Abstract
Epithelial cells of the mammalian intestine are covered with a mucus layer that prevents direct contact with intestinal microbes but also constitutes a substrate for mucus-degrading bacteria. To study the effect of mucus degradation on the host response, germ-free mice were colonized with Akkermansia muciniphila. This anaerobic bacterium belonging to the Verrucomicrobia is specialized in the degradation of mucin, the glycoprotein present in mucus, and found in high numbers in the intestinal tract of human and other mammalian species. Efficient colonization of A. muciniphila was observed with highest numbers in the cecum, where most mucin is produced. In contrast, following colonization by Lactobacillus plantarum, a facultative anaerobe belonging to the Firmicutes that ferments carbohydrates, similar cell-numbers were found at all intestinal sites. Whereas A. muciniphila was located closely associated with the intestinal cells, L. plantarum was exclusively found in the lumen. The global transcriptional host response was determined in intestinal biopsies and revealed a consistent, site-specific, and unique modulation of about 750 genes in mice colonized by A. muciniphila and over 1500 genes after colonization by L. plantarum. Pathway reconstructions showed that colonization by A. muciniphila altered mucosal gene expression profiles toward increased expression of genes involved in immune responses and cell fate determination, while colonization by L. plantarum led to up-regulation of lipid metabolism. These indicate that the colonizers induce host responses that are specific per intestinal location. In conclusion, we propose that A. muciniphila modulates pathways involved in establishing homeostasis for basal metabolism and immune tolerance toward commensal microbiota.
Keywords: Akkermansia muciniphila, mucin, germ-free mice colonization, host responses
Introduction
The human gut is colonized by a complex, diverse, and dynamic community of microbes that continuously interact with the host (Hooper et al., 2002; Kelly et al., 2005). Considerable attention has focused on pathogen recognition at the intestinal epithelium (Cummings and Relman, 2000; Kagnoff and Eckmann, 2001). Remarkably, the host response to intestinal commensals that are abundant in number and diversity have not been studied at a similar level. Our intestinal tract is colonized by thousands of bacterial species, most of which belong to the phyla Firmicutes, Bacteroidetes, Actinobacteria. Proteobacteria and Verrucomicrobia (Zoetendal et al., 2008). Pioneering studies on the impact of commensals on host gene responses have been performed using gnotobiotic germ-free animals (Hooper et al., 2002). It was convincingly shown that following colonization of germ-free mice Bacteroides thetaiotaomicron, a well-characterized member of the intestinal Gram-negative bacteria, activated the host immune system, stimulated angiogenesis (Stappenbeck et al., 2002), and promoted increased fat storage (Backhed et al., 2004). In contrast, murine colonization with Bifidobacterium longum that belongs to the Gram-positive Actinobacteria, showed a different transcriptional response as it resulted in down-regulation of several host genes that were up-regulated by B. thetaiotaomicron during co-colonization of both bacteria (Sonnenburg et al., 2006). Recently, the effect was studied of co-colonization of B. thetaiotaomicron and Eubacterium rectale, a butyrate-producer, belonging to the Firmicutes (Mahowald et al., 2009). This co-colonization induced more significant regulated genes on colonic cells than when species were inoculated alone. Significant regulation of genes involved in cellular growth and proliferation, and cell death were found after co-colonization. These observations were instrumental to demonstrate that (i) there exists a dynamic interaction between host and bacteria that affects homeostasis, (ii) the host response is specific for each bacterium studied, and (iii) intestinal bacteria contribute to host physiology. However, there is a need to expand these studies and address the host response of major intestinal phyla, notably those that are expected to have a direct interaction with their animal host.
Recently, several studies have addressed the response of human cells to Lactobacillus species, lactic acid bacteria that degrade sugars derived from plants and other components of our diet. Lactobacilli are abundant in our early life microbiota while in adults they colonize notably the upper intestinal tract (Heilig et al., 2002). Lactobacilli are abundantly present in a variety of animals and show specific and differential responses at various sites in the murine and human GI tract (Bron et al., 2004; Marco et al., 2007, 2010). Transcriptional analysis of duodenal biopsies from healthy adults exposed to Lactobacillus plantarum showed specific modulation of mucosal gene expression and pathways involved in immune tolerance (van Baarlen et al., 2009). In vitro studies already had suggested a modulation of the immune response by Lactobacillus spp that were found to stimulate polarization of immune T cells toward regulatory T cells (Mohamadzadeh and Klaenhammer, 2008). A molecular mechanism for this was recently provided by the finding that the cell envelope S-layer of L. acidophilus directly signals to the immune system by binding to dendritic cells that are present in the host mucosa (Konstantinov et al., 2008).
A direct interaction between host and bacteria in the intestinal tract is prevented by the presence of a thick mucus layer covering the intestinal cells that protects the epithelium against toxins, acids, and bacterial invasion. Major components of this intestinal mucus are the mucins, heavily glycosylated proteins that form a network via cross-linking of disulfide bridges. Mucin-associated bacteria have been studied previously and are found among the main intestinal phyla (Derrien et al., 2010). Molecular studies based on 16S rRNA sequence analysis indicated that communities that are strongly associated with the colonic mucosa are different from those that are frequently sampled from the feces (Zoetendal et al., 2002; Nielsen et al., 2003; Lepage et al., 2005), with an overrepresentation of bacteria that degrade mucins (Mackie et al., 1999). Recently, we have isolated a strictly anaerobic, Gram-negative bacterial species, Akkermansia muciniphila, that is specialized in the utilization of mucin as a carbon and nitrogen source (Hooper et al., 2002; Derrien et al., 2004). A. muciniphila was the first cultured representative of the Verrucomicrobia and is, so far, the sole representative of this phylum that is present in the human intestinal tract (Derrien et al., 2008). Moreover, its characteristic 16S rRNA signatures have been consistently identified in mucosal clone libraries and may make up over 5% of the retrieved sequences (Eckburg et al., 2005; Wang et al., 2005). Based on analysis of fecal samples, the numbers of A. muciniphila start to increase in newborns and reach a level close to that observed in adults within a year (Collado et al., 2007). This suggests that A. muciniphila is a relatively late colonizer of the human intestine compared to early infant colonizers such as Bifidobacterium and Lactobacillus species (Mackie et al., 1999; Favier et al., 2002). Remarkably, both Bifidobacterium spp. and A. muciniphila made up an important fraction of the microbial cells in pregnant woman but were numerically decreased in those that were overweight (Santacruz et al., 2010). Moreover, recent studies have indicated that Akkermansia-like bacteria are abundantly present in the mucosa of appendices of healthy subjects but strongly reduced during appendicitis (Swidsinski et al., 2011). The same finding was observed in mucosal samples from IBD patients suggesting that A. muciniphila is associated with healthy mucosa (Png et al., 2010).
A recent molecular inventory revealed that Akkermansia species are widely distributed amongst mammals, with a strong predominance in herbivores (Ley et al., 2008). In mice, Akkermansia species may constitute over 1% of the cecal microbial community (Ley et al., 2005; Turnbaugh et al., 2006). In hamsters and pythons their numbers were found to be high and increased during fasting, suggesting a relation with mucus production (Sonoyama et al., 2009; Costello et al., 2010). The frequent presence and high abundance of A. muciniphila in both human and other mammalian intestines underlines the relevance to address the role of this mucin-degrading commensal in the gut as well its impact on the host. As bacterial colonization may induce both general and species-specific responses in the host, we colonized germ-free mice with A. muciniphila and compared and contrasted its distribution, location, and impact on host transcriptional response with that of L. plantarum, a Gram-positive bacterium that principally ferments dietary sugars.
Materials and Methods
Animals
The study protocol was reviewed and approved by the Northern Stockholm Ethics Committee for Animal Experiments. Adult germ-free female NMRI–KI mice (45–65 days) were used (n = 18) for bacterial mono-association. The germ-free animals were inbred for >60 generations at the Laboratory of Medical Microbial Ecology at Karolinska Institute and they were housed in lightweight stainless-steel isolators (Gustafsson, 1959). All mice had free access to a steam-sterilized standard mouse chow (R36; Lactamin, Vadstena, Sweden) and to sterilized water. Artificial light was available between 6 a.m. and 6 p.m.; the temperature was 24 ± 2.2°C, and the humidity was 55 ± 10%. The germ-free status was checked weekly by inoculating fecal samples in different media incubated both aerobically and anaerobically at 20 and 37°C for up to 4 weeks.
Bacteria and growth conditions
Two bacterial strains were used in this study, A. muciniphila MucT (ATTC BAA-835) and L. plantarum WCFS1 (NCIMB 8826). A. muciniphila was grown anaerobically in a basal mucin-based medium as previously described (Derrien et al., 2004) and L. plantarum was grown anaerobically at 37°C in Man–Rogosa–Sharpe broth (MRS; Le Pont de Claix, France).
Mono-association
The germ-free mice were mono-associated using established protocols (Cardona et al., 2001). In brief, 10 ml of cultures in late log phase of A. muciniphila MucT and L. plantarum WCFS1 were centrifuged (4500 rpm, 10 min). Pellets were resuspended in 1 ml of sterile anaerobic phosphate buffer saline (PBS) and dispensed into sterile ampoules which were heat-sealed. The external surface of each ampoule was sterilized with chromsulfuric acid before transfer into respective isolators. Inside the isolators, the ampoules were broken, and 0.2 ml (109 cfu/ml) of the strictly anaerobic A. muciniphila (n = 6) was inoculated intragastrically; L. plantarum (n = 6) was inoculated orally. Germ-free control mice (n = 6) were housed in separate isolators.
Establishment of A. muciniphila in mice
To verify that mice were colonized with A. muciniphila, fecal samples (collected at day 3 and day 7) were diluted in mucin medium, incubated anaerobically at 37°C and inspected daily for growth for 6 days as previously described (Derrien et al., 2004). Exact enumeration of bacteria in intestinal samples was examined by a 16S rRNA quantitative PCR (qPCR) approach. In short, the genomic DNA from pure culture, ileal, cecal, and colonic contents was isolated using the Fast DNA Spin kit (Qbiogene, Inc., Carlsbad, CA, USA). PCR amplification of bacterial 16S rRNA genes was performed on genomic DNA of A. muciniphila using specific primers set AM1 (5′-CCT TGC GGT TGG CTT CAG AT-3′) and AM2 (5′-CAG CAC GTG AAG GTG GGG AC-3′; Collado et al., 2007).
Preparation of specimens
After 7 days of colonization mice were killed by cervical dislocation and terminal ileum, cecum, and ascending colon specimens were sampled. Luminal contents were separated from the epithelium and were kept at −20°C for DNA extraction and bacterial enumeration by qPCR. Tissues were flushed with PBS. For RNA isolation, tissues were immediately preserved in five volumes of RNAlater (Ambion, Austin, TX, USA) and stored at 4°C until use. For histology, biopsies were fixed for 18 h at room temperature in 4% paraformaldehyde, pH 7.3 and subsequently processed for fluorescent in situ hybridization (FISH).
Histology and fluorescent in situ hybridization
After preservation in paraformaldehyde, tissue samples were washed in phosphate buffer, dehydrated in an ethanol gradient, and embedded in paraffin. Five micrometer thick sections were mounted on Superfrost coated slides, dried, and incubated at 37°C for 16 h. For FISH, slides were deparaffinized in xylene and dehydrated in an ethanol gradient. Sections were overlaid with 100 μl hybridization buffer [0.9 M NaCl, 0.02 M Tris–HCl (pH 8.0), 0.01% sodium dodecyl sulfate] containing an oligonucleotide mixture (5 ng/μl) consisting of the A. muciniphila Cy3-labeled MUC-1437 (5′-CCTTGCGGTTGGCTTCAGAT-3′) and total bacterial FITC-labeled EUB-338 (5′-GCTGCCTCCCGTAGGAGT-3′) probes (Biolegio BV, Nijmegen4, The Netherlands). Hybridization was carried out at 50°C for 16 h in a humid chamber. After hybridization, the tissue sections were washed with a washing buffer (0.02 M Tris–HCl pH 8, 0.9 M NaCl) for 10 min at 50°C. Counterstaining was carried out with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich), and the slides were analyzed with a Nikon E600 epifluorescence microscope equipped with appropriate filter sets.
RNA extraction
Total mouse RNA from the intestinal tissue segments was extracted with Trizol® following supplier’s protocol (Trizol reagent, Invitrogen). RNA was purified, treated with DNase, and concentrated using RNeasy mini kit (Qiagen). RNA quantity and quality were assessed spectrophotometrically (ND-1000, NanoDrop Technologies, Wilmington, USA) and Bioanalyzer nano chips (Bioanalyzer 2100; Agilent).
DNA microarray hybridizations and data analysis
Affymetrix GeneChip mouse genome 430 2.0 arrays (Affymetrix) containing 45,000 probe sets for the analysis of around 39,000 transcripts and variants of the approximately 22,000 mouse genes, were used to assess the transcriptional response to A. muciniphila and L. plantarum in the ileum, cecum, and colon. For each group and location, 2 μg of total RNA per mouse was subsequently pooled per group, and 10 μg were used for one cycle cDNA synthesis. Hybridization, washing, and scanning of GeneChip Mouse Genome 430 2.0 Array were done according to the manufacturer’s protocol1. Normalization and probe-level intensities analysis were performed using the multi-mgMOS model (Liu et al., 2005) and a novel intensity-based Bayesian moderated T-statistic (IBMT; (Sartor et al., 2006) was used to identify all genes with a significantly altered transcriptional activity in response to the bacterial treatments. Gene significance cut-offs were cM/sM > 1, and PPLR values > 0.975 or <0.025, corresponding to P-values < 0.05. Gene ontologies describe gene function in relation to a large and growing context of biological knowledge at three levels: biological process, molecular function, or cellular component, using a systematic classification and a formal vocabulary. Differential datasets were analyzed using the ingenuity pathways analysis (IPA) software tool2. Basically, IPA annotations follow the gene ontology (GO) annotation principle, but are based on a proprietary knowledge base of over 1,000,000 protein–protein interactions. GO annotations are used by Ingenuity in order to find overrepresented cellular functions and canonical (conserved in human, mouse, and rat) pathways. The IPA output includes metabolic and signaling pathways with statistical assessment of the significance of their representation being based on Fisher’s Exact Test. Here, this test calculates the probability that genes participate in a given pathway relative to their occurrence in all other pathway annotations. Array data have been submitted to the Gene Expression Omnibus, accession number: GSE18587.
Quantitative reverse transcription PCR
cDNA was synthesized using 500 ng of total RNA employed for microarray analyses using Superscript III reverse transcriptase; random primers according to supplier’s protocol (Invitrogen). qPCR on 6 ng cDNA derived from pooled RNA used for microarray analysis was performed on a Bio-Rad iCycler (Bio-Rad Laboratories) using gene-specific primers (0.2 μM) for selected genes (Table 1) and 1× SYBR Green PCR Master Mix buffer (Bio-rad). Each pair was designed to generate a 100 to 200-bp amplicon were derived from Primerbank3 and primers were purchased from Biolegio BV (Nijmegen, The Netherlands). The specificity of the amplification was verified by melt curve analysis and evaluation of efficiency of PCR amplification. All data were normalized to an internal 18S mRNA control (ΔΔCT analysis), of which the expression did not differ between germ-free and mono-associated mice (data not shown).
Table 1.
Primers sequences used in this study to validate arrays data.
| Gene name | Gene symbol | Function | Accession number | Sequence (5′ → 3′) |
|---|---|---|---|---|
| 18S rRNA | 18S | Ribosome biogenesis and assembly | X00686 | F: CATTCGAACGTCTGCCCTATC R: CCTGCTGCCTTCCTTGGA |
| S100 calcium binding protein g | S100g | Cell adhesion | NM 009789 | F: ATGTGTGCTGAGAAGTCTCCT |
| R: CGCCATTCTTATCCAGCTCCTT | ||||
| Glycosylation dependent adhesion molecule 1 | Glycam1 | Cell adhesion | NM 008134 | F: GTCCTGCTATTTGTCAGTCTTG |
| R: CCTGGTCTTGATTCTCTG | ||||
| Lipoprotein lipase | Lpl | Lipid metabolism | NM 008509 | F: GGGAGTTTGGCTCCAGAGTTT |
| R: TGTGTCTTCAGGGGTCCTTAG | ||||
| Carbonic anhydrase 1 | Car1 | Nitrogen metabolism | BC011223 | F: ACTGGGGATATGGAAGCGAA |
| R: TGCAGGATTATAGGAGATGCTGA | ||||
| Regenerating islet-derived family, member 4 | Reg4 | Vascularization | NM_026328 | F: TGAGCTGGAGTGTCAGTCATA |
| R: CAATCCACACAGGCAGGTTTC | ||||
| Small proline-rich protein 2a | Sppr2a | Vascularization | AV371678 | F: CCTTGTCGTCCTGTCATGT |
| R: GGCATTGCTCATAGCACACTAC |
Results
Akkermansia muciniphila and L. plantarum colonize different intestinal regions of germ-free mice
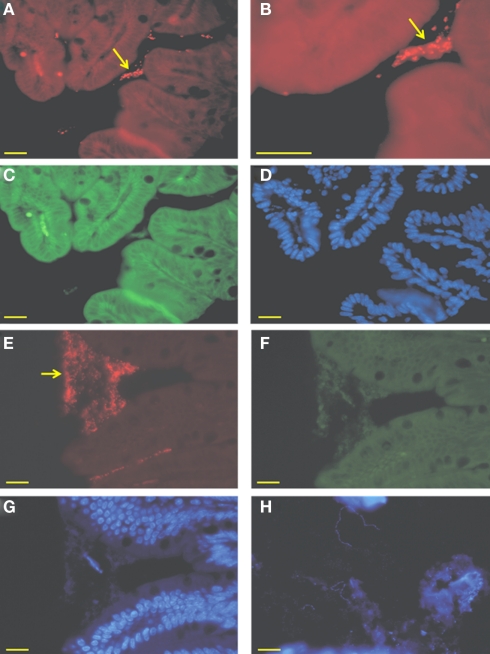
Efficient and reproducible colonization of germ-free mice was obtained by a single intragastric dose of 109 A. muciniphila cells grown anaerobically on mucin-containing medium. To verify that A. muciniphila had colonized the intestinal tract, DNA was extracted from intestinal regions (ileum, cecum, and colon), and used for 16S rRNA qPCR quantification. In all mice that were mono-associated with A. muciniphila, bacterial numbers present in the intestinal lumen reached 1.87 (±2.37) × 108 cells/g of ileal content, 3.12 (±0.7) × 1010 cells/g cecal content, and 1.73 (±0.6) × 109 cells/g colonic content. All A. muciniphila-associated mice were healthy throughout the study period of 7 days. Subsequently, FISH analysis was performed on all three intestinal regions using the specific oligonucleotide probe MUC-1437 that is complementary to part of a hypervariable region of A. muciniphila (Derrien et al., 2008). Aggregations of A. muciniphila cells were found in all sections of formaldehyde fixed samples, within a distance of 50 μm to the epithelium (Figure 1). This analysis indicated that A. muciniphila is closely associated with the mucus layer covering the epithelial cells and support the notion that A. muciniphila is utilizing mucin in situ to sustain growth (Derrien et al., 2004). To further evaluate this correlation between intestinal location and use of carbon sources, we studied the in vivo location of L. plantarum, capable of growing on dietary carbohydrates but a non-mucin-degrader. Based on 16S rRNA-gene qPCR quantification, L. plantarum colonized germ-free mice at levels similar to A. muciniphila, with the exception of the small intestine where the luminal colonization was 10-fold higher (109 cells/g) than in cecum or colon, consistent with previous studies (Marco et al., 2007). DAPI-stained sections from the three intestinal regions showed presence of non-aggregated, long cells with the morphology of L. plantarum cells (Figure 1H). Such cells were not observed in control biopsy sections from germ-free mice, indicating an exclusive luminal location of L. plantarum that contrasts with that of the mucosa-location of A. muciniphila.
Figure 1.
Micrograph of intestinal biopsy samples from mono-colonized mice. (A–D) Ileal biopsies and colonic (E–G) of mice mono-associated with A. muciniphila. Cells were hybridized with the Cy3-labeled MUC-1437, specifically staining A. muciniphila. (A,B,E), aggregates of A. muciniphila cells are zoomed and closely associated with the epithelial cells (B), FITC-labeled EUB-338, targeting all bacteria (C,F) and a DAPI staining (D,G). Arrows indicate aggregates of A. muciniphila. (H) Cecal biopsy from L. plantarum mono-associated mouse stained with DAPI. Bar represents 10 μm.
Colonization by A. muciniphila and L. plantarum induce specific transcriptional responses in the three intestinal regions
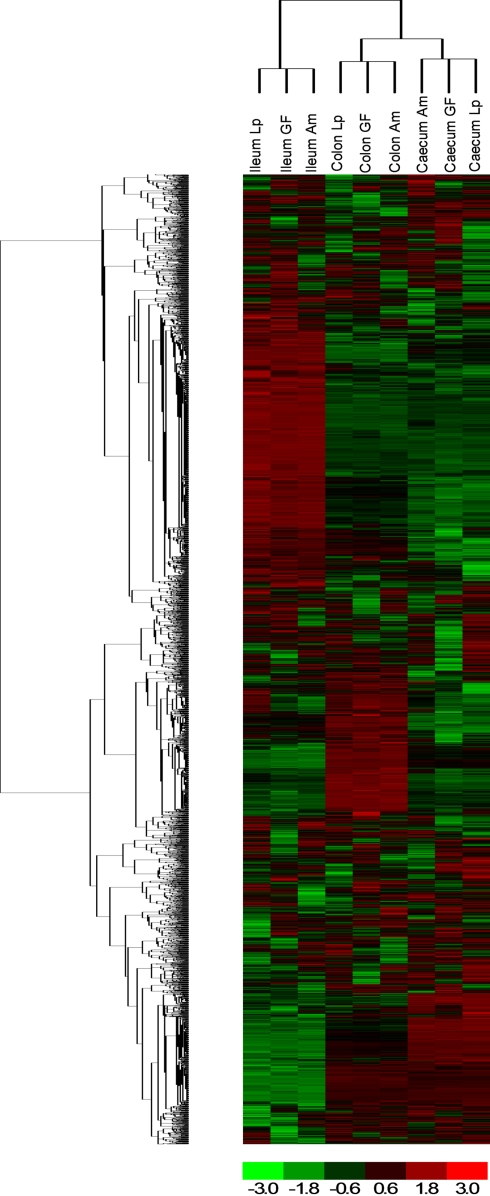
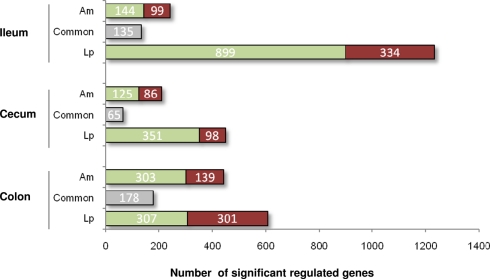
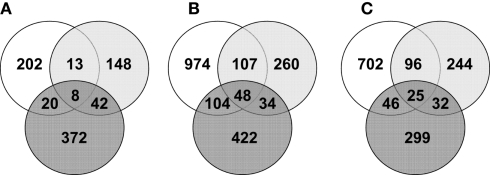
To examine the impact of A. muciniphila and L. plantarum colonization on host mucosal gene expression profiles, microarrays were used to determine the transcriptome of the host before and after colonization using Bioconductor and third-party software (Figure 2). The total number of significantly regulated genes was highest in the ileum of mice mono-associated with L. plantarum (1233) followed by the colon (608) and cecum (449). In A. muciniphila mono-associated mice, the numbers of differentially expressed genes was highest in the colon (442), followed by the ileum (253), and the cecum (211; Figure 3). The percentage of shared genes between A. muciniphila and L. plantarum was highest in the colon (17.0%), followed by the cecum (9.9%), and the ileum (9.1%). After colonization by A. muciniphila, less than 10% of the differentially expressed genes occurred in all three intestinal regions (Figure 4). The percentage of common regulated genes between the cecum and colon was 8.3%, while 4.8% between the ileum and cecum, and 4% between the ileum and colon were shared. In L. plantarum mono-associated mice, percentages followed the same tendency but were twice as high. Together, the expression data and histology show that the mouse intestines undergo region-specific, differential transcriptome changes following colonization by different bacteria. Array data were validated by quantitative reverse transcription PCR (qRT-PCR) for six selected genes harboring diverse fold change in ileum and colon from mono-colonized mice. qRT-PCR confirmed the expression levels for the selected genes (Table 2).
Figure 2.
Hierarchical clustering of genes obtained using D-Chip software. Hierarchical clustering was performed on intensity values generated by Affymetrix MAS 5.0 software, showing differential responses in the three locations (ileum, cecum, and colon) and with bacteria [A. muciniphila (Am) and L. plantarum (Lp) compared to germ-free (GF) mice]. Each column corresponds to one array (pool of six RNA) and each frame corresponds to one probe set. Red corresponds to an up-regulation of the gene expression and green to a down-regulation. Black corresponds to no change.
Figure 3.
Number of significantly up- (red) and down- (green) regulated genes in the ileum, cecum, and colon of mice mono-associated with A. muciniphila and L. plantarum vs. germ-free mice. Gray represents common regulated genes.
Figure 4.
Venn diagrams representing the number of common significantly regulated genes in the ileum (white), cecum (light gray), and colon (dark gray) of mice mono-associated with A. muciniphila (A) and L. plantarum (B) interaction compared to germ-free mice data and L. plantarum compared to A. muciniphila data (C).
Table 2.
Expression levels of selected genes in intestinal samples from A. muciniphila and L. plantarum mono-associated mice compared to the germ-free mice.
| Gene | Location | A. muciniphila | L. plantarum | ||
|---|---|---|---|---|---|
| Arrays | qRT-PCR | Arrays | qRT-PCR | ||
| Reg4 | Colon | −1.07 | −5.02 | −4.29 | −12.01 |
| Sppr2a | Ileum | 1.06 | 1.65 | −3.16 | −4.29 |
| Car1 | Ileum | −2.33 | −3.10 | −29.62 | −7.38 |
| Lpl | Ileum | −1.72 | −1.64 | 2.83 | 1.53 |
| Glycam1 | Colon | 4.50 | 3.14 | 1.59 | 2.08 |
| S100g | Colon | 2.48 | 2.51 | −2.08 | −3.03 |
Assays were performed in triplicate. Data were normalized to 18S ribosomal RNA and results are expressed in fold change.
Colonization by A. muciniphila induces expression of genes involved in regulation of immune responses and homeostasis
Although bacteria are formally seen as “non-self” by the host immune system, colonization of the mammalian gut by bacteria leads in healthy individuals to a more or less neutral, non-inflammatory balance between pro- and anti-inflammatory immune responses, a condition that is known as gut immune homeostasis (Artis, 2008). Since mice colonized by the two different commensal bacteria did not show any sign of inflammation or discomfort, we assumed that the transcriptomes altered after colonization did show part of the transcriptional regulation of immune homeostasis. In order to extract as much biological function and context out of the altered transcriptomes in the mouse intestines after colonization by A. muciniphila and L. plantarum, IPA was used.
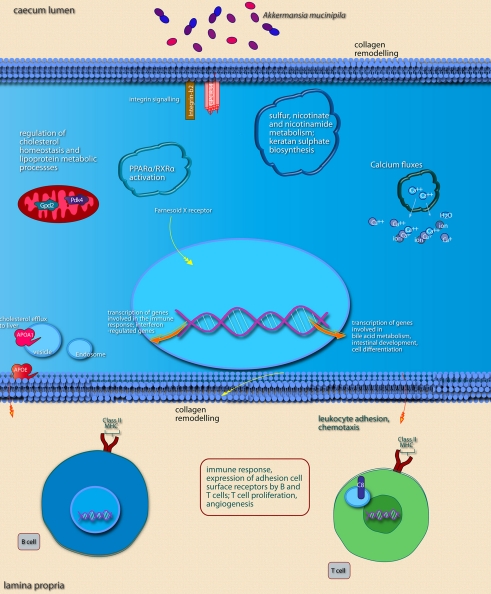
In the cecum of mice mono-associated with A. muciniphila, the region that was most efficiently colonized by this bacterial species, colonization of the mucosa apparently led to an altered expression of genes participating in membrane metabolism and signaling and antigen presentation pathways (Figure 5; Table 3). The major cellular functions that were modulated after cecal colonization by A. muciniphila were involved in the regulation of epithelial homeostasis and cell fate (Table 3). Cecal colonization by A. muciniphila did also result in up-regulation of genes involved in antigen presentation of leukocytes (Figure 5). In the colon, A. muciniphila induced multiple immune response-related pathways, both innate and adaptive (Figure 6; Table 3). Finally, ileal colonization by A. muciniphila led to differential expression of genes involved in metabolic and signaling pathways, mainly via modulation of PPARα-dependent processes. Most of these genes and pathways participate in lipid metabolism, small molecule biochemistry, and metabolic homeostasis (Table 3).
Figure 5.
Main cellular pathways and processes inferred to be modulated or activated after colonization of mouse cecum by A. muciniphila. Only induced pathways are shown. Distinguishing features are the induction of pathways involved in (epithelial) cell membrane modifications, sulfur metabolism, and pathways involved in the maturation of immune cells via induction of surface receptors. Pathways were considered to be induced if (nearly) all of the genes belonging to the pathway were up-regulated. The upper part of the image shows pathways and processes that are expected to occur in epithelial cells as well as pathways and processes with undefined or non-specific localization. Pathways and processes named in text “balloons” were among the most significantly modulated pathways according to IPA canonical pathway analysis. The lower part of the image shows pathways known to be regulated in immune cells of the lamina propria. In order to determine in which cell type and subcellular compartment a gene product was localized, the Entrez Gene GO annotation, Entrez Gene GeneRIFs, and scientific literature information were used. Interacting pathways and processes are grouped together as well as possible, both in terms of annotation as well as within the graphical space limitations.
Table 3.
Canonical pathways in the mouse ileum, cecum, and colon that are more significantly modulated upon colonization by A. muciniphila or L. plantarum.
| Intestinal region | Comparison | |
|---|---|---|
| A. muciniphila vs. GF | L. plantarum vs. GF | |
| Ileum | PPARα–RXRα activation Tryptophan metabolism Serotonin receptor signaling Synthesis and degradation of ketone bodies Dopamine receptor signaling Death receptor signaling | PPARα–RXRα activation LPS/IL-1 mediated inhibition of RXR function Propanoate metabolism Protein ubiquitination pathway Aryl hydrocarbon receptor signaling Pyruvate metabolism |
| Cecum | Keratan sulfate biosynthesis PPARα/RXRα activation Integrin signaling Antigen presentation pathway Sulfur metabolism Nicotinate and nicotinamide metabolism | LPS/IL mediated inhibition of RXR function Aryl hydrocarbon receptor signaling Glutathione metabolism Beta-adrenergic signaling Xenobiotic metabolism signaling p53 signaling |
| Colon | Antigen presentation pathway B cell receptor signaling Leukocyte extravasation signaling T cell receptor signaling IL-4 signaling Complement and coagulation cascades | NRF2-mediated oxidative stress response p38 MAPK signaling Circadian rhythm signaling |
Significance was calculated by Fisher’s Exact Test using ingenuity pathway analysis (IPA). These pathways may be interconnected since they may share common elements, e.g., NF–κB and G-protein coupled receptor (GPCR) signaling can play roles in multiple pathways.
Figure 6.
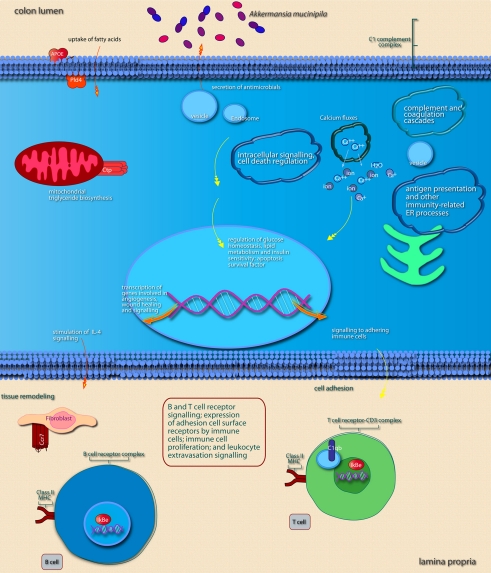
Main cellular pathways and processes inferred to be modulated or activated after colonization of mouse colon by A. muciniphila. Distinguishing features are the induction of pathways involved in intracellular signaling, induction of secretion of (antimicrobial) factors, and of pathways that promote expression of immune cell receptors that mediate adhesion and T or B cell receptor signaling. For further explanation, see legend of Figure 5.
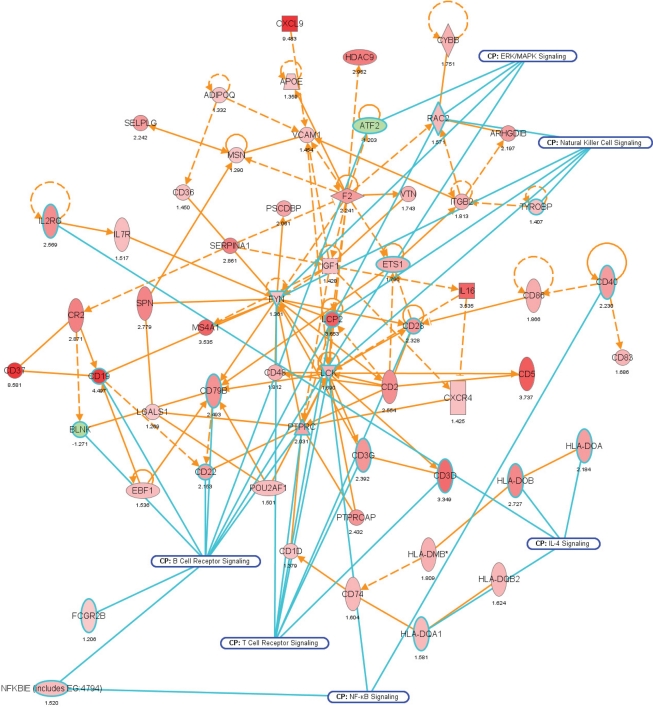
We noted that several categories of those mouse genes that showed the strongest alterations in transcription after intestinal colonization by A. muciniphila were implicated in regulation of the immune response and cell death and proliferation, the latter category especially in the colon (Figure 7). Immune response-associated genes in the colon were involved in chemotaxis and complement cascade, parts of the innate immune response, but also in cell adhesion and the maturation of B and T cells. This might suggest that colonization of the mouse intestine by A. muciniphila modulates the host immune system. Genes involved in vitamin and mineral metabolism were also differentially expressed in the colon, possibly reflecting a modulation of xenobiotic metabolism.
Figure 7.
Network representation of altered cellular functions during colonization of the mouse colon by A. muciniphila for the gene ontology (GO) category “immune response.” The network shows that most of the genes regulating the immune response are up-regulated during colonization. Central regulatory genes encode growth factors (e.g., Igf1), cell surface molecules (Cd19, Cd37), (receptor) phosphatases (Ptprc or Cd45), and protein kinases (Lck, Fyn) that regulate (immune) cell proliferation and differentiation. The protein interaction network was derived from the transcription data using ingenuity pathway analysis (IPA). Nodes indicate proteins, the yellow lines connecting them represent interactions (e.g., protein–protein binding or phosphorylation events) that are mapped by Ingenuity’s knowledge base (see Materials and Methods). Colors indicate altered transcription of the encoding genes, with deeper shades of red indicating stronger up-regulation, and deeper shades of green indicating stronger down-regulation. The genes and their encoded proteins that are used to reconstruct specific networks were selected based on overrepresentation of GO categories (“cellular functions” in IPA) and involvement in significantly (see Materials and Methods) modulated canonical pathways. The overlays with blue radiating lines represent some of these canonical pathways (CP) and are useful to indicate in which cellular pathways differentially regulated genes and their interacting products participate for the given GO categories. Note that in this network exemplifying the genetic regulation of the mouse colonic immune response to A. muciniphila, most of the genes are up-regulated, indicating that the immune response was induced following colonization of these bacteria.
The expression profiles suggest that colonization by A. muciniphila leads to coordinated differential expression of genes involved in metabolic and immune response-regulatory processes. The balance between the involved pathways and processes differs per intestinal region; upon colonization, immune-regulatory processes were most pronounced modulated in the colon, followed by cecum, and ileum (Table 3).
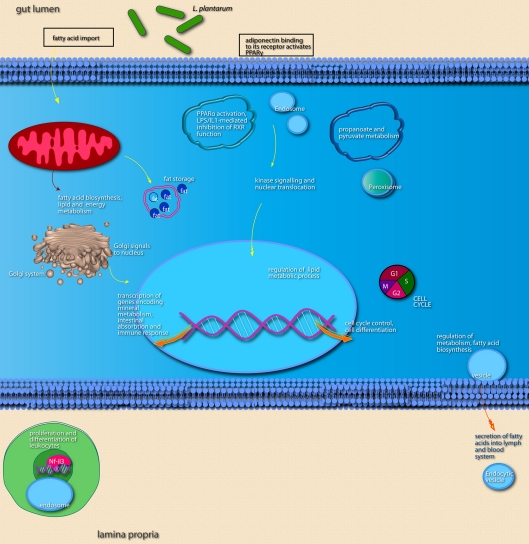
Colonization by L. plantarum stimulates transcription of genes involved in cell signaling, lipid metabolism, and growth and proliferation
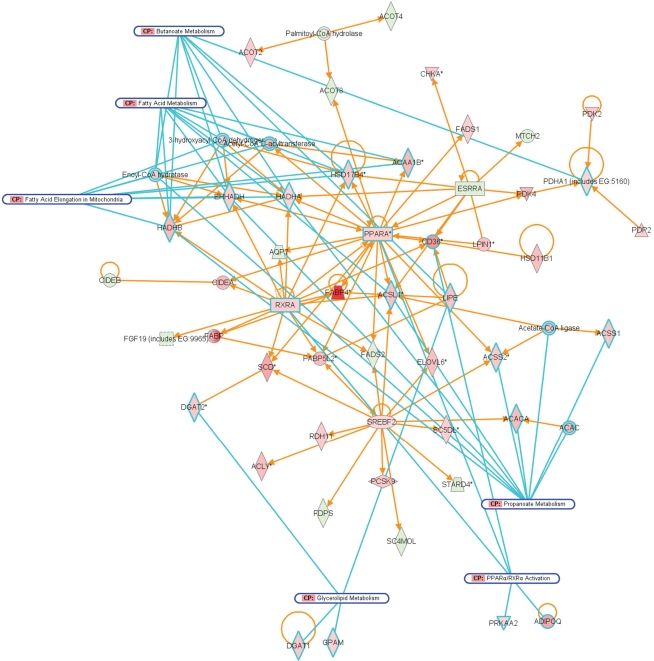
Compared to A. muciniphila, a smaller proportion of genes that were differentially expressed after colonization of the intestines by L. plantarum were involved in immune response. Only a few genes encoding immune-regulatory molecules were differentially expressed; among these were the acute phase-response genes Reg3α and Reg3γ. According to IPA, the only immune response-related pathway that was significantly modulated was the B cell receptor signaling pathway, in the cecum and ileum. In the ileum, colonization by L. plantarum mainly led to regulation of genes participating in activation and regulation of lipid and fatty acid metabolism (Figures 8 and 9; Table 3). In the cecum, genes involved in metabolic and immune response signaling and involved in normal and xenobiotic metabolism were differentially expressed after colonization by L. plantarum. In the colon, genes involved in stress response signaling and tissue development and function were differentially regulated after colonization. Summarizing, after colonization of mouse intestines by L. plantarum, most of the differentially expressed genes and pathways in ileum, cecum, and colon were involved in regulation of lipid and fatty acid metabolism, cellular signaling, molecular transport, and xenobiotic metabolism (Table 3). In the colon, colonization by L. plantarum did also stimulate genes that regulate tissue development.
Figure 8.
Network representation of altered cellular functions during colonization of the mouse ileum by L. plantarum for the category “lipid metabolism.” The network shows that most of the genes that are co-expressed with the major regulators, PPAR and RXR are up-regulated, or up-regulated through the activity of the sterol regulatory element binding transcription factor 2 (Srebf2).
Figure 9.
Main cellular pathways and processes inferred to be modulated or activated after colonization of mouse ileum by L. plantarum. Distinguishing features are the up-regulation of relatively high numbers of genes encoding factors involved in lipid metabolism and of genes encoding transcription factors that regulate intestinal absorptive functions and the immune response.
Bacterial colonization leads to specific expression profiles that lead to tolerance to microbiota
A direct comparison of differential gene expression in response to L. plantarum and. A. muciniphila is of interest since this may show if intestinal colonization of mouse hosts by different bacteria shows common or more specific expression profiles. So far, most of the described comparisons involve colonized and germ-free hosts. These comparisons may not be suitable to find processes that are exclusively altered by one specific bacterial species; specific processes may be obscured by the consequences of microbial colonization per se. For instance, genes involved in antigen presentation pathways were mainly down-regulated in the cecum following colonization by L. plantarum. In contrast, colonization by A. muciniphila tended to up-regulate genes that are involved in antigen presentation pathways (Table 3). In direct comparisons of gene transcription after bacterial colonization, gene expression profiles of mice colons colonized by A. muciniphila showed relatively higher (compared to colonization by L. plantarum) up-regulation of genes participating in immune response signaling and ERK/MAPK signaling. Interestingly, a few genes that encode cell proliferation factors such as epidermal growth factor (Egf), early growth receptor 1 (Egr1), and connective tissue growth factor (Ctgf) were down-regulated in colons colonized by A. muciniphila, compared to expression of the same genes in colons colonized by L. plantarum. Furthermore, ilea colonized by A. muciniphila showed a relatively lower expression of genes involved in lipid and fatty acid metabolism compared to ilea colonized by L. plantarum. In addition, genes involved in cellular growth and developmental pathways were differentially regulated in ilea, with the inference that growth and proliferation were less stimulated in A. muciniphila-colonized ilea compared to ilea colonized by L. plantarum.
The comparative analysis of the response to A. muciniphila and L. plantarum show that colonization of germ-free mice by bacteria that utilize different carbon sources and are located in different sites leads to a differential expression of genes involved in cellular processes and pathways such as the immune response, lipid metabolism, and cell fate. Differential gene expression was not only dependent on the colonizing bacterial species but also on the intestinal region. Despite the up-regulation of genes involved in the immune responses, colonized mice did not develop microscopically visible inflammation nor did they show any sign of discomfort. This suggests that the transcriptional profiles we obtained are involved in the regulation of immune tolerance toward the Gram-negative A. muciniphila and the Gram-positive L. plantarum together with metabolic homeostasis of the colonized intestine. Although colonization by A. muciniphila did lead to a higher number of differentially expressed genes involved in regulation of the immune system, there were no strong indications that these genes participated in driving a pro-inflammatory response (e.g., up-regulation of a pro-inflammatory p50–p56 NF–κB complex (Wietek and O’Neill, 2007). Both in cecum and ileum, after intestinal colonization by A. muciniphila, and compared to GF control mice, immune response-associated genes were not enriched apart from genes involved in the antigen presentation pathway (Table 3). Likely, immune homeostasis had already been reached here, within the 7-days before RNA was extracted, and maintained via immune regulators such as the gene CEBPD (NF–IL6–beta), a regulator of genes involved in immune and inflammatory responses and genes associated with activation and/or differentiation of macrophages. In the colon, where the regulation of immune response-associated genes was highest, over 60 genes, including 16 genes encoding CD antigen markers and 10 genes encoding immune cell membrane receptors were up-regulated, nearly 14% of the total of regulated genes. These increased gene transcripts likely reflect presence of a larger number of immune-competent leukocytes in the lamina propria and are not themselves indicative of a pro-inflammatory immune response. Disproportionate immune responses in the colon may have been directly inhibited through activity of up-regulated serine protease inhibitors (serpins) and indirectly by up-regulation of genes encoding factors that modulate pro-inflammatory signaling, such as IκBε, an inhibitor of nuclear translocation of NF–κB. We compared our expression data with those obtained by profiling wound healing-associated inflammation in mice as reported by Cooper et al. (2004) and found less than 5% similar genes between our colonization experiments and their inflammation datasets. We propose that in mice, establishment of tolerance is achieved fast, especially in the small intestine, and is maintained through the activity of transcription factors in the small intestine, and through protection from overt cytotoxic proteolytic processes and avoidance of pro-inflammatory signaling in the colon.
Discussion
In this study we determined the transcriptional profiles of the mouse intestinal mucosa to examine the balanced changes in the mucosal transcriptome of a germ-free host following colonization by commensal intestinal bacteria. We colonized germ-free mice with bacteria that differed notably in physiology and phylogeny, viz. the Gram-negative, strictly anaerobic mucin-degrading A. muciniphila and the Gram-positive utilizer of dietary carbohydrates L. plantarum, which is incapable of utilizing mucin. Both bacteria colonized the intestinal tract within 7 days after a single inoculation. We used microarray profiling since this has been shown to be a useful tool for studying in vivo interactions between eukaryote hosts and their intestinal bacteria (Di Caro et al., 2005; Ukena et al., 2005; Baken et al., 2006).
Transcriptional changes upon colonization by A. muciniphila involved a large amount of genes participating in the immune response in comparison with germ-free mice or those colonized by L. plantarum. Colonization by L. plantarum, on the other hand, mainly stimulated expression of genes involved in the metabolism. Differentially expressed genes differed depending on bacterial colonizer and intestinal site to such an extent that only few genes were differentially expressed in all three intestinal regions.
The largest changes in gene transcription after colonization by A. muciniphila were observed in the colon; for L. plantarum, the largest changes were found in the ileum. This may be partly explained by the different bacterial numbers reached in the three intestinal regions. L. plantarum reached ten times higher numbers in the ileum compared to A. muciniphila, whereas both bacteria colonized the colon and cecum to the same extent. In the cecum, where the highest bacterial colonization level was observed, similar responses were observed as in the colon. Interestingly, keratan sulfate biosynthesis was among the major regulated pathways in mouse cecal epithelia following colonization by A. muciniphila. This pathway is a feature of epithelial tissues responding to physiological changes such as typically result from development and wounding but it is also involved in mucin production (Funderburgh, 2000). It is possible that these changes are involved in establishing novel biosynthesis levels of epithelial cell components including mucins, since presence of A. muciniphila in the mucus layer may lead to an increased turn-over of, and demand for, extracellular components. The genome of A. muciniphila has been recently elucidated and contains various genes that are predicted to code for secreted sulfatases (Derrien et al., 2010; van Passel et al., 2011), compatible with the fact that mucin serves as carbon, nitrogen, and sulfur source for A. muciniphila.
In the ileum, five canonical pathways that showed significant modulation after colonization by L. plantarum were also modulated following colonization by A. muciniphila. These pathways were mainly involved in regulation of cell signaling and lipid metabolism, small molecule biochemistry, and molecular transport (Table 3). Note that no enrichment of genes that may be involved in the regulation of cellular responses to cellular damage (such as the keratan sulfate biosynthesis pathway) was found in the ileal response to L. plantarum. These comparative differences became more obvious when transcriptional profiles of intestines colonized by A. muciniphila or L. plantarum were compared to each other rather than the response in germ-free animals. Indeed, direct comparisons of genes expressed in intestines colonized by A. muciniphila vs. intestines colonized by L. plantarum revealed that, in general, the intestinal regions colonized by L. plantarum showed enriched expression of genes that participate in cellular lipid metabolism, growth, and proliferation, whereas intestinal regions colonized by A. muciniphila tended to show enriched expression of genes involved in immune responses, death receptor signaling, and responses to cellular compromise. As the L. plantarum is a Gram-positive and A. muciniphila a Gram-negative species, these differential responses could be due to specific components from the bacterial cell envelopes involved in host signaling via cognate receptors. Alternatively, the differential host response could be established because of a more intensive contact of the mouse mucosa with A. muciniphila cells that were found in aggregates at less than 50 μm distance from epithelial cells (Figure 1). Aggregated colonies were also observed in cultures of A. muciniphila growing in a mucin-based medium. In contrast, L. plantarum cells were more randomly distributed in the gut lumen (Figure 1H) and are expected to have far less opportunities for direct contact with the mouse mucosa.
In a recent study, the mouse colonic mucus layer has been investigated for the presence of bacteria (Johansson et al., 2008). Interestingly, no bacteria were present in the proximal 50 μm layer that mainly consisted of Muc2, while bacteria were found in the more accessible outer layer. This may suggest that in a germ-free background, A. muciniphila is able to colonize the inner mucus layer while in conventionalized mice it has to compete with other bacteria. Alternatively, it is possible that the method used for fixing the biopsies in the studies vary and explain the results while it is also possible that A. muciniphila may have escaped earlier detection (Johansson et al., 2008) as a general eubacterial probe was used that only poorly reacts with Akkermansia cells (Derrien et al., 2008).
Overall, our data show that colonization of germ-free mouse intestines by different bacteria leads to differentially altered host transcriptomes with a bias toward balanced immune responses indicative of tolerance for A. muciniphila, and toward a promotion of lipid and fatty acid metabolism, growth, and proliferation for L. plantarum. Since the germ-free mice did not develop any microscopic symptoms of intestinal disease, stress, or any other sign of discomfort following microbial colonization, we conclude that the colonization by these two very different bacteria led to a non-inflammatory, commensal interaction and to intestinal tolerance. In two different studies, human mucosal in vivo transcriptomes were obtained after 6 h exposure of the proximal part of the small intestine to the same L. plantarum strain as used in the present study (Troost et al., 2008). Comparison of these human and mouse transcriptomes obtained after colonization by L. plantarum shows that this bacterial isolate induces similar transcriptional changes (including modulation of genes participating in lipid metabolism, a non-inflammatory immune response, and cellular proliferation) in these two mammalian hosts. This is of great interest since mice are frequently used as a model system to gain a better understanding of the function of the intestinal microbiota. Further studies will be aimed at exploration of the cellular processes described here, and at identification of the bacterial products that mediate the contact (direct contact or soluble factors) with host cells in the intestine.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has been carried out with the financial support of the European Community specific RTD program “Quality of Life and Management of Living Resources” research project EU & Microfunction (QKL1-2001-00135). We acknowledge the technicians at the animal facility in Stockholm and Mechteld Grootte Bromhaar for conducting the microarray processing.
Footnotes
Abbreviations
GF, germ-free; IPA, ingenuity pathway input; qRT-PCR, quantitative RT-PCR.
References
- Artis D. (2008). Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8, 411–420 10.1038/nri2316 [DOI] [PubMed] [Google Scholar]
- Backhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., Semenkovich C. F., Gordon J. I. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baken K., Ezendam J., Gremmer E., de Klerk A., Pennings J., Matthee B., Peijnenburg A., van Loveren H. (2006). Evaluation of immunomodulation by Lactobacillus casei Shirota: immune function, autoimmunity and gene expression. Int. J. Food Microbiol. 112, 8–18 10.1016/j.ijfoodmicro.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Bron P., Grangette C., Mercenier A., de Vos W., Kleerebezem M. (2004). Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186, 5721–5729 10.1128/JB.186.23.7829-7835.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona M., Midtvedt T., Norin E. (2001). Probiotics in gnotobiotic mice: short chain fatty acid production in vitro and in vivo. Scand. J. Lab. Anim. Sci. 28, 75–84 [Google Scholar]
- Collado M. C., Derrien M., Isolauri E., de Vos W. M., Salminen S. (2007). Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73, 7767–7770 10.1128/AEM.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L., Johnson C., Burslem F., Martin P. (2004). Wound healing and inflammation genes revealed by array analysis of “macrophage less” PU.1 null mice. Genome Biol. 6, R5. 10.1186/gb-2004-6-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E. K., Gordon J. I., Secor S. M., Knight R. (2010). Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 4, 1375–1385 10.1038/ismej.2010.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings C. A., Relman D. A. (2000). Using DNA microarrays to study host-microbe interactions. Emerging Infect. Dis. 6, 513–525 10.3201/eid0605.000511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Collado M. C., Ben-Amor K., Salminen S., de Vos W. M. (2008). The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 74, 1646–1648 10.1128/AEM.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., van Passel M., van deBovenkamp J., Schipper R., de Vos W. M., Dekker J. (2010). Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1, 1–15 10.4161/gmic.1.4.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Vaughan E. E., Plugge C. M., de Vos W. M. (2004). Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- Di Caro S., Taob S., Grillo A., Elia C., Gasbarrini G., Sepulveda A., Gasbarrini A. (2005). Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig. Liver Dis. 37, 320–329 10.1016/j.dld.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Eckburg P., Bik E., Bernstein C., Purdom E., Dethlefsen L., Sargent M., Gill S., Nelson K., Relman D. A. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638 10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier C. F., Vaughan E. E., De Vos W. M., Akkermans A. D. L. (2002). Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68, 219–226 10.1128/AEM.68.1.219-226.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburgh J. L. (2000). Mini review keratan sulfate: structure, biosynthesis, and function. Glycobiology 10, 951–958 10.1093/glycob/10.10.951 [DOI] [PubMed] [Google Scholar]
- Gustafsson B. E. (1959). Lightweight stainless steel systems for rearing germfree animals. Ann. N. Y. Acad. Sci. 78, 17–28 10.1111/j.1749-6632.1959.tb53092.x [DOI] [PubMed] [Google Scholar]
- Heilig H., Zoetendal E., Vaughan E., Marteau P., Akkermans A., de Vos W. (2002). Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68, 114–123 10.1128/AEM.68.1.114-123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. V., Midtvedt T., Gordon J. I. (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307 10.1146/annurev.nutr.22.011602.092259 [DOI] [PubMed] [Google Scholar]
- Johansson M. E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. C. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 15064–15069 10.1073/pnas.0709958105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagnoff M. F., Eckmann L. (2001). Analysis of host responses to microbial infection using gene expression profiling. Curr. Opin. Microbiol. 4, 246–250 10.1016/S1369-5274(00)00198-3 [DOI] [PubMed] [Google Scholar]
- Kelly D., Conway S., Aminov R. (2005). Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 26, 326–333 10.1016/j.it.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Konstantinov S., Smidt H., de Vos W., Bruijns S., Singh S., Valence F., Molle D., Lortal S., Altermann E., Klaenhammer T., van Kooyk Y. (2008). S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U.S.A. 105, 19474–19479 10.1073/pnas.0810305105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P., Seksik P., Sutren M., de la Cochetiere M.-F., Jian R., Marteau P., Dore J. (2005). Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 1, 473–480 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., Gordon J. I. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Milo M., Lawrence N. D., Rattray M. (2005). A tractable probabilistic model for Affymetrix probe-level analysis across multiple chips. Bioinformatics 21, 3637–3644 10.1093/bioinformatics/bth437 [DOI] [PubMed] [Google Scholar]
- Mackie R., Sghir A., Gaskins H. (1999). Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69, 1035S–1045S [DOI] [PubMed] [Google Scholar]
- Mahowald M. A., Rey F. E., Seedorf H., Turnbaugh P. J., Fulton R. S., Wollam A., Shah N., Wang C., Magrini V., Wilson R. K., Cantarel B. L., Coutinho P. M., Henrissat B., Crock L. W., Russell A., Verberkmoes N. C., Hettich R. L., Gordon J. I. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U.S.A. 106, 5859–5864 10.1073/pnas.0901529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco M. L., Bongers R. S., de Vos W. M., Kleerebezem M. (2007). Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl. Environ. Microbiol. 73, 124–132 10.1128/AEM.01475-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco M. L., de Vries M. C., Wels M., Molenaar D., Mangell P., Ahrne S., de Vos W. M., Vaughan E. E., Kleerebezem M. (2010). Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 4, 1481–1484 10.1038/ismej.2010.61 [DOI] [PubMed] [Google Scholar]
- Mohamadzadeh M., Klaenhammer T. R. (2008). Specific Lactobacillus species differentially activate Toll-like receptors and downstream signals in dendritic cells. Expert Rev. Vaccines 7, 1155–1164 10.1586/14760584.7.2.163 [DOI] [PubMed] [Google Scholar]
- Nielsen D. S., Moller P. L., Rosenfeldt V., Paerregaard A., Michaelsen K. F., Jakobsen M. (2003). Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon. Appl. Environ. Microbiol. 69, 7545–7548 10.1128/AEM.69.2.861-868.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png C. W., Linden S. K., Gilshenan K. S., Zoetendal E. G., McSweeney C. S., Sly L. I., McGuckin M. A., Florin T. H. J. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105, 2420–2428 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- Santacruz A., Collado M. C., García-Valdés L., Segura M. T., Martín-Lagos J. A., Anjos T., Martí-Romero M., Lopez R. M., Florido J., Campoy C., Sanz Y. (2010). Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 104, 83–92 10.1017/S0007114510000176 [DOI] [PubMed] [Google Scholar]
- Sartor M., Tomlinson C., Wesselkamper S., Sivaganesan S., Leikauf G., Medvedovic M. (2006). Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics 7, 538. 10.1186/1471-2105-7-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J., Chen C., Gordon J. (2006). Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4, e413. 10.1371/journal.pbio.0040413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoyama K., Fujiwara R., Takemura N., Ogasawara T., Watanabe J., Ito H., Morita T. (2009). Response of gut microbiota to fasting and hibernation in Syrian hamsters. Appl. Environ. Microbiol. 75, 6451–6456 10.1128/AEM.00692-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck T. S., Hooper L. V., Gordon J. I. (2002). Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U.S.A. 99, 15451–15455 10.1073/pnas.202604299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Dörffel Y., Loening-Baucke V., Theissig F., Rückert J. C., Ismail M., Rau W. A., Gaschler D., Weizenegger M., Kühn S., Schilling J., Dörffel W. V. (2011). Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60, 34–40 10.1136/gut.2011.239301.68 [DOI] [PubMed] [Google Scholar]
- Troost F., van Baarlen P., Lindsey P., Kodde A., de Vos W., Kleerebezem M., Brummer R.-J. (2008). Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics 9, 374. 10.1186/1471-2164-9-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1131 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Ukena S., Westendorf A., Hansen W., Rohde M., Geffers R., Coldewey S., Suerbaum S., Buer J., Gunzer F. (2005). The host response to the probiotic Escherichia coli strain Nissle 1917: specific up-regulation of the proinflammatory chemokine MCP-1. BMC Med. Genet. 6, 43. 10.1186/1471-2156-6-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Baarlen P., Troost F., van Hemert S., van der Meer C., de Vos W., de Groot P., Hooiveld G., Brummer R., Kleerebezem M. (2009). Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. U.S.A. 106, 2371–2376 10.1073/pnas.0809919106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel M. W., Kant R., Zoetendal E. G., Plugge C. M., Derrien M., Malfatti S. A., Chain P. S., Woyke T., Palva A., de Vos W. M., Smidt H. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6, e16876. 10.1371/journal.pone.0016876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Ahrné S., Jeppsson B., Molin G. (2005). Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54, 219. 10.1016/j.femsec.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Wietek C., O’Neill L. (2007). Diversity and regulation in the NF-kappaB system. Trends Biochem. Sci. 32, 311–319 10.1016/j.tibs.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Zoetendal E., Rajilic-Stojanovic M., de Vos W. M. (2008). High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615 10.1136/gut.2007.133603 [DOI] [PubMed] [Google Scholar]
- Zoetendal E. G., von Wright A., Vilpponen-Salmela T., Ben-Amor K., Akkermans A. D. L., de Vos W. M. (2002). Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68, 3401–3407 10.1128/AEM.68.9.4225-4232.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]