryo, Xvent-2 is a repressor, but on the ventral side, an activator. We explored the function of Xvent-2 on in the ventral embryo and identified a putative XRE in the PV.1A promoter. Recently, the Xvent-1B promoter was shown to be activated by cooperation between Xvent-2 and GATA-2 [15], and several putative binding elements were identified; however, a specific cis-acting element for Xvent-2 was not identified in the Xvent-1B promoter. We identified a specific XRE between −136 and −103 bp in the PV.1A promoter, but this motif is not sufficient for complete transcriptional activation of PV.1A and may function primarily as an enhancer of BMP-4 signaling.
In the developing Xenopus embryo, ventral-specific genes are repressed within dorsal or organizer tissues. This repression may be mediated though several mechanisms. First, activin-like signaling on the dorsal side induces the expression of BMP-4 antagonists such as Chordin, Noggin, and Follistatin. These antagonists are secreted extracellularly and bind to BMP-4, preventing it from accessing its receptor. In the absence of BMP-4 signaling, its downstream target genes cannot be transcribed.
Alternatively, ventral genes are repressed dorsally by dorsal-specific proteins such as Goosecoid and AP-1 through binding of dorsal-specific repressors to consensus sites in promoters of ventral-specific genes. PV.1A, but not Xvent-2, is repressed by Goosecoid (data not shown). We are currently biochemically investigating the mechanisms by which PV.1A is repressed within dorsal and organizer tissues using the PV.1A promoter to identify negative cis-acting elements that are bound and regulated by dorsal-specific proteins.
In this study, we investigated the transcriptional regulation of PV.1A and identified cis-acting elements, including BRE, ORE, and XRE, in its promoter sequences. Additionally, we found that the dorsal-specific transcription factors Goosecoid and AP-1 exert negative effects on PV.1A reporter activity through BMP-4 signaling. Our results indicate that the complex transcriptional control exerted over downstream effectors of BMP-4 occurs through interplay between positive and negative regulators of transcription.
Supporting Information
Bioinformatics analysis of the 5′-flanking region of PV.1A. (A) Comparison among the 5′-flanking sequences of PV.1A, Xbr-1a/Xvent-2, and Xvent-2B indicates a high level of identity within BMP-4 response elements. BRE, ORE, and XRE indicate BMP-4, Oaz-, and Xvent-2 response elements, respectively. (B) The schematic diagram illustrates serial-deletion promoter constructs. Serially deleted and site-directed mutant constructs were subcloned into the pGL2-basic plasmid. Arrowheads indicate positions of site-directed mutagenesis.
(JPG)
BRE binding assays. The BMP-4 response element was confirmed by EMSA using probes against triple-repeat BRE (A, B) or the TATA-box region of the promoter (C, D). The protein extracts indicated were obtained from in vitro translated proteins (A, B) or uninjected animal caps (C, D). Unlabeled competitor probe was added at 100-fold excess (C) or the concentration indicated (D). Asterisks indicate specific bands.
(JPG)
Smad binding within the 5′-flanking region of PV.1A. (A, B) The -180 WT construct (20 pg/embryo) were co-injected with the RNAs indicated (0.5 ng/embryo) into 2-cell stage embryos. Animal caps were dissected from injected embryos at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity as described in Materials and Methods. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
(JPG)
Repressive effects of dorsal-specific protein binding to the 5′-flanking region of PV.1A. The -2525 PV.1A promoter construct (20 pg/embryo) was co-injected with BMP-4 (0.5 ng/embryo) and Goosecoid (0.1 ng/embryo) or AP-1 (0.5 ng/embryo) into 2-cell stage embryos. Animal caps were dissected from injected embryos at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity as described in Materials and Methods. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
(JPG)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Basic Science Research Program (KRF-2005-C00115 and KRF-2005-015-C00390) and by the Priority Research Centers Program (2010-0029642) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Piccolo S, Sasai Y, Lu B, De Robertis EM. Dorsoventral patterning in Xenopus: inhibition of ventral signals by direct binding of chordin to BMP-4. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 3.Fainsod A, Deissler K, Yelin R, Marom K, Epstein M, et al. The dorsalizing and neural inducing gene follistatin is an antagonist of BMP-4. Mech Dev. 1997;63:39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- 4.Harland RM. The transforming growth factor beta family and induction of the vertebrate mesoderm: bone morphogenetic proteins are ventral inducers. Proc Natl Acad Sci U S A. 1994;91:10243–10246. doi: 10.1073/pnas.91.22.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemmati-Brivanlou A, Melton D. Vertebrate neural induction. Annu Rev Neurosci. 1997;20:43–60. doi: 10.1146/annurev.neuro.20.1.43. [DOI] [PubMed] [Google Scholar]
- 6.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 7.Savage C, Das P, Finelli AL, Townsend SR, Sun CY, et al. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc Natl Acad Sci U S A. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. Antagonizing the Spemann organizer: role of the homeobox gene Xvent-1. Embo J. 1995;14:6268–6279. doi: 10.1002/j.1460-2075.1995.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ault KT, Dirksen ML, Jamrich M. A novel homeobox gene PV.1 mediates induction of ventral mesoderm in Xenopus embryos. Proc Natl Acad Sci U S A. 1996;93:6415–6420. doi: 10.1073/pnas.93.13.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, et al. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- 11.Maeda R, Kobayashi A, Sekine R, Lin JJ, Kung H, et al. Xmsx-1 modifies mesodermal tissue pattern along dorsoventral axis in Xenopus laevis embryo. Development. 1997;124:2553–2560. doi: 10.1242/dev.124.13.2553. [DOI] [PubMed] [Google Scholar]
- 12.Schuler-Metz A, Knochel S, Kaufmann E, Knochel W. The homeodomain transcription factor Xvent-2 mediates autocatalytic regulation of BMP-4 expression in Xenopus embryos. J Biol Chem. 2000;275:34365–34374. doi: 10.1074/jbc.M003915200. [DOI] [PubMed] [Google Scholar]
- 13.Melby AE, Clements WK, Kimelman D. Regulation of dorsal gene expression in Xenopus by the ventralizing homeodomain gene Vox. Dev Biol. 1999;211:293–305. doi: 10.1006/dbio.1999.9296. [DOI] [PubMed] [Google Scholar]
- 14.Trindade M, Tada M, Smith JC. DNA-binding specificity and embryological function of Xom (Xvent-2). Dev Biol. 1999;216:442–456. doi: 10.1006/dbio.1999.9507. [DOI] [PubMed] [Google Scholar]
- 15.Friedle H, Knochel W. Cooperative interaction of Xvent-2 and GATA-2 in the activation of the ventral homeobox gene Xvent-1B. J Biol Chem. 2002;277:23872–23881. doi: 10.1074/jbc.M201831200. [DOI] [PubMed] [Google Scholar]
- 16.Lee HS, Park MJ, Lee SY, Hwang YS, Lee H, et al. Transcriptional regulation of Xbr-1a/Xvent-2 homeobox gene: analysis of its promoter region. Biochem Biophys Res Commun. 2002;298:815–823. doi: 10.1016/s0006-291x(02)02570-6. [DOI] [PubMed] [Google Scholar]
- 17.Onichtchouk D, Glinka A, Niehrs C. Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development. 1998;125:1447–1456. doi: 10.1242/dev.125.8.1447. [DOI] [PubMed] [Google Scholar]
- 18.Papalopulu N, Kintner C. A Xenopus gene, Xbr-1, defines a novel class of homeobox genes and is expressed in the dorsal ciliary margin of the eye. Dev Biol. 1996;174:104–114. doi: 10.1006/dbio.1996.0055. [DOI] [PubMed] [Google Scholar]
- 19.Ladher R, Mohun TJ, Smith JC, Snape AM. Xom: a Xenopus homeobox gene that mediates the early effects of BMP-4. Development. 1996;122:2385–2394. doi: 10.1242/dev.122.8.2385. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt JE, von Dassow G, Kimelman D. Regulation of dorsal-ventral patterning: the ventralizing effects of the novel Xenopus homeobox gene Vox. Development. 1996;122:1711–1721. doi: 10.1242/dev.122.6.1711. [DOI] [PubMed] [Google Scholar]
- 21.Rastegar S, Friedle H, Frommer G, Knochel W. Transcriptional regulation of Xvent homeobox genes. Mech Dev. 1999;81:139–149. doi: 10.1016/s0925-4773(98)00239-1. [DOI] [PubMed] [Google Scholar]
- 22.Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, et al. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 23.Henningfeld KA, Friedle H, Rastegar S, Knochel W. Autoregulation of Xvent-2B; direct interaction and functional cooperation of Xvent-2 and Smad1. J Biol Chem. 2002;277:2097–2103. doi: 10.1074/jbc.M108524200. [DOI] [PubMed] [Google Scholar]
- 24.Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu CC, Yamada G, Blum M. Retinoic acid teratogenicity: the role of goosecoid and BMP-4. Cell Mol Biol (Noisy-le-grand) 1999;45:617–629. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bioinformatics analysis of the 5′-flanking region of PV.1A. (A) Comparison among the 5′-flanking sequences of PV.1A, Xbr-1a/Xvent-2, and Xvent-2B indicates a high level of identity within BMP-4 response elements. BRE, ORE, and XRE indicate BMP-4, Oaz-, and Xvent-2 response elements, respectively. (B) The schematic diagram illustrates serial-deletion promoter constructs. Serially deleted and site-directed mutant constructs were subcloned into the pGL2-basic plasmid. Arrowheads indicate positions of site-directed mutagenesis.
(JPG)
BRE binding assays. The BMP-4 response element was confirmed by EMSA using probes against triple-repeat BRE (A, B) or the TATA-box region of the promoter (C, D). The protein extracts indicated were obtained from in vitro translated proteins (A, B) or uninjected animal caps (C, D). Unlabeled competitor probe was added at 100-fold excess (C) or the concentration indicated (D). Asterisks indicate specific bands.
(JPG)
Smad binding within the 5′-flanking region of PV.1A. (A, B) The -180 WT construct (20 pg/embryo) were co-injected with the RNAs indicated (0.5 ng/embryo) into 2-cell stage embryos. Animal caps were dissected from injected embryos at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity as described in Materials and Methods. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
(JPG)
Repressive effects of dorsal-specific protein binding to the 5′-flanking region of PV.1A. The -2525 PV.1A promoter construct (20 pg/embryo) was co-injected with BMP-4 (0.5 ng/embryo) and Goosecoid (0.1 ng/embryo) or AP-1 (0.5 ng/embryo) into 2-cell stage embryos. Animal caps were dissected from injected embryos at stage 8.5 and incubated until stage 11 in 0.5X MBS for measurement of luciferase activity as described in Materials and Methods. Experiments were repeated three times using independent sample sets. Data are shown as mean ± SD.
(JPG)