Abstract
Translation of genetic information encoded in messenger RNAs into polypeptide sequences is carried out by ribosomes in all organisms. When a full protein is synthesized, a stop codon positioned in the ribosomal A site signals termination of translation and protein release. Translation termination depends on class I release factors. Recently, atomic-resolution crystal structures were determined for bacterial 70S ribosome termination complexes bound with release factors RF1 or RF2. In combination with recent biochemical studies, the structures resolve long-standing questions about translation termination. They bring insights into the mechanisms of recognition of all three stop codons, peptidyl-tRNA hydrolysis, and coordination of stop-codon recognition with peptidyl-tRNA hydrolysis. In this review, the structural aspects of these mechanisms are discussed.
Keywords: 70S ribosome, RF1, RF2, crystal structures, release factors
INTRODUCTION
Ribosomes are the cellular machines responsible for translating genetic information into polypeptide sequences. During the step-wise movement of a messenger RNA through the ribosome, amino acids are incorporated into the elongating polypeptide chain. The fidelity of amino acids incorporation depends on base-pair complementarities between sense codons and anticodons of aminoacyl-tRNAs. A critical event in the translation of each protein is termination, which determines the length of a protein. Translation termination is encoded by nonsense, or stop, codons (Brenner et al. 1965, 1967). In the majority of species, three out of 64 codons (UAA, UAG, and UGA) are used to signal translation termination. Unlike recognition of sense codons, stop-codon recognition does not depend on tRNAs. Instead, termination of translation is aided by proteins called class I release factors (RF) (Capecchi 1967; Vogel et al. 1969; Caskey et al. 1971). In eubacteria, translation termination is mediated by release factor RF1 in response to a UAG or UAA stop codon, and by RF2 in response to a UGA or UAA codon. RF1 and RF2 sequences are homologous (Nakamura et al. 1995; Ito et al. 1996); their three-dimensional structures are similar to each other (Vestergaard et al. 2001; Shin et al. 2004). In eukaryotes and archaea, translation termination at all three stop codons is catalyzed by eRF1 and aRF1, respectively. eRF1 and aRF1 are homologous to each other but share little sequence homology with their bacterial counterparts (Frolova et al. 1994; Dontsova et al. 2000; Song et al. 2000). Mitochondrial protein mtRF1L (mtRF1a/HMRF1L), a homolog of bacterial RF1, was recently identified as a release factor mediating peptide release in response to stop codons UAA and UAG in mitochondria (Soleimanpour-Lichaei et al. 2007; Nozaki et al. 2008). The function of a second tentative mitochondrial release factor mtRF1 and the use of AGA and AGG triplets as reassigned stop codons in mitochondria are currently being debated (Nozaki et al. 2008; Temperley et al. 2010; Young et al. 2010a). In addition to class I release factors, bacterial and eukaryotic translation termination systems involve class II release factors, RF3 and eRF3, respectively. Unlike class I release factors, they belong to the GTPase family and act by assisting class I release factors in a GTP-dependent manner. Bacterial RF3 was shown to promote dissociation of RF1 or RF2 from the ribosome upon peptidyl-tRNA hydrolysis (Freistroffer et al. 1997). The role of eRF3 is less understood. Unlike its bacterial counterpart, eRF3 forms a stable complex with class I release factor eRF1 (Stansfield et al. 1995; Zhouravleva et al. 1995; Frolova et al. 1996) and is hypothesized to be involved in a termination proofreading mechanism in eukaryotes (Buckingham et al. 1997; Salas-Marco and Bedwell 2004), to stimulate the efficiency of eRF1 (Zhouravleva et al. 1995; Mitkevich et al. 2006) and to promote eRF1 recycling (Zavialov et al. 2001). eRF3 is essential for cell viability (Kushnirov et al. 1988; Wilson and Culbertson 1988), while RF3 is not (Grentzmann et al. 1994; Mikuni et al. 1994).
There are parallels and differences between the functions of class I release factors and aminoacyl-tRNAs. The functions of both macromolecules depend on recognition of specific codons placed in the A site of the small ribosomal subunit. Codon recognition results in a nucleophilic reaction in the peptidyl-transferase center (PTC) of the large ribosomal subunit, namely, the transpeptidation reaction in the case of sense-codon recognition by an aminoacyl-tRNA, or hydrolysis of peptidyl-tRNA upon RF-mediated recognition of a stop codon. On the mechanistic level, however, peptide release promoted by release factors is markedly different from polypeptide chain elongation. The discovery of protein release factors implied that the principle of codon recognition via base-pairing may be violated, unless stop codons were recognized by base-pairing with ribosomal RNA, as was suggested by several studies (Dalgarno and Shine 1973; Shine and Dalgarno 1974; Arkov and Murgola 1999; Ivanov et al. 2001). Photocrosslinking studies revealed that release factors bind in the vicinity of stop codons (Tate et al. 1990; Brown and Tate 1994). These results were consistent with the idea that an “RF anticodon,” rather than a complementary rRNA trinucleotide, can be used to decipher stop codons (Caskey 1980). On the basis of mutational studies, the PxT and SPF motifs, conserved in RF1 and RF2, respectively, were proposed to serve as “tripeptide anticodons” (Ito et al. 2000). Amino acids located in other regions of release factors, however, were also proposed to be involved in stop-codon recognition (Ito et al. 1998). Cryo-EM and ∼6 Å X-ray studies of translation termination 70S complexes (Klaholz et al. 2003; Rawat et al. 2003, 2006 Petry et al. 2005) have demonstrated that RF1 or RF2 bind to the ribosomal A site and that their PxT or SPF motifs, respectively, are positioned to interact with the stop codons. The mechanism of stop- codon recognition, however, remained unclear.
Another aspect in which translation termination differs from elongation is that, upon stop-codon recognition, the ester bond of peptidyl-tRNA is cleaved by hydrolysis. In contrast, the hydrolysis of peptidyl-tRNAs during elongation must be avoided to prevent formation of incompletely translated proteins. Early proposals regarding the mechanism of the hydrolysis reaction posed that, during termination, the ribosomal peptidyl-transferase center is “perturbed” or converted into an esterase (Tompkins et al. 1970; Caskey et al. 1971; Caskey 1980) to allow the cleavage of the peptidyl-tRNA. Subsequent studies implicated the direct involvement of release factors in the hydrolysis reaction. The universally conserved GGQ motif was identified as critical for the function of both bacterial and eukaryotic release factors. Mutations of glycine residues in the GGQ motif rendered release factors inactive (Frolova et al. 1999; Shaw and Green 2007). The universal conservation of the glutamine of the GGQ motif invited proposals that its side chain is critical for positioning and/or activation of a water molecule for nucleophilic attack (Song et al. 2000; Trobro and Aqvist 2007, 2009). Mutational studies, however, demonstrated that substitutions of the glutamine with bulkier or shorter side chains lead to only modest defects in peptide release (Dincbas-Renqvist et al. 2000; Seit Nebi et al. 2000; Seit-Nebi et al. 2001; Shaw and Green 2007; Korostelev et al. 2008), indicating that the side chain group is not required for catalysis. In the cryo-EM and low-resolution X-ray structures, the GGQ motif is bound in the peptidyl-transferase center (Klaholz et al. 2003; Rawat et al. 2003, 2006 Petry et al. 2005). Due to insufficient resolution of the structures, the role of this interaction remained unclear.
The third aspect in which the elongation and translation termination mechanisms differ involves the use of proofreading for achieving a low frequency of errors. Selection of cognate aminoacyl-tRNAs by the ribosome depends on an error-correction mechanism, which includes an irreversible GTP hydrolysis step (Thompson 1988; Rodnina et al. 1996). In contrast, the accuracy of stop-codon recognition by bacterial class I release factors was shown to be independent of proofreading (Freistroffer et al. 2000).
Following more than four decades of research on translation termination, three fundamental questions remained to be answered: (1) What is the mechanism of stop-codon recognition? Are stop-codon nucleotides recognized directly by the release factors, by the ribosome, or by some combination of the two (Youngman et al. 2007)? (2) What is the mechanism of hydrolysis of the peptidyl-tRNA ester linkage? Are release factors directly involved in catalysis, or do they convert the ribosomal peptidyl-transferase center to an esterase? (3) How, in the absence of proofreading, is premature termination kept to low levels similar to or even lower than those of amino acid misincorporation (Jorgensen et al. 1993; Freistroffer et al. 2000; Wohlgemuth et al. 2010)?
Recently, crystal structures of Thermus thermophilus 70S ribosome complexes bound with RF1 in response to UAA or UAG codons and RF2 in response to UAA or UGA codons were determined at 3.0–3.6 Å resolution (Korostelev et al. 2008, 2010; Laurberg et al. 2008; Weixlbaumer et al. 2008). The structures of these complexes, formed in the presence of deacylated tRNAs, and the structure of RF2 bound to the 70S ribosome in the presence of a peptidyl-tRNA analog (Jin et al. 2010) represent the product and substrate states, respectively, of the peptidyl-tRNA ester bond hydrolysis reaction. They provide insights into the mechanisms of recognition of all three stop codons, peptidyl-tRNA hydrolysis, and coordination of stop-codon recognition with ester bond hydrolysis. The structural aspects of these mechanisms that have emerged from the recent crystallographic and biochemical studies are discussed in this review.
POSITION AND CONFORMATION OF RELEASE FACTOR BOUND TO 70S RIBOSOME
Release factors bind to the A site on the 70S ribosome (Korostelev et al. 2008; Laurberg et al. 2008; Weixlbaumer et al. 2008). The binding of a release factor does not induce large-scale conformational changes on the ribosome, as the overall conformation of the ribosome in translation termination complexes is analogous to that of a 70S complex in which the A site is vacant (Jenner et al. 2010). The structural similarity of the 70S structures bound with RF1 in the presence and in the absence of E-site tRNA (Laurberg et al. 2008; Korostelev et al. 2010) suggests that occupancy of the ribosomal E site is not likely to affect the hydrolysis of peptidyl-tRNA mediated by release factors.
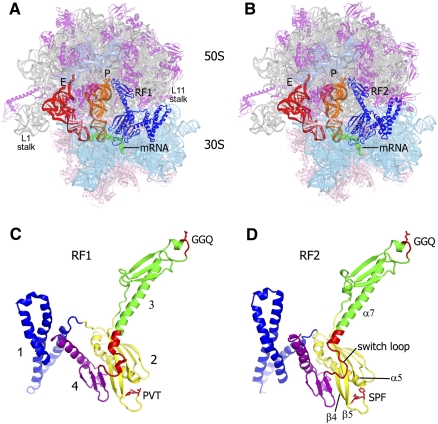
The shapes of release factors are similar in the RF1- and RF2-bound termination complexes (Fig. 1) but very different from the crystal structures of the isolated factors. Bacterial release factors consist of four domains (Fig. 1C,D). Domain 1 of a release factor is bound in the vicinity of the ribosomal GTPase-associated center. In the RF2-bound structures of the 70S termination complexes (Korostelev et al. 2008; Weixlbaumer et al. 2008), domain 1 interacts with the L11 stalk; there is no such interaction in the RF1-bound structures (Korostelev et al. 2008; Laurberg et al. 2008). This structural dissimilarity is consistent with functional differences between RF1- and RF2-mediated peptide release from L11-depleted ribosomes observed in vitro by Boukaz et al. (2006); in another study, however, mutations or deletion of L11 were shown to equally reduce the activities of RF1 and RF2 (Sato et al. 2006). The functional importance of the structural difference between RF1- and RF2-bound 70S complexes in the L11 region, therefore, remains to be explored.
FIGURE 1.
Crystal structures of the 70S translation termination complexes bound with RF1 and RF2. (A,B) 3.2-Å and 3.0-Å structures of the 70S termination complexes bound with RF1 and RF2 (blue) in response to the UAA stop codon (mRNA is shown in green) and in the presence of deacylated P- (orange) and E-site (red) tRNAs. 23S rRNA is shown in gray, 5S rRNA in teal, 50S subunit proteins in magenta, 16S rRNA in cyan, and 30S subunit proteins in pink. (C,D) The structures of RF1 and RF2 in their ribosome-bound conformation, rotated ∼180° from A and B; the structures are colored according to their four-domain organization; GGQ, PVT, and SPF motifs and the switch loop are shown in red.
Domains 2 and 4 form a compact superdomain (Fig. 1C,D), which binds to the 30S subunit and is involved in stop-codon recognition. Domain 3 spans between the functional centers of the small and large ribosomal subunit; helix α7 of domain 3 bridges the decoding head of the release factor with the universally conserved GGQ motif implicated in the catalysis of peptidyl-tRNA in the peptidyl-transferase center (Fig. 1).
RECOGNITION OF STOP CODONS BY CLASS I RELEASE FACTORS
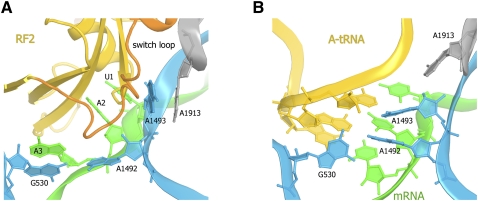
Atomic-resolution structures of RF1 and RF2 bound to the 70S ribosome reveal that stop codons are recognized directly by release factors. Similarly to the base-pairing mechanism of sense codons, the recognition of stop codons is achieved through a network of hydrogen bonds between mRNA nucleotides and the reading head of domain 2 of either release factor. Unlike tRNAs, release factors interact not only with the Watson-Crick edges but also with Hoogsteen edges of codon nucleotides. The codon conformation in RF-bound complexes (Fig. 2A) differs from that in A-site tRNA bound complexes (Fig. 2B) in that the third nucleotide of a stop codon is unstacked from the first two codons and is recognized by a release factor separately from the first two nucleotides. The conformation and role of the ribosomal decoding center in the translation termination complexes are distinct from those in elongation complexes. The universally conserved nucleotides G530, A1492, and A1493 of 16S rRNA, which were shown to participate in sense-codon discrimination by aminoacyl-tRNAs (Ogle et al. 2001), are not directly involved in stop-codon recognition (Fig. 2). Instead, they participate in stabilizing the catalytically active conformation of the release factor and thus appear to be involved in the coordination of stop-codon recognition with peptidyl-tRNA hydrolysis (discussed in a separate section of this review).
FIGURE 2.
Differences in the conformations of the decoding center between 70S ribosome complexes bound with (A) RF2 (Korostelev et al. 2008) and (B) a tRNA cognate to the A-site codon (Selmer et al. 2006). RF2 and tRNA are shown in yellow, the switch loop of a release factor is shown in orange, 23S rRNA nucleotide A1913 is shown in gray, 16S rRNA nucleotides G530, A1492, and A1493 are shown in cyan, and mRNA is shown in green.
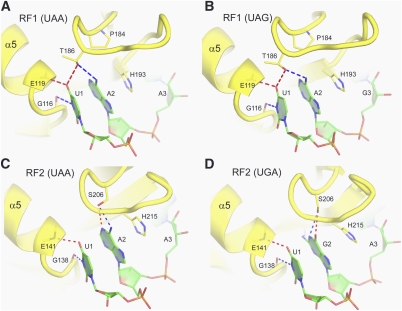
More than a dozen amino acids of the release factor are positioned to examine the A-site codon. The codon reading head of the release factor comprises the N-terminal end of helix α5 and the conserved recognition loop [aa 181–195 (185–199) and 201–217 (200–216) of T. thermophilus (Escherichia coli) RF1 and RF2, respectively] formed between the β4 and β5 strands of the central β-sheet of domain 2 (Figs. 1C,D, 3).
FIGURE 3.
Interactions of the first two stop-codon nucleotides with release factors RF1 and RF2. (A) Recognition of U1 and A2 of the UAA codon by RF1 (Laurberg et al. 2008) and (B) the UAG codon by RF1 (Korostelev et al. 2010). (C) Recognition of U1 and A2 of the UAA codon by RF2 (Korostelev et al. 2008). (D) Recognition of U1 and G2 of the UGA codon by RF2 (Weixlbaumer et al. 2008). Release factors are shown in yellow, and mRNA is shown in green. (Adapted, with modifications, from Korostelev et al. 2010.)
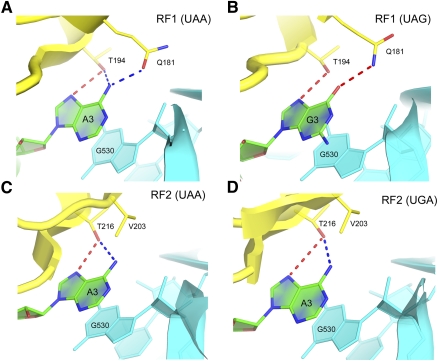
Three elements of the reading head are responsible for recognition of the three stop-codon nucleotides. The nucleotides of a stop codon will be referred to as U1, A2 or G2, and A3 or G3. (1) The N-terminal tip of helix α5 recognizes U1 via formation of specific H-bonds from the backbone of α5 (Fig. 3). Notably, the hydrogen-bonding pattern between the polypeptide backbone and U1 resembles that of a canonical A:U Watson-Crick base-pairing interaction. This interaction, identical in the cases of both RF1 (Fig. 3A,B) and RF2 (Fig. 3C,D), accounts for the universal occurrence of U in the first position of all three stop codons. (2) Conserved amino acids of the recognition loop, including the PxT and SPF motifs of RF1 and RF2, respectively, define the specificity of release factors for the second nucleotide. Release factor specificity relies on the side chains of Thr186 (190 in E. coli) of the PxT motif in RF1 and Ser206 (205) of the SPF motif in RF2. The β-hydroxyl group of Thr186 hydrogen bonds with the first and second bases of the stop codon (Fig. 3A,B); by donating a hydrogen bond to the 4-keto oxygen of U1, the hydroxyl group of Thr186 is, therefore, constrained to accept a hydrogen bond from the N6-amino group of A2, defining the specificity of RF1 for adenine at the second position (Laurberg et al. 2008; Korostelev et al. 2010). Unlike Thr186 of RF1, the side chain of Ser206 of RF2 does not interact with U1 but is positioned to hydrogen-bond with the Watson-Crick face of either A or G at the second position of a stop codon (Fig. 3C,D). Thr186 of the PVT motif and Ser206 of the SPF motif, however, do not appear to be the sole determinants of release factors specificity toward the second nucleotide. Substituting the PxT motif of RF1 with the SPF motif of RF2 fails to modify RF1 specificity but instead renders RF1 inactive with any of the stop codons (Young et al. 2010b). In contrast, substitution of the whole 13-residue recognition loop of RF1 with that from RF2 rendered RF1 active in response to either A or G the second position of a stop codon. These results demonstrate that stop-codon specificity in this region is achieved through the network of interactions provided not only by the PxT and SPF motifs but also by other elements of the recognition loop. The latter likely include Glu187 (RF1) and Asp209 (RF2) (Young et al. 2010b), which are positioned to interact with the Hoogsteen face of the second nucleotide. (3) The N- and C-terminal ends of the recognition loop define the specificity for the third nucleotide located in the G530 pocket. Recognition of the third stop-codon nucleotide by both RF1 and RF2 occurs separately from the first two nucleotides, in the G530 pocket of the decoding center (Fig. 2A). A3 or G3 is sandwiched between G530 of 16S rRNA and Ile192 of RF1 or Arg214 of RF2. In both release factors, the universally conserved threonine (Thr194 in RF1 and Thr216 in RF2) at the C-terminal end of the recognition loop is positioned to examine the Hoogsteen edge of the third stop-codon nucleotide. Its side-chain hydroxyl group donates a hydrogen bond to N7 of the purine ring of A3 or G3 and is also able to accept a hydrogen bond from the N6 amino group of A3 (Fig. 4). Gln181, which is conserved at the N-terminal end of the recognition loop in all RF1 sequences, H-bonds with the 6-position of the third codon base. By flipping the amide group of the Gln181 side chain, RF1 can accept an H-bond from the N6-amino group of adenine or donate a hydrogen bond to the O6-keto group of guanine (Fig. 4A,B), thus explaining the ability of RF1 to recognize either A or G in the third position. In RF2, which specifically recognizes A3, a hydrophobic residue is located in the place of Gln181 (Fig. 4C,D). Since the position of the hydrophobic side chain (Val203 in T. thermophilus RF2) would prevent H-bonding of the O6 of guanine to water, discrimination against guanine in the third position is likely achieved, at least in part, as a result of a free-energy penalty due to desolvation of guanine (Korostelev et al. 2008).
FIGURE 4.
Interactions of the third stop-codon nucleotide in the 70S translation termination complexes bound with RF1 and RF2. Recognition of the third nucleotide takes place in a G530 pocket of the decoding center. (A,B) Rotation of the conserved Gln181 side chain of RF1 allows it to form hydrogen bonds with either A3 or G3 of the UAA or UAG codons, respectively (Laurberg et al. 2008; Korostelev et al. 2010). (C,D) In RF2, Gln181 is replaced by Val203, restricting the recognition specificity of RF2 to A3. Release factors are shown in yellow, mRNA is shown in green, and 16S rRNA in cyan. (Adapted, with modifications, from Korostelev et al. 2010.)
RELEASE FACTORS DIRECTLY PARTICIPATE IN CATALYSIS OF PEPTIDYL-tRNA HYDROLYSIS
During protein synthesis, a peptidyl-tRNA is protected from hydrolysis in a compact pocket of the peptidyl-transferase center (Schmeing et al. 2005b). Upon recognition of a stop codon by a release factor, the ester bond bridging the peptidyl moiety with the terminal nucleotide A76 of peptidyl-tRNA is hydrolyzed. The main questions concerning the peptidyl-tRNA hydrolysis are how release factors promote the entry of a water molecule into the PTC and whether release factors contribute to catalysis directly by interacting with the substrates, transition-state intermediate, and/or products of the reaction.
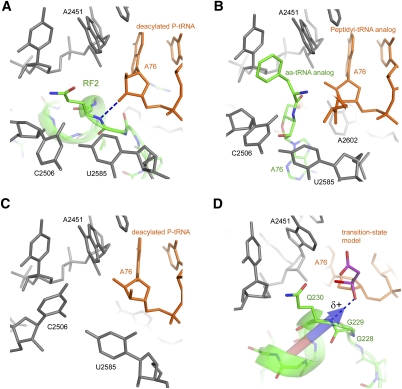
Crystal structures of 70S translation termination complexes (Korostelev et al. 2008, 2010; Laurberg et al. 2008; Weixlbaumer et al. 2008; Jin et al. 2010) reveal that the tip of domain 3 “crowned” with the conserved GGQ motif is inserted into the PTC and contacts the nucleotides of 23S rRNA and P-site tRNA (Fig. 5). The conformations of 23S rRNA nucleotides U2506 and U2585, which are sensitive to the occupancies of the 50S A and P sites (Schmeing et al. 2005b), are similar to those observed in ribosome complexes, in which the A-site aminoacyl-binding pocket is occupied. Namely, U2506 and U2585 are retracted from the A-site binding pocket when the pocket is occupied by the release factor, aminoacyl-tRNA (Voorhees et al. 2009), or a transition-state analog (Schmeing et al. 2005a). The position of the side chain of the GGQ motif glutamine overlaps with that of the ester and aminoacyl groups of an aminoacyl-tRNA bound to the A site (Fig. 5A,B). The similarity between the tightly packed PTC conformations of the RF- and A-site-tRNA-bound complexes suggests two possible scenarios to allow entry of a water molecule into the PTC upon release factor binding. One possibility is that a unique conformation of the peptidyl-transferase center exists transiently in the course of domain 3 accommodation, in which the PTC is opened and the passage of a water molecule from solution is allowed. There is an alternative scenario. Examination of high-resolution crystal structures of unbound release factors (Song et al. 2000; Vestergaard et al. 2001) reveals that ordered water molecules are bound to the GGQ region. It is, therefore, possible that a stably bound water molecule is transported into the PTC with the release factor. In contrast, such transport by an aminoacyl-tRNA is unlikely since its ester bond is protected from interaction with water by EF-Tu (Nissen et al. 1995).
FIGURE 5.
Conformation of the peptidyl-transferase center depends on the occupancy of the A site. The conformations of the PTC are similar in ribosome complexes, in which the A site is occupied by (A) a release factor (Korostelev et al. 2008) and (B) aminoacyl-tRNA (Voorhees et al. 2009). (C) C2506 and U2585 are found in a different conformation when the 50S subunit A-site is vacant (Selmer et al. 2006). (D) Superposition of a peptidyl-transferase transition-state analog complexed with the 50S subunit (Schmeing et al. 2005a) on the structure of the 70S termination complex (Laurberg et al. 2008). The main-chain amide of Gln230 is positioned to H-bond with the oxyanion of the transition state. The dipole moment of the a-helix likely increases the partial positive charge on the Gln230 backbone NH group and the ability of the NH group to stabilize the negatively charged transition-state intermediate. The dipole moment vector was calculated at Protein Dipole Moments Server (Felder et al. 2007).
The recent biochemical and crystallographic studies suggest that release factors directly participate in catalysis of peptidyl-tRNA hydrolysis. The universally conserved GGQ motif of release factors has been implicated in catalysis of the hydrolysis reaction by multiple studies (Frolova et al. 1999; Seit-Nebi et al. 2001; Mora et al. 2003; Shaw and Green 2007; Trobro and Aqvist 2007). In some studies, the side chain of the glutamine (Gln230 in T. thermophilus RF1 or Gln235 in E.coli RF1) was proposed to be directly involved in catalysis (Song et al. 2000; Trobro and Aqvist 2007, 2009). In the crystal structures of release factors bound to the ribosome in the presence of a deacylated P-site tRNA, representing a product of the hydrolysis reaction, the glutamine side chain points away from the 3′-OH of ribose 76, the likely position of the scissile bond (Korostelev et al. 2008; Laurberg et al. 2008). In the structure of RF2 bound to the ribosome in complex with an aminoacyl-tRNA analog, representing a substrate-bound state, the side chain amide group of the glutamine is placed >4 Å away from the putative nucleophilic water molecule (Jin et al. 2010), in disagreement with the proposal that the glutamine side chain is critical for orienting a water molecule for a nucleophilic attack (Song et al. 2000). These observations, together with the results of mutational and biochemical studies, which showed that only modest peptide release defects are conferred by some mutants of Gln230 (Seit Nebi et al. 2000; Seit-Nebi et al. 2001; Shaw and Green 2007), allow the possibility of direct participation of the glutamine side chain in catalysis to be ruled out. Instead, the side chain likely defines release-factor specificity for water by excluding nucleophiles other than water from the reaction (Shaw and Green 2007) and optimizes the binding affinity of release factors. Methylation of the glutamine at the δ-N position, which was proposed to be important for the efficiency of translation termination (Dincbas-Renqvist et al. 2000; Mora et al. 2007), also appears to contribute to the affinity of release factor binding to the ribosome rather than to the catalytic activity of release factors (discussed in Korostelev et al. 2010).
Unlike the side chain of the glutamine, its backbone amide NH group is positioned to participate in catalysis via hydrogen bonding to the leaving 3′-hydroxyl group of A76 of the P-site tRNA (Laurberg et al. 2008). Superposition of the 70S-RF1 complex structure with that of a peptidyl-transferase transition-state analog complexed with the 50S subunit (Schmeing et al. 2005a) suggests that the oxyanion of the transition-state tetrahedral intermediate can also be stabilized by a hydrogen bond with the backbone amide group of Gln230 (Laurberg et al. 2008). Interestingly, the backbone amide group is located at the tip of a short α-helical element, whose dipole moment likely increases the partial positive charge on the hydrogen atom, further emphasizing the possible role of this group in stabilizing the negative charge of the transition state oxyanion (Fig. 5D).
Participation of the backbone amide group in catalysis is not unprecedented; proteases, esterases, and GTPases have been shown to employ backbone amide NH groups for transition-state and/or leaving group stabilization (Maegley et al. 1996; Jaeger et al. 1999; Wilmouth et al. 2001). Substitution of Gln230 with proline, in which the hydrogen-bonding ability of the backbone NH group is eliminated, results in complete loss of peptide release activity (Korostelev et al. 2008). Since binding to the ribosome was not compromised by the proline substitution and since the proline substitution in the case of Gln230 is unlikely to perturb the conformations of the release factor or PTC (Korostelev et al. 2008), the loss of peptide release activity is likely due to loss of the backbone NH group. Together, the structural and biochemical data strongly suggest that the backbone NH group of the glutamine of the GGQ motif plays a catalytic role via stabilization of the leaving group and/or the tetrahedral transition-state intermediate of the hydrolysis reaction. The crystal structures allow rationalization of the requirement for the two neighboring glycine residues of the GGQ motif, mutations of which confer up to 104-fold slower rates of peptide release (Frolova et al. 1999; Shaw and Green 2007). The glycines adopt backbone conformations disallowed for larger amino acids, resulting in optimal placement of the catalytic NH group proximal to the leaving group and/or the transition-state intermediate. Substitution of any of the glycines with a larger amino acid would affect the polypeptide backbone conformation and change the position of the backbone amide group in the active site (Laurberg et al. 2008), in keeping with the inactivation of release factors by corresponding mutations (Frolova et al. 1999; Shaw and Green 2007).
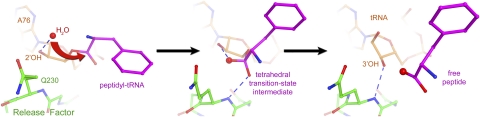
In order to assess the role of the ribosomal PTC in the hydrolysis reaction, it is worth taking into account the analogy between peptidyl-tRNA hydrolysis and transpeptidation. In both reactions, the carbonyl carbon atom of the ester bond of a peptidyl-tRNA in the P site is attacked by a nucleophile. In the case of transpeptidation, the nucleophile is represented by the α-amino group of the aminoacyl-tRNA located in the A site, while, in the case of translation termination, the ester bond is attacked by a water molecule. It is, therefore, reasonable to suggest that the peptidyl-tRNA hydrolysis reaction employs a proton shuttle mechanism (Dorner et al. 2002, 2003; Weinger et al. 2004), according to which the 2′-OH group of A76 of the P-site tRNA facilitates proton transfer to the 3′-OH group of the same nucleotide during the peptidyl transfer reaction (for review, see Rodnina et al. 2006). In fact, the 2′-hydroxyl of A76 of peptidyl-tRNA was shown to be essential for the peptidyl-tRNA hydrolysis reaction (Brunelle et al. 2008; Zaher et al. 2011). The crystal structures of the translation termination complexes support the proton shuttle model (Jin et al. 2010; Korostelev et al. 2010) and suggest that the 2′-hydroxyl group of A76 is positioned to participate in catalysis by interacting with the nucleophilic water molecule and likely with the transition state (Fig. 6). A high-resolution structure of a ribosome-bound transition-state analog of the peptidyl-tRNA hydrolysis reaction in the presence of a release factor is required to fill this gap in our structural understanding of the reaction pathway.
FIGURE 6.
Scheme for the mechanism of the peptidyl-tRNA hydrolysis reaction. (Left panel) Nucleophilic attack. The nucleophilic water molecule is positioned for nucleophilic attack at H-bonding distance of the 2′-OH of ribose 76 of the peptidyl-tRNA (Jin et al. 2010). (Center panel) Transition-state stabilization. The oxyanion of the developing tetrahedral transition state is stabilized by the hydrogen bond with the backbone amide NH group of Gln230. (Right panel) Product stabilization. Following hydrolysis, the 3′-hydroxyl leaving group of the deacylated P-site tRNA H-bonds with the backbone amide NH group of Gln230. Crystal structures of RF2-bound 70S complexes in the presence of a peptidyl-tRNA analog (Jin et al. 2010) and deacylated tRNA (Korostelev et al. 2008) were used in the left and right panels, respectively. The transition state (center panel) was modeled by superimposing the 23S rRNA structure of the RF2 termination complex (Korostelev et al. 2008) with the structure of a 50S subunit containing a transition-state analog (Schmeing et al. 2005a). (Adapted, with modifications, from Korostelev et al. 2010.)
In summary, the mechanism of peptidyl-tRNA hydrolysis resembles that of transpeptidation in that the 2′-OH group of A76 directly participates in catalysis. The critical difference between the two mechanisms is that, unlike the peptidyl transfer reaction, peptidyl-tRNA hydrolysis is dependent on direct participation of a protein, which provides a catalytic backbone amide group of the glutamine from the GGQ motif. The universal conservation of the GGQ motif suggests that the catalytic mechanism employed by class I release factors is conserved among bacterial, mitochondrial, archaeal, and eukaryotic translation systems.
STOP-CODON RECOGNITION AND PEPTIDYL-tRNA HYDROLYSIS ARE COORDINATED VIA CONFORMATIONAL SWITCH IN RELEASE FACTOR
Class I release factors are high-fidelity enzymes. While translation termination in bacteria does not involve proofreading (Freistroffer et al. 2000), the error rates of peptide release (10−3–10−6) are comparable to those of sense-codon decoding (Jorgensen et al. 1993; Freistroffer et al. 2000; Wohlgemuth et al. 2010). In order to achieve this low error frequency, hydrolysis of peptidyl-tRNA has to be strictly coordinated with stop-codon recognition. This coordination is likely accomplished by preventing the docking of domain 3 into the peptidyl-transferase center prior to recognition of a stop codon. In this model, a release factor initially interacts with the ribosome in a catalytically inactive conformation. Upon stop-codon recognition, a conformational change would occur resulting in the docking of the GGQ motif into the PTC (Fig. 7). The idea that release factors bind to the ribosome in a catalytically inactive state is supported by structural and molecular dynamics studies of free RFs. RF1 and RF2 crystallize in a compact conformation (Vestergaard et al. 2001; Shin et al. 2004) in which the distance between the codon-reading head and GGQ motif is ∼50 Å shorter than that in the 70S-RF complexes. As SAXS studies demonstrate, release factors sample “compact” and “open” conformations in solution, possibly corresponding to those release factors adopt upon crystallization in the free and ribosome-bound states, respectively (Vestergaard et al. 2005; Zoldak et al. 2007). A molecular-dynamics study, however, suggests that the stable “open” conformation of RF2 in solution would be catalytically inactive since the distance between the SPF and GGQ motifs is ∼20 Å shorter than that found in the translation-termination complexes (Ma and Nussinov 2004).
FIGURE 7.
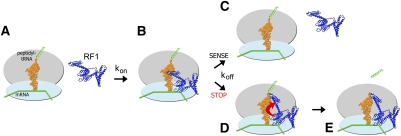
Proposed mechanism for coordination of peptidyl-tRNA hydrolysis with stop-codon recognition via a conformational switch in class I release factors. (A,B) Initially, the release factor binds to the ribosome in a catalytically inactive conformation. (C) If a sense codon is located in the A site, the release factor quickly dissociates (Hetrick et al. 2009). (D) If the release factor recognizes a stop codon in the A site, its switch loop along with domain 3 and the decoding center rearrange. Interaction between the switch loop and the switch-loop binding pocket in the decoding center results in tight binding of the release factor to the ribosome. (E) In this catalytically competent conformation, the GGQ motif is inserted in the peptidyl-transferase center and is capable of contributing to catalysis of peptidyl-tRNA ester bond hydrolysis.
By comparing crystal structures of unbound release factors with those bound to the 70S ribosome, Laurberg et al. (2008) proposed that the loop connecting domains 3 and 4 of the release factors functions as a switch between the inactive and catalytically competent conformations of the protein. The switch loop packs in a pocket in the decoding center formed by ribosomal protein S12 and the universally conserved nucleotides A1492 and A1493 of helix 44 of 16S rRNA and A1913 of helix 69 of 23S rRNA (Fig. 2). The formation of this switch-loop binding pocket is coupled with conformational rearrangements in the decoding center upon recognition of a stop codon by either RF1 or RF2 (Korostelev et al. 2008, 2010; Laurberg et al. 2008; Weixlbaumer et al. 2008). Thus, interaction of the switch loop with the decoding center of a ribosome bearing a stop codon in the A site stabilizes the catalytic conformation of the release factor. This interaction likely serves to coordinate peptidyl-tRNA cleavage with stop-codon recognition.
Recent biochemical and mutational studies emphasize the importance of the conformational switch for translation termination fidelity. By using hydroxyl-radical probing, He and Green (2010) have shown that the reading head of RF1 contacts the decoding center of the ribosome bearing either a stop or sense codon. The hydroxyl-radical-generated cleavage of 23S rRNA in the vicinity of the GGQ motif, however, was more pronounced in the case of stop-codon complexes rather than sense-codon complexes, suggesting that, in the case of a sense-codon ribosome complex, domain 3 would be mispositioned. Consistent with these results, a mutational study revealed that disruption of the interaction between the switch loop and helix 69 of 23S rRNA, which is not part of the codon-recognition interaction, results in a more than 103-fold decrease in the rate of peptide release (Korostelev et al. 2010). Hetrick and colleagues (Hetrick et al. 2009) demonstrated that kon rates, with which RF1 associates with the ribosomes, are similar, whether a stop or sense codon is present in the A site. In contrast, koff rates, with which the release factor dissociates from the ribosome, are several orders of magnitude lower in the case of stop codons. While the fluorescence-based assay used in the study did not reveal additional steps in the binding kinetics of RF1, these dramatic differences are consistent with the idea that, upon stop-codon recognition, the release factor undergoes conformational rearrangements resulting in an increased affinity of RF binding to the ribosome.
EVOLUTIONARY INSIGHTS FROM MECHANISM OF TRANSLATION TERMINATION
Ribosomal RNA forms the core of the ribosome. The nucleotide sequences and three-dimensional structural organization of rRNA are conserved among organisms of all kingdoms of life (Woese et al. 1975; Gutell et al. 1985; Ban et al. 2000; Wimberly et al. 2000; Yusupov et al. 2001; Schuwirth et al. 2005; Ben-Shem et al. 2010). In contrast, most ribosomal proteins are located at the periphery of the ribosome; their amino acid sequences and tertiary structures are more divergent. These and other considerations invited the proposal that ribosomal proteins have evolved subsequently to rRNA, resulting in the fine-tuning of a translational RNA-based apparatus (Noller and Woese 1981; Noller 1999, 2004, 2006; Klein et al. 2004; Bokov and Steinberg 2009). Interestingly, the release factors appear to have evolved to completely substitute, rather than to fine-tune, an RNA-based peptide release machinery of an early RNA World. The following clues suggest that the bacterial and eukaryotic translation termination mechanisms have developed independently and replaced an RNA-based mechanism of translation termination as discussed below: (1) the ability of deacylated tRNAs to hydrolyze peptidyl-tRNAs on the ribosome in a codon-dependent manner; (2) independent evolution of class I release factors; and (3) the involvement of the polypeptide backbone of the release factors in the mechanism of peptide release.
Taking into account the mechanistic similarity between the translation termination and elongation processes, it is tempting to think of a “release RNA,” which could have functioned in an early RNA World translation apparatus. In fact, deacylated tRNAs have been shown to promote peptide release in a codon-dependent manner (Caskey et al. 1971), albeit at lower rates than those observed for release factors (Zavialov et al. 2002). Interestingly, the 3′-OH group of A76 of a deacylated tRNA bound to the A site nearly superimposes with the backbone NH group of the glutamine from the GGQ motif of the release factors (Laurberg et al. 2008). This suggests the involvement of the 3′-hydroxyl or a water molecule coordinated by the 3′-hydroxyl in a catalytic mechanism for peptide release similar to that mediated by class I release factors (Laurberg et al. 2008; Simonović and Steitz 2008). A potential disadvantage of employing such release RNAs in early translation systems could have been the inability to control against a high frequency of premature termination; since deacylated tRNAs do not bind to EF-Tu, the GTP-dependent proofreading mechanism, which greatly increases the fidelity of the aminoacyl-tRNA selection process (Pape et al. 1999; Gromadski and Rodnina 2004), could not be brought into play. The low efficiency and accuracy of “release RNAs” could thus have forced the evolution of alternative translation termination catalysts (Noller 2010), resulting in the robust protein-based translation termination systems present today.
The mechanism of bacterial translation termination substantially differs from that in eukaryotes. While the assignment of stop codons is the same in bacterial and eukaryotic organisms, there is no obvious sequence and structural conservation between bacterial and eukaryotic class I release factors. The overall domain organization and shape of bacterial release factors differ from those of their eukaryotic counterparts (Song et al. 2000; Vestergaard et al. 2001). The stop-codon recognition mechanisms of bacterial and eukaryotic release factors are likely to be distinct, as the reading heads of bacterial RF1 and RF2 bear no obvious structural resemblance to the putative codon-recognition regions of eRF1 (Bertram et al. 2000; Song et al. 2000; Frolova et al. 2002; Laurberg et al. 2008). Moreover, the eukaryotic translation termination mechanism differs from that in bacteria in that it is thought to depend on proofreading (Buckingham et al. 1997; Salas-Marco and Bedwell 2004) and rate enhancement (Alkalaeva et al. 2006) mediated by the class II release factor eRF3, to which eRF1 is stably bound (Stansfield et al. 1995; Zhouravleva et al. 1995; Frolova et al. 1996). These substantial dissimilarities suggest that bacterial and eukaryotic peptide release systems have evolved independently. The absence of a common protein release factor ancestor implies that the essential function of translation termination in early “RNA World” ribosomes may have been carried out by a mechanism that was distinct from those in present-day translation systems.
All of the central aspects of translation termination mediated by RF1 and RF2 depend on the elements of the polypeptide backbones of the release factors (Korostelev et al. 2008, 2010; Laurberg et al. 2008; Weixlbaumer et al. 2008). First, strong discrimination for uridine as the first base of the stop codons is accomplished through a base pair-like interaction of U1 with the backbone of the tip of helix α5 (Fig. 3). Second, the backbone amide NH group of the glutamine of the GGQ motif is positioned to play an essential catalytic role in the hydrolysis of peptidyl-tRNA (Figs. 4, 5). Finally, coordination of peptidyl-tRNA hydrolysis with stop-codon recognition is achieved via the switch loop, whose polypeptide backbone length, rather than the identities of its side chains, is critical for peptide release efficiency (Korostelev et al. 2010). The involvement of the polypeptide backbone in the release factor function suggests that these functional aspects may have arisen from interactions of the ribosome with simpler polypeptides of less determined sequence, in an evolutionary departure from a primitive RNA-based translation machinery.
CONCLUDING REMARKS: TERMINATING STUDIES ON TRANSLATION TERMINATION WOULD BE PREMATURE
The recent crystallographic structures along with biochemical and mutational studies have substantially expanded our understanding of the mechanism of translation termination. Some gaps, however, remain to be filled. First, higher-resolution structures of translation-termination complexes are required to provide finer details of the mechanism of stop-codon recognition; for example, the possibility of the critical involvement of water molecules, suggested by a recent molecular dynamics study (Sund et al. 2010), needs to be explored. Second, the mechanism by which a water molecule is delivered to the PTC upon stop-codon recognition remains unclear. Third, a crystal structure of a transition-state analog of the peptidyl-tRNA hydrolysis reaction on the ribosome that would allow visualization of interactions critical for the catalysis is lacking. Fourth, the proposed mechanism of coordination of stop-codon recognition with peptidyl-tRNA hydrolysis via a conformational switch has to be probed by visualizing the conformational dynamics of the release factors upon binding to the ribosome bearing a stop or sense codon in its A site. Finally, the structural understanding of translation termination in eukaryotes is lagging behind that of bacteria. The emergence of the crystal structure of a eukaryotic 80S ribosome (Ben-Shem et al. 2010) will likely pave the path to crystallographic studies on 80S translation termination complexes. While some aspects of eukaryotic peptide release, such as the involvement of the GGQ motif in the catalysis, will likely turn out to be similar to those of bacterial translation termination, it will be of great interest to gain the structural knowledge of the mechanisms of stop-codon recognition and coordination of codon recognition with peptidyl-tRNA hydrolysis by the eRF1-eRF3 system. Eukaryotic translation termination is of special interest, not only because its mechanism is, in many aspects, different from the bacterial one, but also because it is involved in key cellular regulatory mechanisms, such as mRNA quality surveillance and post-transcriptional gene expression modulation via nonsense mediated decay (for review, see Maquat 2004; Neu-Yilik and Kulozik 2008). These mechanisms depend on interactions of the eRF1-eRF3 translation termination system with the NMD factors (Czaplinski et al. 1998; Wang et al. 2001) and poly(A)-binding protein (Hoshino et al. 1999). Translation termination appears to be involved in other regulatory pathways as well, e.g., during viral response. Here, eRF1 and eRF3 have been suggested to directly interact with viral reverse transcriptase (Orlova et al. 2003), RNase L (Le Roy et al. 2005) and the RNase L inhibitor Rli1, or ABCE1 (Khoshnevis et al. 2010), which was recently proposed to act as the archaeal and eukaryotic ribosome recycling factor (Pisarev et al. 2010; Barthelme et al. 2011). The structural bases for understanding the interactions of the eRF1-eRF3 system with these and other proteins (Baierlein and Krebber 2010) and, possibly, RNA molecules remain to be elucidated.
ACKNOWLEDGMENTS
I thank Harry F. Noller and Dmitri N. Ermolenko for many stimulating discussions and comments on this manuscript. I also thank Alexei V. Korennykh and Sean Ryder for critically reading the manuscript. Crystal structures were downloaded from RCSB Protein Data Bank (http://www.rcsb.org). Figures were rendered using Pymol (DeLano 2002).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2733411.
REFERENCES
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Arkov AL, Murgola EJ 1999. Ribosomal RNAs in translation termination: Facts and hypotheses. Biochemistry (Mosc) 64: 1354–1359 [PubMed] [Google Scholar]
- Baierlein C, Krebber H 2010. Translation termination: New factors and insights. RNA Biol 7: 548–550 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R 2011. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci 108: 3228–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M 2010. Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I 2000. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6: 1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457: 977–980 [DOI] [PubMed] [Google Scholar]
- Bouakaz L, Bouakaz E, Murgola EJ, Ehrenberg M, Sanyal S 2006. The role of ribosomal protein L11 in class I release factor-mediated translation termination and translational accuracy. J Biol Chem 281: 4548–4556 [DOI] [PubMed] [Google Scholar]
- Brenner S, Stretton AO, Kaplan S 1965. Genetic code: The “nonsense” triplets for chain termination and their suppression. Nature 206: 994–998 [DOI] [PubMed] [Google Scholar]
- Brenner S, Barnett L, Katz ER, Crick FH 1967. UGA: A third nonsense triplet in the genetic code. Nature 213: 449–450 [DOI] [PubMed] [Google Scholar]
- Brown CM, Tate WP 1994. Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J Biol Chem 269: 33164–33170 [PubMed] [Google Scholar]
- Brunelle JL, Shaw JJ, Youngman EM, Green R 2008. Peptide release on the ribosome depends critically on the 2′-OH of the peptidyl-tRNA substrate. RNA 14: 1526–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham RH, Grentzmann G, Kisselev L 1997. Polypeptide chain release factors. Mol Microbiol 24: 449–456 [DOI] [PubMed] [Google Scholar]
- Capecchi MR 1967. Polypeptide chain termination in vitro: Isolation of a release factor. Proc Natl Acad Sci 58: 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey CT 1980. Peptide chain termination. Trends Biochem Sci 5: 234–237 [Google Scholar]
- Caskey CT, Beaudet AL, Scolnick EM, Rosman M 1971. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci 68: 3163–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno L, Shine J 1973. Conserved terminal sequence in 18SrRNA may represent terminator anticodons. Nat New Biol 245: 261–262 [DOI] [PubMed] [Google Scholar]
- DeLano WL 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA [Google Scholar]
- Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M 2000. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J 19: 6900–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontsova M, Frolova L, Vassilieva J, Piendl W, Kisselev L, Garber M 2000. Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett 472: 213–216 [DOI] [PubMed] [Google Scholar]
- Dorner S, Polacek N, Schulmeister U, Panuschka C, Barta A 2002. Molecular aspects of the ribosomal peptidyl transferase. Biochem Soc Trans 30: 1131–1136 [DOI] [PubMed] [Google Scholar]
- Dorner S, Panuschka C, Schmid W, Barta A 2003. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res 31: 6536–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder CE, Prilusky J, Silman I, Sussman JL 2007. A server and database for dipole moments of proteins. Nucleic Acids Res 35: W512–W521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M 1997. Release factor RF3 in E coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J 16: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M 2000. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci 97: 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, et al. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703 [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341 [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Seit-Nebi A, Kisselev L 2002. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA 8: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Brechemier-Baey D, Heurgue V, Mora L, Buckingham RH 1994. Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc Natl Acad Sci 91: 5848–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Gutell RR, Weiser B, Woese CR, Noller HF 1985. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol 32: 155–216 [DOI] [PubMed] [Google Scholar]
- He SL, Green R 2010. Visualization of codon-dependent conformational rearrangements during translation termination. Nat Struct Mol Biol 17: 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B, Lee K, Joseph S 2009. Kinetics of stop codon recognition by release factor 1. Biochemistry 48: 11178–11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680 [DOI] [PubMed] [Google Scholar]
- Ito K, Ebihara K, Uno M, Nakamura Y 1996. Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc Natl Acad Sci 93: 5443–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y 1998. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc Natl Acad Sci 95: 8165–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y 2000. A tripeptide “anticodon” deciphers stop codons in messenger RNA. Nature 403: 680–684 [DOI] [PubMed] [Google Scholar]
- Ivanov V, Beniaminov A, Mikheyev A, Minyat E 2001. A mechanism for stop codon recognition by the ribosome: A bioinformatic approach. RNA 7: 1683–1692 [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Dijkstra BW, Reetz MT 1999. Bacterial biocatalysts: Molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53: 315–351 [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17: 555–560 [DOI] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V 2010. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci 107: 8593–8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen F, Adamski FM, Tate WP, Kurland CG 1993. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol 230: 41–50 [DOI] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H 2010. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep 11: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M 2003. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421: 90–94 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol 340: 141–177 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Zhu J, Asahara H, Noller HF 2010. Recognition of the amber UAG stop codon by release factor RF1. EMBO J 29: 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Ter-Avanesyan MD, Telckov MV, Surguchov AP, Smirnov VN, Inge-Vechtomov SG 1988. Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene 66: 45–54 [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857 [DOI] [PubMed] [Google Scholar]
- Le Roy F, Salehzada T, Bisbal C, Dougherty JP, Peltz SW 2005. A newly discovered function for RNase L in regulating translation termination. Nat Struct Mol Biol 12: 505–512 [DOI] [PubMed] [Google Scholar]
- Ma B, Nussinov R 2004. Release factors eRF1 and RF2: A universal mechanism controls the large conformational changes. J Biol Chem 279: 53875–53885 [DOI] [PubMed] [Google Scholar]
- Maegley KA, Admiraal SJ, Herschlag D 1996. Ras-catalyzed hydrolysis of GTP: A new perspective from model studies. Proc Natl Acad Sci 93: 8160–8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE 2004. Nonsense-mediated mRNA decay: Splicing, translation, and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Mikuni O, Ito K, Moffat J, Matsumura K, McCaughan K, Nobukuni T, Tate W, Nakamura Y 1994. Identification of the prfC gene, which encodes peptide-chain-release factor 3 of Escherichia coli. Proc Natl Acad Sci 91: 5798–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL 2006. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res 34: 3947–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH 2003. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol 47: 267–275 [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH 2007. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J Biol Chem 282: 35638–35645 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito K, Matsumura K, Kawazu Y, Ebihara K 1995. Regulation of translation termination: Conserved structural motifs in bacterial and eukaryotic polypeptide release factors. Biochem Cell Biol 73: 1113–1122 [DOI] [PubMed] [Google Scholar]
- Neu-Yilik G, Kulozik AE 2008. NMD: Multitasking between mRNA surveillance and modulation of gene expression. Adv Genet 62: 185–243 [DOI] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270: 1464–1472 [DOI] [PubMed] [Google Scholar]
- Noller HF 1999. On the origin of the ribosome: Coevolution of subdomains of tRNA and rRNA. In The RNA world, 2nd ed (ed. Gesteland R et al.), pp. 197–219 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Noller HF 2004. The driving force for molecular evolution of translation. RNA 10: 1833–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF 2006. Evolution of ribosomes and translation from an RNA world. In The RNA world, 3rd ed (ed. Gesteland R et al.), pp. 287–307 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Noller HF 2010. Evolution of protein synthesis from an RNA World. Cold Spring Harb Perspect Biol a003681. doi: 10.1101/cshperspect.a003681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Woese CR 1981. Secondary structure of 16S ribosomal RNA. Science 212: 403–411 [DOI] [PubMed] [Google Scholar]
- Nozaki Y, Matsunaga N, Ishizawa T, Ueda T, Takeuchi N 2008. HMRF1L is a human mitochondrial translation release factor involved in the decoding of the termination codons UAA and UAG. Genes Cells 13: 429–438 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Orlova M, Yueh A, Leung J, Goff SP 2003. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell 115: 319–331 [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FV IV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V 2005. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV 2010. The role of ABCE1 in eukaryotic post-termination ribosomal recycling. Mol Cell 37: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat UBS, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J 2003. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421: 87–90 [DOI] [PubMed] [Google Scholar]
- Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J 2006. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol 357: 1144–1153 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Pape T, Fricke R, Kuhn L, Wintermeyer W 1996. Initial binding of the elongation factor Tu.GTP.aminoacyl-tRNA complex preceding codon recognition on the ribosome. J Biol Chem 271: 646–652 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Beringer M, Wintermeyer W 2006. Mechanism of peptide bond formation on the ribosome. Q Rev Biophys 39: 203–225 [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24: 7769–7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Ito K, Nakamura Y 2006. Ribosomal protein L11 mutations in two functional domains equally affect release factors 1 and 2 activity. Mol Microbiol 60: 108–120 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA 2005a. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell 20: 437–448 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA 2005b. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438: 520–524 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 A resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Seit Nebi A, Frolova L, Ivanova N, Poltaraus A, Kiselev L 2000. Mutation of a glutamine residue in the universal tripeptide GGQ in human eRF1 termination factor does not cause complete loss of its activity. Mol Biol (Mosk) 34: 899–900 [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L 2001. Class-1 translation termination factors: Invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res 29: 3982–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Shaw JJ, Green R 2007. Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell 28: 458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim SH 2004. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol 341: 227–239 [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci 71: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonović M, Steitz TA 2008. Peptidyl-CCA deacylation on the ribosome promoted by induced fit and the O3′-hydroxyl group of A76 of the unacylated A-site tRNA. RNA 14: 2372–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimanpour-Lichaei HR, Kuhl I, Gaisne M, Passos JF, Wydro M, Rorbach J, Temperley R, Bonnefoy N, Tate W, Lightowlers R, et al. 2007. mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol Cell 27: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D 2000. The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14: 4365–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sund J, Ander M, Aqvist J 2010. Principles of stop-codon reading on the ribosome. Nature 465: 947–950 [DOI] [PubMed] [Google Scholar]
- Tate W, Greuer B, Brimacombe R 1990. Codon recognition in polypeptide chain termination: Site directed crosslinking of termination codon to Escherichia coli release factor 2. Nucleic Acids Res 18: 6537–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM 2010. Hungry codons promote frameshifting in human mitochondrial ribosomes. Science 327: 301 doi: 10.1126/science.1180674 [DOI] [PubMed] [Google Scholar]
- Thompson RC 1988. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem Sci 13: 91–93 [DOI] [PubMed] [Google Scholar]
- Tompkins RK, Scolnick EM, Caskey CT 1970. Peptide chain termination. VII. The ribosomal and release factor requirements for peptide release. Proc Natl Acad Sci 65: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S, Aqvist J 2007. A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell 27: 758–766 [DOI] [PubMed] [Google Scholar]
- Trobro S, Aqvist J 2009. Mechanism of the translation termination reaction on the ribosome. Biochemistry 48: 11296–11303 [DOI] [PubMed] [Google Scholar]
- Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M 2001. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell 8: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Vestergaard B, Sanyal S, Roessle M, Mora L, Buckingham RH, Kastrup JS, Gajhede M, Svergun DI, Ehrenberg M 2005. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol Cell 20: 929–938 [DOI] [PubMed] [Google Scholar]
- Vogel Z, Zamir A, Elson D 1969. The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry 8: 5161–5168 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20: 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA 2004. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol 11: 1101–1106 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmouth RC, Edman K, Neutze R, Wright PA, Clifton IJ, Schneider TR, Schofield CJ, Hajdu J 2001. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat Struct Biol 8: 689–694 [DOI] [PubMed] [Google Scholar]
- Wilson PG, Culbertson MR 1988. SUF12 suppressor protein of yeast. A fusion protein related to the EF-1 family of elongation factors. J Mol Biol 199: 559–573 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Woese CR, Fox GE, Zablen L, Uchida T, Bonen L, Pechman K, Lewis BJ, Stahl D 1975. Conservation of primary structure in 16S ribosomal RNA. Nature 254: 83–86 [DOI] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Rodnina MV 2010. Optimization of speed and accuracy of decoding in translation. EMBO J 29: 3701–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Edgar CD, Murphy J, Fredebohm J, Poole ES, Tate WP 2010a. Bioinformatic, structural, and functional analyses support release factor-like MTRF1 as a protein able to decode nonstandard stop codons beginning with adenine in vertebrate mitochondria. RNA 16: 1146–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Edgar CD, Poole ES, Tate WP 2010b. The codon specificity of eubacterial release factors is determined by the sequence and size of the recognition loop. RNA 16: 1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, He SL, Nikstad LJ, Green R 2007. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell 28: 533–543 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Shaw JJ, Strobel SA, Green R 2011. The 2′-OH group of the peptidyl-tRNA stabilizes an active conformation of the ribosomal PTC. EMBO J doi: 10.1038/emboj.2011.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M 2001. A post-termination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Mora L, Buckingham RH, Ehrenberg M 2002. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell 10: 789–798 [DOI] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14: 4065–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoldak G, Redecke L, Svergun DI, Konarev PV, Voertler CS, Dobbek H, Sedlak E, Sprinzl M 2007. Release factors 2 from Escherichia coli and Thermus thermophilus: Structural, spectroscopic, and microcalorimetric studies. Nucleic Acids Res 35: 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]