Abstract
Gene expression in eukaryotic cells is profoundly influenced by the post-transcriptional processing of mRNAs, including the splicing of introns in the nucleus and both nuclear and cytoplasmic degradation pathways. These processes have the potential to affect both the steady-state levels and the kinetics of changes to levels of intron-containing transcripts. Here we report the use of a splicing isoform-specific microarray platform to investigate the effects of diverse stress conditions on pre-mRNA processing. Interestingly, we find that diverse stresses cause distinct patterns of changes at this level. The responses we observed are most dramatic for the RPGs and can be categorized into three major classes. The first is characterized by accumulation of RPG pre-mRNA and is seen in multiple types of amino acid starvation regimes; the magnitude of splicing inhibition correlates with the severity of the stress. The second class is characterized by a rapid decrease in both pre- and mature RPG mRNA and is seen in many stresses that inactivate the TORC1 kinase complex. These decreases depend on nuclear turnover of the intron-containing pre-RNAs. The third class is characterized by a decrease in RPG pre-mRNA, with only a modest reduction in the mature species; this response is observed in hyperosmotic and cation-toxic stresses. We show that casein kinase 2 (CK2) makes important contributions to the changes in pre-mRNA processing, particularly for the first two classes of stress responses. In total, our data suggest that complex post-transcriptional programs cooperate to fine-tune expression of intron-containing transcripts in budding yeast.
Keywords: pre-mRNA processing, splicing, stress responses, budding yeast, gene expression, casein kinase 2
INTRODUCTION
Regulation of ribosomal protein gene (RPG) expression is of great importance to overall cellular metabolism; up to 50% of all RNA polymerase II transcription in budding yeast is dedicated to making RPG mRNAs in nutrient-rich conditions (Warner 1999). Accordingly, RPG expression is known to be tightly regulated both transcriptionally and post-transcriptionally. Transcriptional regulation involves multiple transcription factors, kinases, and chromatin modifications (Rohde and Cardenas 2003; Marion et al. 2004; Martin et al. 2004; Zhao et al. 2006). Measurements of mRNA half-lives for all yeast transcripts reveal that the half-lives of the RPGs are strikingly similar to one another, suggesting that this is a regulated property (Wang et al. 2002). Moreover, RPGs generally have shorter than average 5′ UTRs, which is believed to contribute to their high translational efficiency, and longer than average poly(A) tails, which is believed to contribute to their stability (David et al. 2006; Beilharz and Preiss 2007). Despite widespread intron loss from the Saccharomyces cerevisiae genome, introns have been retained in a majority of ribosomal protein genes, and RPGs contain over one-third of the remaining S. cerevisiae introns (Fisk et al. 2006; Stajich et al. 2007). This bias in intron distribution suggests that the presence of introns in ribosomal protein genes could also contribute to their regulation. Indeed, we showed previously that pre-mRNAs for the RPGs accumulate in response to treatment with the amino acid starvation-mimicking drug, 3-amino 1,2,4-triazole (3-AT) (Pleiss et al. 2007a).
Translation initiation is inhibited in response to 3-AT, and prolonged treatment leads eventually to a decrease in ribosomal protein gene expression (Moehle and Hinnebusch 1991; Natarajan et al. 2001). Many other stresses that down-regulate translation are also known to cause a decrease in ribosomal protein gene expression, presumably because the vast energy resources normally directed toward ribosome biogenesis under favorable growth conditions must be redirected toward the relevant stress response (Li et al. 1999; Gasch et al. 2000).
While it has been suggested that the stability of the RPG transcripts is specifically altered in response to some stresses (Grigull et al. 2004; Molin et al. 2009; Romero-Santacreu et al. 2009), the best-characterized stress-responsive regulation of RPG expression occurs at the level of transcription. A central nutrient- and stress-sensitive regulator of RPG transcription is the TORC1 (Target Of Rapamycin Complex 1) kinase complex, which phosphorylates multiple substrates in favorable conditions to promote RPG expression (for review, see Wullschleger et al. 2006; Zaman et al. 2008). Under a variety of stressful conditions, including depletion of glucose, nitrogen, phosphorus, heat shock, and hyperosmotic stress, the TORC1 complex is inactivated, leading to a strong decrease in RPG expression (Urban et al. 2007). Additional signaling pathways are known to contribute to the responses to these translation-inhibitory stresses. Protein kinase A, Ire1, Hog1, Pho85, and Gcn2 contribute to unique aspects of the responses to glucose starvation, endoplasmic reticulum stress, hyperosmotic stress, phosphate starvation, and amino acid starvation, respectively, and in many cases, examples of crosstalk among these pathways and between these pathways and the TORC1 pathway, have also been identified (Patil and Walter 2001; Wykoff and O'Shea 2001; Wilson and Roach 2002; Cherkasova and Hinnebusch 2003; Patil and Walter 2001; Schroder et al. 2004; Hohmann 2009).
In addition to inhibiting RPG expression, inactivation of TORC1 leads to entry into a G1-like quiescent state, referred to as G0 or stationary phase (Pedruzzi et al. 2003). Entry into G0 is protective under stressful conditions—mutations or conditions that prevent it cause decreased long-term survival (Swinnen et al. 2006; Boer et al. 2008; Talarek et al. 2010). In short, a complex network of stress-responsive signaling mediates appropriate responses to many stresses, which are characterized by some common features such as transcriptional down-regulation of ribosomal protein genes and entry into G0, and some features unique to each individual stress.
In addition to TORC1, there are other, less well-characterized signaling complexes that are likely to contribute to regulation under stressful conditions. One such kinase complex with proposed roles in nutrient sensing is casein kinase 2 (CK2). In yeast, CK2 consists of two regulatory subunits (Ckb1 and Ckb2) and two catalytic subunits (Cka1 and Cka2), and appears to be highly promiscuous within the cell, with dozens of reported substrates (Bidwai et al. 1994; Olsten and Litchfield 2004). They include subunits of all three RNA polymerases, chromatin remodeling ATPases, splicing factors, transcription factors, ribosome biogenesis factors, mRNA-binding proteins, tRNA synthetases, ribosomal proteins, and translation factors (Sawa et al. 2004; Allen 2008; Deng et al. 2008; Dermody et al. 2008). Unlike many other kinases involved in stress-signaling pathways, CK2 has no known direct interactions with the TORC1 complex. In general, its regulation is very poorly understood—the catalytic subunits have high constitutive activity in vitro, and the roles of the regulatory subunits in directing this activity toward appropriate substrates are unknown (Prudent et al. 2010). That CK2 appears to have substrates involved in many aspects of regulating protein biosynthetic capacity places it in a unique position to contribute to translation-inhibitory stress responses.
We wished to determine the contribution of the presence of introns in stress-regulated transcripts to the overall integrated stress response of the cell. We used a custom microarray platform (previously described in Pleiss et al. 2007b) to specifically measure changes in the amounts of pre-mRNA, mature mRNA, and total mRNA for the majority of intron-containing genes in yeast following exposure to a wide range of translation-inhibitory stresses. This data set allowed us to contextualize the role of regulation at the level of pre-mRNA processing, including both splicing and turnover, within a large arena of growth conditions and responses, and offered novel insights into mechanisms by which this regulation is imposed and integrated with other stress response pathways.
RESULTS
A microarray platform for analyzing pre-mRNA processing in response to stress
We used a previously described splicing-sensitive microarray platform that allowed for specific measurements of pre-mRNA (using a probe against the intron sequence), mature mRNA (using a probe across the junction sequence), and total mRNA (using a probe against the second exon sequence) to assay changes in relative levels of these species during stress (Pleiss et al. 2007b). Yeast were exposed to a range of stresses, and culture aliquots were collected at 5-, 10-, 20-, and 40-min time points after the induction of the stress. These time points were selected in an effort to capture both rapid changes to processing of pre-mRNA, and later changes to steady-state levels of mature mRNA. Total RNA was extracted, cDNA was synthesized and labeled with fluorophores, and samples were competitively hybridized to microarrays, all as previously described. In stress treatments in which media was exchanged, each sample was hybridized against a mock-treated sample. For heat shock and stress treatments involving the addition of a drug, samples were hybridized against a pretreatment sample.
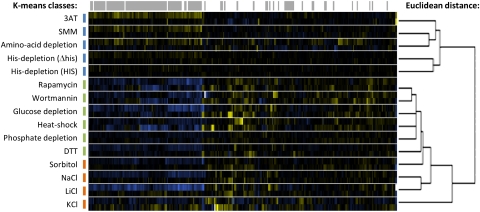
The relative abundances (as ratios of the treated samples to the untreated references) of the pre-mRNA, mature mRNA, and total mRNA species for each intron-containing gene over each stress time course were measured by dye-flipped replicate microarrays (full data set available online in the Gene Expression Omnibus database: http://www.ncbi.nlm.nih.gov/geo/ GEO accession GSE28919. Also see the web supplement to this manuscript: http://biochemistry.ucsf.edu/labs/guthrie/bergkesselWhitworth2011.html). To visualize the results, the total, pre-mRNA, and mature mRNA values for all of the time points for each transcript were plotted on the same row of a heat map, colored according to the log2-transformed ratio values from the microarrays, and the rows of the heat map were ordered by a hierarchical clustering algorithm (Supplemental Figs. S1–S15). In order to analyze the relationships between each of the different stress responses, as determined by changes in pre-mRNA and mature mRNA levels, the time course data for each stress response were first summarized by taking the time point exhibiting the greatest overall change in the intron and junction feature responses. These values were then clustered, using both hierarchical and k-means clustering (Fig. 1; see Materials and Methods for more details). Hierarchical clustering assesses the similarity of each stress to every other stress in the data set and was used to construct a dendrogram displaying these relationships. K-means clustering, on the other hand, fits the stresses to a predefined number of classes, minimizing within-class variability. In all k-means analyses presented below, solutions were calculated for the full range of possible numbers of classes, and the data shown represent the solutions with the smallest number of classes that provided meaningful subdivisions of the data.
FIGURE 1.
Comparison of pre- and mature mRNA level changes across diverse environmental stresses by clustering analysis. Stress treatments, named at left, are ordered vertically by hierarchical clustering using Euclidean distance (dendrogram shown at the right of the figure). K-means clustering was also used to group responses into three distinct subclasses, highlighted by colored bars. Genes were also ordered by hierarchical clustering along the horizontal axis. For each stress response, changes in pre-mRNA levels (top row) and mature mRNA levels (bottom row) are shown in the central heatmap (saturated blue indicates at least a 5.7-fold (22.5) decrease in abundance, saturated yellow at least a 5.7-fold increase). The values shown represent the largest absolute value of change observed over the time course of each stress treatment for each gene. Gray bars across the top of the heatmap mark the locations of RPG transcripts.
We focused on stresses previously reported to affect translation, RNA processing, or both. Following our earlier demonstration that treatment with 3-AT induces the accumulation of RPG pre-mRNA, we wanted to test other formats of amino acid starvation. 3-AT induces an amino acid starvation response when cells are grown in synthetic media lacking histidine by inhibiting an enzyme in the histidine biosynthetic pathway. In addition to repeating the 3-AT treatment using the same time course and cell collection protocol used for the rest of the stress treatments in this study, we also simply removed histidine by filtering a culture growing in synthetic complete media, washing the cells with synthetic media lacking only histidine and resuspending them in that media. To create a situation in which this stress would be more stringent, we repeated the histidine depletion using a strain that was auxotrophic for histidine. We also examined the response when all amino acids, uracil, and adenine were removed by replacing synthetic complete media with media containing only yeast nitrogen base, ammonium sulfate, and glucose. Finally, we used another drug, sulfometuron methyl (SMM), which, like 3-AT, inhibits an amino acid biosynthetic pathway and induces the general amino acid control pathway. SMM inhibits an enzyme in the biosynthetic pathways for leucine, isoleucine and valine, and thus elicits an amino acid starvation response when added to a culture growing in media lacking isoleucine and valine (Jia et al. 2000; Hinnebusch 2005).
We tested two additional starvation stresses that have been shown to at least transiently inactivate TORC1. We starved cells for phosphate by replacing synthetic complete media with media lacking potassium phosphate, and for a carbon source by replacing rich media with rich media lacking glucose. We also tested the effects of TORC1 inactivation directly by treating cells with two drugs that inhibit this complex: rapamycin and wortmannin (Alarcon et al. 1999).
Conditions leading to the accumulation of unfolded or misfolded proteins in the endoplasmic reticulum are also known to down-regulate translation and to induce the unfolded protein response (UPR) (Patil and Walter 2001). We tested two stresses that induce the UPR: treatment with dithiothreitol (DTT), a strong reducing agent, and a temperature shift from 30°C to 42°C (heat shock). Heat shock has previously been suggested to broadly inhibit pre-mRNA splicing (Yost and Lindquist 1991; Vogel et al. 1995; Bracken and Bond 1999).
Salt stress is also known to strongly down-regulate translation (Gasch et al. 2000). Treatment with 500 mM sodium chloride (NaCl), commonly used to elicit a hyperosmotic stress response, is both a hyperosmotic stress and a cation-toxic stress. We tested this concentration of NaCl and also used lithium chloride (LiCl) at a concentration that is primarily cation toxic (300 mM) and potassium chloride (KCl) at a concentration that is primarily hyperosmotic-stress inducing (1 M). We also treated cells with 1 M sorbitol, a hyperosmotic stress, but not a salt (Garcia et al. 1997).
Stress responses cluster into three groups
The first class defined by clustering analysis (Fig. 1; blue bars in k-means classes) shows behavior similar to that previously described as the pre-mRNA processing response to amino acid starvation—accumulation of the pre-mRNA for a majority of the RPGs (Pleiss et al. 2007a). 3-AT treatment, SMM treatment, and depletion of all amino acids form one branch within this group, and depletion of just histidine, in both wild-type and histidine auxotroph strains, forms another branch. Closer inspection of the full time courses for each of these stresses shows that accumulation of RPG pre-mRNA at the earliest (5- and 10-min) time points is a common characteristic of all of the amino acid starvation responses (Fig. 2). Furthermore, the magnitude of the accumulation seems to scale with the severity of the stress.
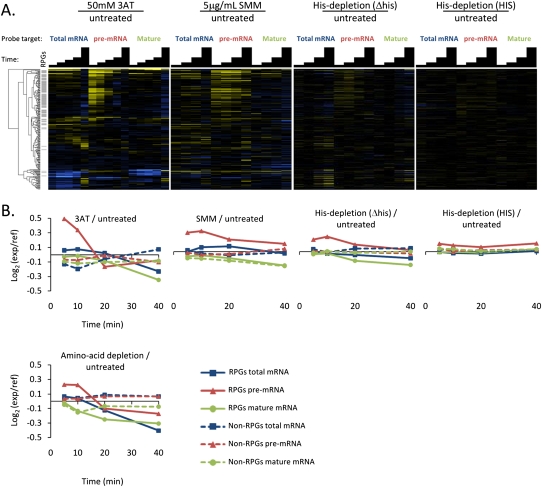
FIGURE 2.
Accumulation of RPG pre-mRNAs under amino acid starvation conditions. (A) Time-resolved splicing profiles resulting from comparison of wild-type cells treated with amino acid biosynthesis inhibitors or shifted into amino acid-depleted media, compared with a mock treatment. Samples were collected at 5, 10, 15, 20, and 40 min (3-AT) or at 5, 10, 20, 40 min (all others) following treatment. Transcripts are arranged on the vertical axis by hierarchical clustering (dendrogram shown at left), and gray bars highlight the positions of RPGs within this cluster. In the heatmap, saturated blue indicates at least a 5.7-fold decrease and saturated yellow at least a 5.7-fold increase in transcript abundance. (B) Averaged RPG and non-RPG transcript behaviors across data sets shown in A.
Depletion of just histidine from the growth media elicits a much more subtle response than the other treatments. Washing histidine away likely fails to completely remove all histidine from the cell, and instead, causes a slow decrease in the available pools of histidine. In the wild-type strain, up-regulation of the histidine biosynthetic pathway may be nearly able to keep up with the depletion of histidine from environmental sources. The perturbation to translation is thus mild and transient. Accumulation of the RPG pre-mRNA is barely discernible in the heat map representation of the data, but plotting the average log2-transformed ratio for each feature type (Fig. 2B; RPG pre-, mature, and total mRNA, and non-RPG pre-, mature, and total mRNA) shows that the RPG pre-mRNA levels are in fact modestly elevated, while the other features are essentially unchanged.
When histidine is depleted from a histidine auxotroph, the response is somewhat more pronounced, presumably because the histidine shortage caused by washing histidine out of the media cannot be successfully relieved by up-regulation of the histidine biosynthetic pathway in this strain. Treatments with 3-AT or SMM cause inhibition of biosynthetic pathways that are already engaged as the sole sources of histidine or isoleucine and valine, respectively, and are expected to cause a sudden sharp drop in the available pools of those amino acids. The perturbation to translation is thus more dramatic, and a strong accumulation of RPG pre-mRNAs is observed throughout the early portion of the time course, while amino acid biosynthetic genes are being up-regulated.
Depletion of all amino acids (plus uracil and adenine) causes a degree of accumulation of RPG pre-mRNA similar to that observed for histidine depletion in a histidine auxotroph in the early time points of the time course. The wild-type strain used in this experiment is, in fact, auxotrophic for leucine, uracil, and lysine, so the similarity of the magnitude and kinetics of the RPG pre-mRNA accumulation in this treatment to that seen when a histidine auxotroph is depleted for histidine is perhaps unsurprising. However, the behaviors of the mature and total mRNA species diverge significantly in the later time points of the time course, as these species decrease substantially in abundance following depletion of all amino acids. The loss of RPG mature mRNA at later time points is a pronounced feature of the responses that lead to inactivation of TORC1 (see Fig. 3 and results below), and may reflect crosstalk between the amino acid starvation and TORC1 pathways (Staschke et al. 2010). Amino acids can also be utilized as a nitrogen source, so depletion of all amino acids may also trigger some activation of nitrogen starvation pathways that lead to TORC1 inactivation (for review, see Zaman et al. 2008).
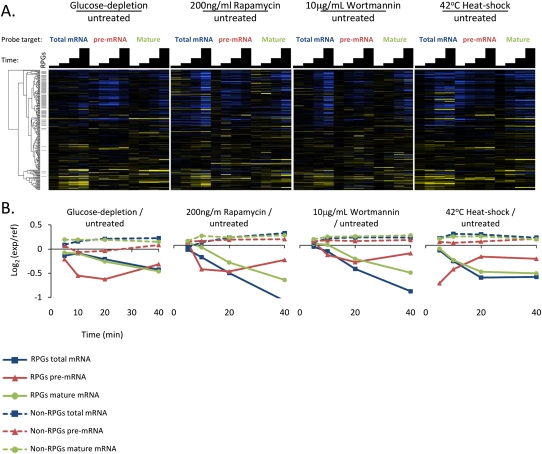
FIGURE 3.
Rapid loss of RPG pre- and mature mRNAs following glucose starvation, heat shock, and direct TORC1 inhibition. (A) Time-resolved splicing profiles are shown from comparisons of wild-type cells in treated and untreated samples across a time course of 5, 10, 20, 40 min. Transcripts are arranged on the vertical axis in the same order as in Figure 2 to facilitate comparison, and gray bars highlight the positions of RPGs within this cluster. In the heatmap, saturated blue indicates at least a 5.7-fold decrease and saturated yellow at least a 5.7-fold increase in transcript abundance. (B) Averaged RPG and non-RPG transcript behaviors across data sets shown in A.
The second class of responses identified by clustering analysis is characterized by decreases in all three species of RPG transcripts, and includes glucose and phosphate depletion, drugs that inhibit TORC1, heat shock, and DTT treatment (green bars in Fig. 1). The pre-mRNAs decrease more rapidly than the mature and total mRNA species, with large decreases apparent at the first time point, while the mature and total only show substantial decreases at the 20- and 40-min time points (Fig. 3). Interestingly, heat shock appears to induce the most rapid and dramatic loss of RPG mRNA transcripts. Previous work had suggested that heat shock leads to inhibition of all pre-mRNA splicing and widespread accumulation of pre-mRNAs (Yost and Lindquist 1991; Vogel et al. 1995; Bracken and Bond 1999). However, this conclusion was made based on the behavior of a small number of transcripts. Our results show that while a few transcripts do indeed accumulate pre-mRNA in response to heat shock (Fig. 3A; Fig. 8B, below) the majority of RPG pre-mRNAs are instead rapidly degraded. The magnitudes of the changes observed in response to phosphate starvation were small, but the patterns of changes were similar enough to those observed in other TORC1-inactivating stresses for it to cluster with this group.
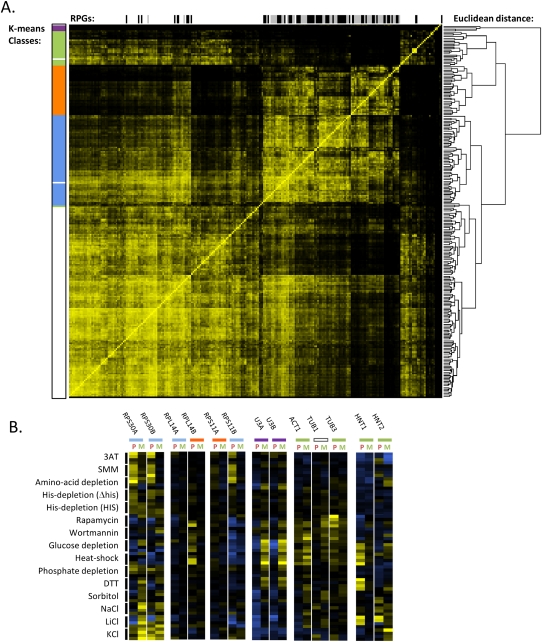
FIGURE 8.
Analysis of relatedness of transcript behaviors across all environmental stress conditions. (A) Pairwise distance analysis was used to cluster transcripts symmetrically across the horizontal and vertical axes. In the central heatmap, brighter yellows represent greater degrees of correlation in transcript behaviors across the stress response data set (small Euclidean distances). The dendrogram to the right of the heatmap shows hierarchical clustering of transcript correlations. The colored bars to the left of the heatmap highlight transcripts that fall into each of the five transcript subclasses identified by k-means clustering. Across the top of the heatmap, light-gray bars show the location of small ribosomal subunit transcripts, and black bars show the location of large ribosomal subunit transcripts. (B) Stress response profiles of select transcripts. Gene names are listed across the top, and colored bars refer to the k-means classes in A. For each stress, the four time points (5, 10, 20, and 40 min) are displayed vertically in the four adjacent rows indicated by each black bar.
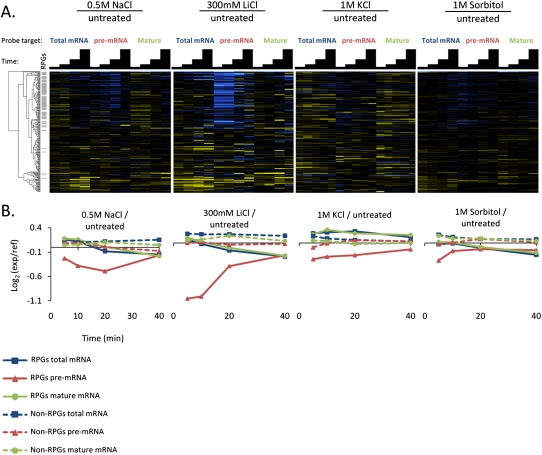
The third class of response identified by the clustering analysis included the ionic and hyperosmotic stresses (Fig. 1, red bars). These responses are characterized by a rapid loss of RPG pre-mRNA, but relatively smaller decreases in the mature mRNAs than in the second class (Fig. 4). For the cation-toxic stresses (NaCl and LiCl), the loss of RPG pre-mRNA is more dramatic than for the hyperosmotic stresses. In the KCl treatment, the levels of total and mature mRNAs for the RPGs actually increase over the time course. A sample taken at 60 min showed that the total and mature mRNAs for the RPGs did eventually decrease below the level at which they started. The response to sorbitol is milder in magnitude than the responses to the salt stresses, but still shows a decrease in RPG pre-mRNAs at the early time points, coupled with very small decreases in the mature and total mRNAs only late in the time course.
FIGURE 4.
Rapid loss of RPG pre-mRNAs in response to cation toxicity and hyperosmotic stresses. (A) Time-resolved splicing profiles are shown from comparisons of wild-type cells in treated and untreated samples across a time course of 5, 10, 20, and 40 min. Transcripts are arranged on the vertical axis in the same order as in Figure 2 to facilitate comparison, and gray bars highlight the positions of RPGs within this cluster. In the heatmap, saturated blue indicates at least a 5.7-fold decrease, and saturated yellow at least a 5.7-fold increase in transcript abundance. (B) Averaged RPG and non-RPG transcript behaviors across data sets shown in A.
Hyperosmotic and cationic stresses lead to transient inactivation of TORC1 (Urban et al. 2007), and the expected concomitant down-regulation of RPG transcription is reflected by the loss of RPG pre-mRNA. The persistence of the mature and total mRNA species for the RPGs, relative to the other TORC1-inactivating stresses in the previous class of responses, could reflect an increase in the stability of the cytoplasmic pool of mature mRNAs for these genes, or a higher proportion of the pre-mRNAs being spliced to mature transcripts rather than going directly to decay, or both.
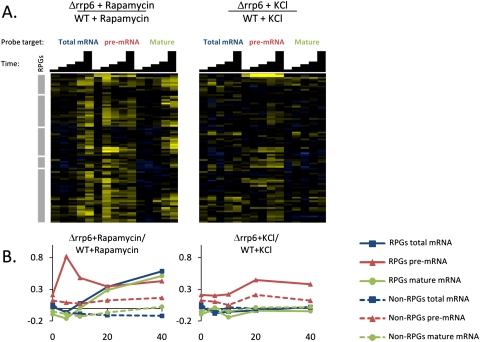
We wanted to further pursue the question of whether the mechanisms contributing to the loss of RPG pre-mRNA are distinct for the second and third classes of stress. Changes in three rates contribute to changes in the steady-state levels of pre-mRNAs: the rate of appearance of new pre-mRNAs due to transcription, the rate of disappearance of pre-mRNAs due to splicing, and the rate of disappearance of pre-mRNAs due to degradation. To determine the contribution of active nuclear degradation to the loss of RPG pre-mRNAs in these two classes of stress, we repeated the rapamycin and KCl treatments in a Δrrp6 strain, and competitively hybridized these samples directly against samples from the wild-type strain (Fig. 5). Rrp6 is a nucleus-specific component of the exosome that contributes to nuclear decay of aberrant mRNAs (Schmid and Jensen 2008; Honorine et al. 2010). At steady state, levels of many pre-mRNAs are mildly elevated in a Δrrp6 strain compared with a wild-type strain (see the time 0 column in Fig. 5).
FIGURE 5.
The nuclear exosome component Rrp6 contributes to the decrease in RPG transcripts in rapamycin treatment. (A) Time-resolved splicing profiles are shown from comparisons of Δrrp6 cells to wild-type cells both treated with either rapamycin or KCl, with samples across a time course of 0, 5, 10, 20, and 40 min after the beginning of the treatment. Transcripts were arranged on the vertical axis by hierarchical clustering, and the major branch containing the majority of the RPGs is shown. Gray bars highlight the positions of RPGs within this cluster. In the heatmap, saturated blue indicates at least a 5.7- fold decrease, and saturated yellow at least a 5.7-fold increase in transcript abundance. (B) Averaged RPG and non-RPG transcript behaviors across data sets shown in A.
However, upon rapamycin treatment, the levels of most RPG pre-mRNAs become dramatically higher in the Δrrp6 strain compared with the wild type subjected to the same treatment. At later time points, the levels of mature RPG transcripts are also higher in the Δrrp6 strain, suggesting that, in the wild-type strain, the nuclear degradation of pre-mRNAs at early points in the time course contributes substantially to the decrease in mature transcript observed at later time points. In contrast, in the KCl treatment, most transcripts show little additional accumulation over the time course, although a small subset of RPG pre-mRNAs show mild increases in the Δrrp6 strain at the 20- and 40-min time points. Thus, it appears that nuclear degradation of RPG pre-mRNAs does not account for most of the decreases observed during KCl treatment, especially at early time points. Instead, the decreases in this stress could be due to a decreased rate of new transcription, or an increased rate of pre-mRNA splicing. Previous work has suggested that RPG transcription is, in fact, down-regulated during hyperosmotic stress (Gasch et al. 2000), and that the RPG transcripts are extremely efficiently cotranscriptionally spliced in nonstress conditions (Carrillo Oesterreich et al. 2010). Based on these findings, we believe it most likely that the decreases observed in the RPG pre-mRNA in the KCl treatment are due to decreased transcription with little change in splicing efficiency, and not due to increased splicing efficiency.
Although quantitative analysis of the behavior of the splicing isoform-specific species in various stresses yields three distinct classes of response, it is also clear that each response has some unique characteristics, and that some characteristics are shared across classes of response. The hierarchical clustering algorithm defined two major branches in a dendrogram representing all of the stresses (Fig. 1, right): one that contained stresses causing increases in the pre-mRNAs for at least some RPGs, and another that contained all of the stresses that displayed decreases in the pre-mRNAs for the RPGs. The stresses in the first branch are not known to lead directly to TORC1 inactivation or cell cycle arrest, while the second branch included the stresses expected to inactivate TORC1. The classification of the responses by both clustering algorithms is largely driven by the behavior of the RPGs, as they represent ∼40% of the transcripts analyzed, and their abundances were generally coordinately and dramatically affected by the stresses we tested. We were interested in gaining further insight into which additional signaling pathways might contribute to the observed changes in RPG pre-mRNA processing. Our previous work had shown that Gcn2, the kinase canonically responsible for sensing amino acid starvation and signaling to the translation machinery (for review, see Hinnebusch 2005) was not required for the RPG pre-mRNA accumulation response to amino acid starvation (Pleiss et al. 2007a). We undertook a candidate screening approach to find other kinases that might be involved in mediating responses to stress at the level of pre-mRNA splicing.
CK2 is involved in mediating stress responses at the level of pre-mRNA processing
We focused initially on identifying factors that were required for the accumulation of pre-mRNA in response to amino acid starvation. Because direct inactivation of TORC1 by rapamycin or wortmannin clustered with a variety of stresses known to cause TORC1 inactivation, and away from amino acid starvation stresses, we sought a candidate that did not have known direct interactions with TORC1 signaling. CK2 satisfied this condition, and, additionally, was shown to genetically interact with several splicing factors in yeast (Wilmes et al. 2008).
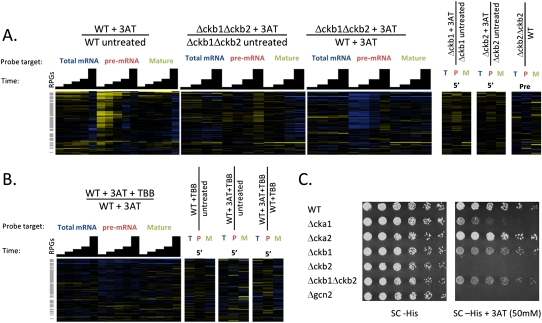
CK2 consists of two catalytic subunits (Cka1 and Cka2) and two regulatory subunits (Ckb1 and Ckb2). Deletion of any single subunit does not cause an obvious growth phenotype, and deletion of both regulatory subunits yields an epistatic (positive) genetic interaction in a quantitative epistasis mini-array profile (Collins et al. 2007). We thus started by testing the ability of a Δckb1Δckb2 double regulatory subunit mutant to respond at the level of pre-mRNA splicing to amino acid starvation. Strikingly, we found that the accumulation of RPG pre-mRNA following 3-AT treatment was almost completely abolished in this mutant (Fig. 6A). We next looked at the effects of deleting each of the two regulatory subunits individually, and found that while each of the deletions diminished the accumulation of RPG pre-mRNA following 3-AT treatment relative to the wild type, the Δckb2 mutant had a stronger effect than did the Δckb1 mutant.
FIGURE 6.
CK2 is required for inhibition of RPG pre-mRNA splicing following amino acid starvation. (A) Splicing profiles are shown for wild-type or CK2-deficient cells (Δckb1Δckb2) undergoing amino acid starvation induced by 3-AT compared with an untreated control. (B) Splicing profiles are shown for wild-type cells undergoing amino acid starvation induced by 3-AT in either the presence or absence of a CK2 inhibitor, TBB. In both A and B, transcripts are arranged on the vertical axis by hierarchical clustering, gray bars highlight the positions of RPGs, and in the heatmap, saturated blue indicates at least a 5.7-fold decrease in transcript abundance and saturated yellow at least a 5.7-fold increase. (C) Growth phenotypes for CK2 mutants under control (SC-HIS) and amino acid starvation conditions (SC-HIS + 3AT).
We also examined the effects of pretreating cells with a drug that inhibits CK2 activity, 4,5,6,7- Tetrabromo-2-azabenzimidazole (TBB) (Pagano et al. 2008). Inhibiting the catalytic activity of CK2 strongly abrogates the accumulation of RPG pre-mRNAs in response to 3-AT as well (Fig. 6B). Deletion of the CK2 regulatory subunits and treatment with TBB both cause some mild changes to steady-state levels of pre-mRNA, mature mRNA, and total mRNA for the RPGs, but these changes do not phenocopy the changes observed in response to 3-AT (Fig. 6A,B). This suggests that normal CK2 activity constitutively influences transcription, splicing, and/or turnover of the RPG transcripts, but that the loss of 3-AT response observed when CK2 is perturbed is not because the perturbation itself causes cells to be in an amino acid starvation response state constitutively.
Finally, we asked whether the contribution made by CK2 to the amino acid starvation response at the level of pre-mRNA processing might correlate with a fitness phenotype under amino acid starvation conditions. We spotted serial dilutions of strains carrying mutations for each of the CK2 subunits, plus the regulatory subunit double mutant, onto plates containing 50 mM 3-AT. For reference, a strain carrying a deletion of Gcn2, the kinase involved in the translational response to amino acid starvation, was also plated. The Δckb2 strain showed a strong fitness defect under conditions of exposure to 3-AT. Interestingly, the Δckb1 strain showed a milder fitness defect, and the double regulatory subunit delete showed a defect similar to the milder Δckb1 defect. The Δcka1 strain also showed an intermediate fitness defect, while the Δcka2 strain grew as well as the wild type (Fig. 6C). The fitness defect of the catalytic subunit delete is perhaps consistent with the pre-mRNA processing defect caused by inhibiting the catalytic activity of CK2 with TBB. These results suggest that the CK2 subunits may have specialized functions with regard to the amino acid starvation response.
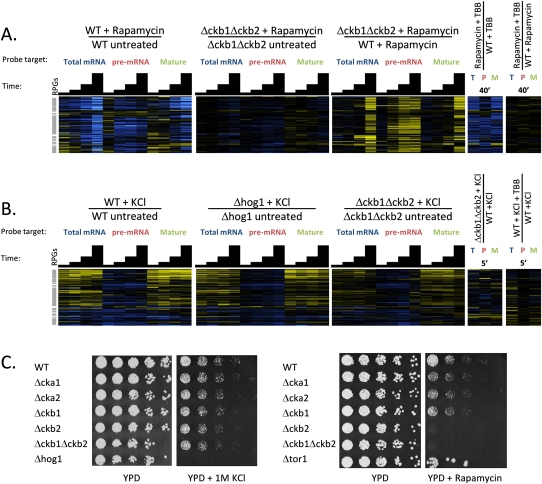
Because the response to rapamycin also involves a change in processing of the RPG pre-mRNAs, leading to their Rrp6-mediated degradation, we asked whether CK2 contributes to this response as well. We subjected the Δckb1Δckb2 double mutant to the rapamycin stress time course, and found that the loss of RPG pre-mRNA was diminished in this mutant compared with the wild type (Fig. 7A). We also pretreated the wild-type strain with TBB before subjecting it to rapamycin treatment, and in contrast to the result with the regulatory subunit double mutant, found that inhibiting the catalytic activity of CK2 had little effect on the response to rapamycin treatment (Fig. 7A). We also assayed each of the CK2 subunit deletes for fitness defects by spotting the strains onto plates containing rapamycin. Interestingly, the pattern of sensitivity to rapamycin was distinct from that observed for 3-AT. Deletion of CKB2 caused extreme sensitivity that was not suppressed by deletion of CKB1 in the double mutant. No other subunit deletion caused substantial sensitivity to rapamycin, consistent with the observation that deletion of both regulatory subunits, but not inhibition of the catalytic activity of CK2, diminished the response to rapamycin at the level of RPG pre-mRNA processing.
FIGURE 7.
Effects of CK2 inhibiton on other stress response classes. (A) Splicing profiles of cells in control or rapamycin treatment carrying caseine kinase 2 deletion mutations or treated with a CK2 inhibitor (TBB). (B) Splicing profiles of cells undergoing osmotic stress induced by KCl, carrying deletions in HOG1 or CK2, or treated with the CK2 inhibitor TBB. In both A and B, transcripts are arranged on the vertical axis by hierarchical clustering, gray bars highlight the positions of RPGs, and in the heatmap, saturated blue indicates at least a 5.7-fold decrease in transcript abundance and saturated yellow at least a 5.7-fold increase. (C) Growth phenotypes for CK2 mutants under control (YPD) and KCl or rapamycin treatment conditions.
To determine which signaling pathways might contribute to the loss of RPG pre-mRNA in hyperosmotic stress, we started by exposing Δhog1 cells to a KCl treatment time course. Hog1 is the mitogen-activated kinase known to be responsible for transcriptional induction of several genes involved in the response to high osmolarity. However, we found the response to KCl at the level of RPG pre-mRNA processing to be virtually unperturbed by deletion of this kinase (Fig. 7B), reminiscent of the lack of requirement for Gcn2 in the pre-mRNA processing response to amino acid starvation. We also tested whether CK2 contributes to this stress response by measuring the response in the Δckb1Δckb2 strain and in cells pretreated with TBB, and found that neither perturbation of CK2 function affected the behavior of the RPG transcripts during KCl treatment (Fig. 7B). Consistent with this observation, none of the CK2 subunit deletions caused a substantial fitness defect when grown on plates containing 1 M KCl (Fig. 7C).
Several groups of transcripts are differentially regulated at the level of pre-mRNA processing
Thus far, we have focused on the behavior of the RPG transcripts as a group, because a majority of these transcripts behave coordinately across most of the stresses we tested, and these behaviors dominate quantitative analysis of stress-induced changes to pre-mRNA processing as a whole. In order to look for subgroups of RPGs behaving distinctly under stress, as well as for groups of non-RPG transcripts that might be coordinately regulated at the level of pre-mRNA processing, we again used hierarchical and k-means clustering. This time, instead of analyzing the behavior of each stress across all transcripts, we subjected to clustering the behavior of each transcript across all stresses. Hierarchical clustering produced a complex dendrogram (Fig. 8A, right), and k-means clustering produced five groups of transcripts (Fig. 8A, left). Two of these (orange and blue) comprised subgroups of RPGs, two other small groups (purple and green) contained primarily non-RPG transcripts, and the fifth group contained the rest of the transcripts (white). The larger RPG subgroup (blue) contains most of the RPG transcripts that show strong pre-mRNA accumulation in response to the amino acid starvation stresses and strong loss of pre-mRNA in response to TORC1-inactivating stresses.
Interestingly, χ2 analysis of paralog pairs of the RPGs revealed that both members of a paralog pair were less likely than expected by chance to be present in the same subgroup of RPGs (P < 10-4). Examples of paralog pairs that separate into the two different RPG subgroups include RPL14A (blue) and B (orange), and RPS11A (orange) and B (blue) (Fig. 8B). In contrast, one of the small clusters (purple) contains both paralogous copies of the U3 snoRNA. These transcripts seem to behave oppositely to the majority of the RPG transcripts in many of the stresses tested (Fig. 8B). While the RPGs are largely being degraded in stringent stresses, the levels of mature and total U3 snoRNA go up, and the levels of the pre-RNA for these transcripts go down.
The other non-RPG cluster, represented by green in Figure 8A, contains several transcripts whose pre-mRNA and mature mRNA levels change dramatically in some stresses, but are not easily classified as having a coherent set of biological functions. Nonetheless, some interesting patterns can be observed. The ACT1 gene, which has traditionally been used as a splicing reporter, shows increased levels of both the pre-mRNA and the mature mRNA in many stresses. For example, heat shock, which has previously been reported to cause accumulation of the ACT1 pre-mRNA, does indeed do so in our hands, but it also leads to an increase in the mature message. TUB1 and TUB3 are paralogs encoding α-tubulin. Like many of the RPG paralogs, they fall into different k-means clusters on the basis of their behavior in stress (Fig. 8B). HNT1 and HNT2 belong to the histidine triad family of nucleotide-binding proteins. They both fall into the green cluster of transcripts in this analysis, and interestingly, show strong pre-mRNA accumulation in some of the stresses where RPG pre-mRNAs are rapidly lost. This further illustrates that regulation at the level of pre-mRNA processing is dynamic and transcript specific, with a range of behaviors observed for different transcripts under the same stress condition.
DISCUSSION
We present here a broad survey of pre-mRNA processing changes in response to stress. All of the types of amino acid starvation that we tested led to a transient accumulation of RPG pre-mRNA, and the magnitude of the accumulation scaled with the severity of the stress. Unlike amino acid starvation, stresses that led to TORC1 inactivation caused a rapid loss of RPG pre-mRNA, and could be further subdivided into two categories: one category that is characterized by a strong decrease in the mature mRNAs for the RPGs following a decrease in the RPG pre-mRNAs, and another category that is characterized by relative stabilization of the mature RPG mRNAs despite decreases in the pre-mRNAs. We showed that the nuclear exosome component Rrp6 was required for the RPG mRNA decreases seen in response to rapamycin, which fell into the first class of stresses leading to TORC1 inactivation, but not for the decreases seen in response to KCl, a member of the second class. We also showed that perturbation of CK2 has different effects on different stress responses. Finally, we can observe different patterns of change for a variety of non-RPG transcripts. In total, these results suggest that regulation at the level of pre-mRNA processing contributes specifically and dynamically to the regulation of the gene expression program in the face of a wide range of environmental challenges.
Fine-tuning regulation of ribosomal protein gene expression and ribosome biogenesis
By far the most striking effects observed in this study were on RPG transcripts, suggesting that changes to pre-mRNA processing can contribute to regulation of these transcripts in the context of stresses that inhibit translation at different levels and over different periods of time. It seems likely that changes to pre-mRNA processing could allow for modulation of both the initial kinetics of the down-regulation and also the kinetics of the recovery from the stress response.
In the case of amino acid starvation, translation is at least transiently down-regulated at the level of initiation, which contributes to the Gcn4-dependent induction of amino acid biosynthetic genes and also presumably conserves cellular energy resources while amino acid biosynthesis is being up-regulated (Hinnebusch 2005). It has been shown, however, that amino acid starvation does not induce a protracted growth arrest. Even starvation of an auxotroph for the amino acid it cannot make fails to induce a G0 arrest, despite a long-term loss of viability, while starvation of the same strain for glucose, nitrogen, or phosphate does induce a protective G0 growth arrest (Boer et al. 2008). For a wild-type yeast strain, a decrease in the environmental availability of one or several specific amino acids in the presence of sufficient carbon, nitrogen, sulfate, and phosphate sources should not require a growth arrest, as the missing amino acids can be synthesized. The regulation at the level of pre-mRNA processing of the RPGs that we have observed, in which splicing is inhibited and pre-mRNAs transiently accumulate, could provide a transient inhibition of ribosomal protein biogenesis consistent with the transient translational inhibition caused by amino acid starvation. The increasing accumulation of RPG pre-mRNA during the first 10 min of the stress suggests that transcription of the RPGs is ongoing. In the mildest form of this stress, histidine depletion, the RPG pre-mRNA accumulation was very modest and transient indeed, and no decreases in the mature pools of RPG mRNA were detected, suggesting that transcription of the RPGs was largely unperturbed. The capacity to transiently down-regulate RPG expression by a mechanism that allows RPG promoters to remain active could allow for a faster resumption of maximal growth rate following alleviation of the amino acid shortage.
In contrast, the common characteristic of the other two groups of stresses was a decrease in the pre-mRNAs for the RPGs. In the second class of stress responses (including heat shock, rapamycin, wortmannin, and glucose starvation), a rapid decrease in pre-mRNA levels for the RPGs was followed by a strong decrease in the mature mRNA levels. The requirement for Rrp6 in this response suggests that active degradation of the pre-mRNA contributes substantially both to the early loss of pre-mRNA and the later loss of mature mRNA. Interestingly, when rapamycin treatment was combined with 3-AT treatment, the response closely approximated the response to rapamycin alone, suggesting that this more stringent response at the level of RPG pre-mRNA processing (leading to nuclear turnover of nascent transcripts) is dominant over the transient amino acid starvation response (Supplemental Fig. S22). This epistatic behavior suggests that the two responses, although very different, impinge on the same process, and is consistent with the observation that both responses require the Ckb2 subunit of CK2. The pattern of RPG pre-mRNA loss observed for this class of stresses is indicative of preparation for a protracted arrest, and also a very rapid downshift in ribosome production, which may be appropriate when energy resources are at a premium or when protein folding resources are overtaxed.
For all of the hyperosmotic and ionic stresses, the loss of mature mRNAs is mild compared with the loss of pre-mRNAs for the RPGs, over a wide range of magnitudes of pre-mRNA loss. This may be due to stabilization of the cytoplasmic pools of mature mRNA, and is consistent with recent studies that have shown decreased decay rates specifically for RPG mRNAs in hyperosmotic and ionic stresses, even as these messages were being transcriptionally repressed (Molin et al. 2009; Romero-Santacreu et al. 2009). Although the cationic LiCl stress caused intermediate phenotypes in these assays (Supplemental Fig. S23), neither Rrp6 nor CK2 was required for the response to KCl, suggesting that the response does not involve active turnover of the RPG pre-mRNA, and that the regulation of this response is distinct from the regulation of the amino acid starvation and rapamycin responses. We also found that combining the KCl and 3-AT stresses yielded a response that was distinct from the responses to either individual stress (Supplemental Fig. S22). This nonepistatic behavior supports the model that amino acid starvation and KCl affect RPG transcripts via distinct pathways. In any case, a mechanism of RPG repression whereby new transcription is halted, but the messages that have already been transcribed continue to be processed and/or stabilized to varying degrees, could allow the cell to prepare for a protracted growth arrest while still transiently maintaining needed translational capacity for adapting to the stress.
Consistent with the idea that regulation at the level of pre-mRNA processing may contribute to both induction of and recovery from a stress response, we also observed interesting behavior in the pre- and mature RNA species for the two copies of U3, the spliced snoRNA that helps direct rRNA cleavage, and is therefore essential in ribosome biogenesis. In the stresses that most strongly down-regulate the RPGs at both the pre- and mature mRNA levels, the U3 transcripts showed increases for the total and mature species and decreases for the pre-RNA. Thus, U3 appears to be up-regulated at the same time the cell is broadly down-regulating ribosome biogenesis. While the requirement for new ribosomes is much diminished during a G0 arrest, translation does not stop completely. It has been shown that after the initial rapid and dramatic polysome collapse during glucose starvation, translation can in fact resume (Ashe et al. 2000). The cell may need to up-regulate U3 in preparation for resumed protein synthesis. Interestingly, the salt stresses just show a decrease of the unspliced U3 message, without a corresponding increase in the mature message, and the amino acid starvation stresses show little change in the abundance of the U3 transcript species. Thus, it appears that regulation at the level of pre-mRNA processing may also differentially influence the levels of a ribosome biogenesis factor in different stresses.
Another observation potentially consistent with ongoing but differentially regulated synthesis of ribosomes under stress conditions is the tendency of paralogous pairs of ribosomal protein genes to show differential behavior across stress treatments. Different regulatory elements governing expression of the RPG paralogs could allow for ongoing expression of each required ribosomal protein under very different regulatory regimes. For example, in some cases it is known that one of the paralogs is more highly expressed under rich nutrient conditions (Komili et al. 2007), and the two paralogous genes encoding a ribosomal protein often show different genetic interactions (Haarer et al. 2007). It may be that expression of the other paralog is important under less optimal conditions, when expression of the normally more highly expressed paralog is shut down. Notably, we also observed distinct behavior across the stress treatments for the TUB1 and TUB3 transcripts (paralogous copies of the gene encoding α tubulin). This suggests that maintaining paralogous copies of highly expressed genes, which can be differentially regulated both at the level of transcription and at the level of pre-mRNA processing, may be a broadly utilized strategy.
CK2 has stress-specific effects on pre-mRNA processing
We have shown here that perturbing CK2 in various ways alters the response at the level of pre-mRNA processing to some stresses, and causes sensitivity to those same stresses. CK2 is known to phsophorylate a large number of substrates with roles in transcription and splicing, but its regulation is not well understood. Our results suggest that one role for CK2 may be the modulation of pre-mRNA processing dynamics of ribosomal protein genes under different environmental conditions, and give some broad insight into possible regulatory mechanisms. The Ckb2 regulatory subunit appears to have a distinct role in the responses to both amino acid starvation and rapamycin. Deletion of CKB2 caused significantly more sensitivity to these stresses than deletion of any of the other subunits. This is consistent with the recent finding that CKB2, but not any other CK2 subunit, was among a cluster of genes that, when deleted, decreased competitive fitness under both phosphate and leucine starvation conditions (Gresham et al. 2011). In mammalian cells, the CK2 β regulatory subunit appears to be capable of associating with and regulating other kinases besides CK2—it has been reported to activate A-Raf, Chk1, and Wee1, while it appears to inhibit c-Mos (for review, see Bibby and Litchfield 2005). Alternate kinase associations for either of the regulatory subunits have not yet been identified in yeast, but our results suggest that, at least under some stress conditions, Ckb2 may have an important independent function.
The pattern of sensitivity observed for 3-AT is intriguing—deletion of CKB2 alone causes the strongest growth inhibition, while deletion of CKB1 on top of the CKB2 deletion suppresses some of that sensitivity. Deletion of CKB1 alone or CKA1 alone causes a similar degree of sensitivity as the Δckb1Δckb2 double delete. This is distinct from the pattern observed with rapamycin, where deletion of CKB2 caused a high degree of sensitivity whether or not CKB1 was also deleted. This pattern of sensitivity may suggest that Ckb1 can also function independently of the CK2 holoenzyme, and possibly antagonistically to Ckb2 in the 3-AT response. The fact that the Δckb1Δckb2 double delete and the Δcka1 delete show some sensitivity, suggests that the CK2 holoenzyme likely also plays some role in the 3-AT response. The multisubunit nature of CK2, and the apparent ability of its four subunits to act both independently and also as a heterotetrameric holoenzyme, suggests a model in which a wide range of possible combinations of subunit associations allows for integration of diverse inputs affecting the behavior of CK2, and also a diverse range of outputs that in turn affect multiple aspects of gene expression. The fact that its subunits appear to play independent roles in addition to contributing to activity of the holoenzyme suggests the intriguing possibility that different functional roles may compete for CK2 subunits. Such a model is reminiscent of previous work suggesting that CK2 might participate in coordinating synthesis of RPG mRNA and rRNA. This work showed that excess rRNA processing factors could compete the RPG transcription factor Ifh1 away from active transcription complexes containing Rap1 and Fhl1, thus down-regulating RPG transcription. The complex containing Ifh1 and the rRNA processing factors Utp22 and Rrp7 also contained at least some subunits of CK2 (Rudra et al. 2007).
Regulation of non-RPG transcripts at the level of pre-mRNA processing
One motivation for measuring pre-mRNA and mature mRNA species for a majority of the intron-containing genes across a wide variety of stresses was to determine whether other groups of transcripts might show clear coregulation at the level of pre-mRNA processing. The behavior of the RPGs dominated the patterns of change to pre-mRNA processing in all stresses tested, but clustering analysis of transcript behavior across all stresses did identify some additional transcripts whose levels of pre-mRNA and mature mRNA change in a variety of stresses. These transcripts did not fall into easily identifiable biological categories. Since we intentionally chose stresses known to down-regulate translation, it is not surprising that most of the effects we observed were on ribosomal protein genes. Thus, there may well be functionally coherent groups of transcripts that are coordinately regulated at the level of pre-mRNA splicing in response to other types of stress. For example, meiotic genes are also over-represented among the intron-containing genes in yeast, and have been shown to be regulated both at the level of transcription and at the level of splicing (Munding et al. 2010). While we could not measure expression of meiotic genes in the haploid cells we used for these experiments, it is interesting to note that the conditions that induce meiosis in S. cerevisiae diploids involve extreme nutrient deprivation—depletion of both carbon and nitrogen sources for a long period of time. Regulation at the level of pre-mRNA processing may in fact play a role at multiple stages in a response to ongoing nutrient deprivation of varying degrees of severity—inhibition of RPG splicing in response to a transient and mild starvation, active degradation of RPG pre-mRNAs in response to a more severe or more protracted starvation, and induction and splicing of meiotic genes when a diploid is exposed to protracted, severe starvation. In each case, the capacity to regulate expression of the relevant genes at the level of pre-mRNA processing may contribute to fine-tuning the kinetics of changes to gene expression.
Another potential function for regulation at the level of pre-mRNA processing may be to dampen changes in expression levels, which would be difficult to detect using the measurements in this study. Some intron-containing genes, such as GLC7, ACT1, and YRA1, are known to be toxic when overexpressed, and the presence of an intron in these genes may help prevent excess transcripts from being expressed (Liu et al. 1992; Rodriguez-Navarro et al. 2002). In the case of YRA1, the intron is required for an autoregulatory mechanism by which excess Yra1 protein prevents splicing of the YRA1 transcript (Preker et al. 2002; Preker and Guthrie 2006). For the ACT1 transcript, we see that many stresses lead to increases in both the pre-mRNA and the mature transcript. One explanation for this pattern could be that under conditions of transcriptional up-regulation, splicing can become rate-limiting, thus dampening the increase in expression of a protein for which dosage is sensitively regulated.
Our work has demonstrated that the presence of introns in RPGs provides distinct opportunities for fine-tuning the regulation of their expression under a wide range of stress conditions. The broad scope of this investigation has allowed us to gain insight into the contribution of regulation at the level of pre-mRNA processing to the overall regulation of gene expression in budding yeast. The capacity for diverse regulatory responses has far-ranging implications for the potential of this process to contribute substantially to control of both the magnitude and kinetics of responses. The high level of conservation of the pre-mRNA processing machinery across all eukaryotes suggests that mechanistic insights gained here, implicating CK2 in regulating stress-responsive changes to pre-mRNA processing, are likely to be broadly applicable. We expect that future investigations utilizing this tractable model will continue to yield insight into the mechanism of regulation at the level of pre-mRNA processing.
MATERIALS AND METHODS
Strains and growth conditions
The strain yMB1 (Pleiss et al. 2007a) was used as the wild-type strain in all stress experiments, and was constructed by repairing the HIS3 locus in the BY4742 strain (Brachmann et al. 1998) by homologous recombination of the full HIS3 open reading frame into the partially deleted locus. The BY4742 strain is isogenic except for the HIS3 locus, and was used as the histidine auxotroph in the histidine depletion experiments. The Δckb1 Δckb2 strain was constructed by crossing the MATa Δckb1::KAN strain from the deletion collection (Giaever et al. 2002) to the MATα Δckb2::NAT strain carrying SGA markers (Fiedler et al. 2009). From this cross, spores carrying both deletions, but none of the SGA markers, were selected, and the HIS3 locus was subsequently repaired as in yMB1, yielding the following genotype: MATa Δckb1::KAN Δckb2::NAT leu2Δ0 ura3Δ0. The Δrrp6, Δcka1, Δcka2, Δckb1, and Δckb2 strains were BY4741 strains from the deletion collection in which the HIS3 locus was repaired as described above, and the Δhog1 strain was a BY4741 strain from the deletion collection (Giaever et al. 2002).
In general, for each stress condition, a 50-mL culture was grown to mid-log phase (OD600 0.3–0.7) in the appropriate starting medium at 30°C, and then the stress was applied as described below. Synthetic complete media is defined as: 6.7 g/L of yeast nitrogen base (containing 5 g/L of ammonium sulfate, 1 g/L of monopotassium phosphate, and 0.2 g/L of inositol, among other vitamins and nutrients, from Difco unless otherwise noted), the following supplemented amino acids and purines (all from Sigma): 15 mg/L of valine; 20 mg/L of arginine, adenine, histidine, methionine, tryptophan, and uracil; 30 mg/L of lysine, tyrosine, and isoleucine; 50 mg/L of phenylalanine; 60 mg/L of leucine; and 200 mg/L of threonine, and 20 g/L of glucose. Rich media is defined as 10 g/L of bacto yeast extract (Difco), 20 g/L of bacto peptone (Difco), and 20 g/L of glucose. Except in the case of heat shock, growth after the stress application was also at 30°C. A total of 10–15 mL aliquots of culture were collected by centrifugation at 5, 10, 20, and 40 min after application of the stress, and cell pellets were immediately frozen in liquid nitrogen.
3-Amino-1,2,4-triazole (3-AT) treatment
Yeast were grown to mid-log phase in synthetic media lacking histidine, and then 3-AT (Sigma) was added to a final concentration of 50 mM.
Amino acid depletion
Yeast were grown to mid-log phase in synthetic complete media and then collected on a 95-mm nitrocellulose filter (Whatmann) by vacuum filtration. Cells were washed with 500 mL of media containing only yeast nitrogen base plus ammonium sulfate and glucose, and were then resuspended in 50 mL of this media.
Histidine depletion
Yeast were grown to mid-log phase in synthetic complete media, and then collected on a 95-mm nitrocellulose filter by vacuum filtration. Cells were washed with 500 mL of synthetic media lacking histidine and then resuspended in 50 mL of this media.
Sulfometuron methyl (SMM) treatment
Yeast were grown to mid-log phase in synthetic media lacking isoleucine and valine, and then SMM (Sigma) from a stock solution of 1 mg/mL of SMM in acetone was added to a final concentration of 5 μg/mL.
Phosphate depletion
Yeast were grown to mid-log phase in synthetic complete media and then collected on a 95-mm nitrocellulose filter by vacuum filtration. Cells were washed with 500 mL of synthetic media made using yeast nitrogen base that lacked monopotassium phosphate (MP Biomedicals) and were then resuspended in 500 mL of this media.
Glucose depletion
Yeast were grown to mid-log phase in rich media, and then collected on a 95-mm nitrocellulose filter by vacuum filtration. Cells were washed with 500 mL of rich media lacking glucose and were then resuspended in 50 mL of this media.
Rapamycin treatment
Yeast were grown to mid-log phase in rich media, and then rapamycin (Calbiochem) from a stock solution of 1 mg/mL in 90% ethanol, 10% Tween-20 was added to a final concentration of 200 ng/mL.
Wortmannin treatment
Yeast were grown to mid-log phase in rich media, and then wortmannin (Sigma) from a stock solution of 1 mg/mL in DMSO was added to a final concentration of 10 μg/mL.
Dithiothreitol (DTT) treatment
Yeast were grown to mid-log phase in rich media, and then DTT (Sigma) from a stock solution of 1 M in water was added to a final concentration of 10 mM.
Heat shock
Yeast were grown to mid-log phase in rich media, and then were shifted to a 42°C water bath for 5 min with manual shaking. Following the collection of the aliquot for the 5-min time point, the culture was incubated on a rotary shaker at 42°C for the remainder of the time course.
Lithium chloride (LiCl) treatment
Yeast were grown to mid-log phase in rich media, and then collected on a 95-mm nitrocellulose filter by vacuum filtration. Cells were washed with 500 mL of rich media containing 300 mM LiCl and were then resuspended in 50 mL of this media.
Potassium chloride (KCl) treatment
Yeast were grown to mid-log phase in rich media, and then collected on a 95-mm nitrocellulose filter by vacuum filtration. Cells were washed with 500 mL of rich media containing 1 M KCl and were then resuspended in 50 mL of this media.
Sodium chloride (NaCl) treatment
Yeast were grown to mid-log phase in rich media, and then collected on a 95-mm nitrocellulose filter by vacuum filtration. Cells were washed with 500 mL of rich media containing 500 mM NaCl and were then resuspended in 50 mL of this media.
Sorbitol treatment
Yeast were grown to mid-log phase in rich media, and then collected on a 95-mm nitrocellulose filter by vacuum filtration. Cells were washed with 500 mL of rich media containing 1 M sorbitol and were then resuspended in 50 mL of this media.
4,5,6,7-Tetrabromo-2-azabenzimidazole (TBB) treatment
For TBB treatment alone, yeast were grown to mid-log phase in rich media, and then TBB (Sigma), from a stock solution of 10 mM in DMSO, was added to a final concentration of 20 μM. For pretreatment with TBB, yeast were grown and stress was applied as described above, but TBB was added to a final concentration of 20 μM 20 min prior to the application of the stress.
Growth assays
Yeast were grown to mid-log phase and were then diluted to an OD600 nm of 0.1. From this, fourfold serial dilutions were made and spotted on agar plates containing the indicated nutrients and/or drugs. Plates were incubated at 30°C for 2–4 d.
Microarrays
RNA was isolated from the frozen cell pellets by hot acid phenol extraction as has been described previously (Schmitt et al. 1990). Briefly, pellets were resuspended and mixed vigorously in equal volumes of acid phenol and aqueous buffer, then incubated for 15 min at 65°C, mixing vigorously every 2–3 min. An equal volume of chloroform was added and the mixture transferred to a 2-mL phase-lock tube (eppendorf). The mixture was additionally extracted with an equal volume of phenol:chloroform:isoamyl alcohol and then an equal volume of chloroform. The aqueous phase was transferred to a new microcentrifuge tube and sodium acetate was added to a final concentration of 300 mM. An equal volume of isopropyl alcohol was added, and the RNA was allowed to precipitate for at least 2 h at −20°C. The precipitated RNA was washed once with 70% ethanol, dried, and resuspended in water. A total of 50 μg of RNA was used for each cDNA synthesis reaction to generate cDNA for dye-flipped replicate arrays. cDNA synthesis, clean-up, and labeling was carried out in 96-well plate format, and hybridization to a custom splicing microarray platform was performed as described previously (Pleiss et al. 2007b).
Microarray image analysis and preprocessing
Microarray images were acquired using an Axon Instruments GenePix 4000B scanner, reading at wavelengths of 635 and 532 nm (Molecular Devices, http://www.moleculardevices.com). Image analysis was performed using Axon Instruments GenePix Pro version 6.0. Ratio values derived from the median pixel intensities for the 635- and 532-nm images of each spot were used to represent probe behaviors in data preprocessing analysis. A standardized qualitative assessment of array quality was performed using the Bioconductor arrayQuality package, version 1.4.0 s (Gentleman et al. 2004). Spot ratio data were log2 transformed and normalized within each array using print-tip-based loess regression implemented in the Bioconductor MArray package (Wang et al. 2002). As all experimental contrasts are represented in the data set at least twice (as dye-flipped replicates) and as many as six times for biological replicates, both within-array and between-array data replication were analyzed for data quality. Array pairs with lower than 80% correlation values were discarded from any further analysis. Additionally, arrays showing exceptional within-array variability (measured by mean replicate variance >2 SD above average across data set) were discarded from further analysis. The final microarray data set represents 594 unique hybridizations covering each experimental contrast with between two- and sixfold replication. Prior to higher order analysis, within- and between-array replicates on high-quality arrays were averaged. Files containing the preprocessed and averaged data for all experiments are available online at http://biochemistry.ucsf.edu/labs/guthrie/bergkesselWhitworth2011.html.
Clustering
Hierarchical clustering was performed using the R standard library distance matrix computation (Becker et al. 1988; Borg and Groenen 1997). After preprocessing, data were clustered without centering or filtering. Unless otherwise noted, distance calculations were performed using Euclidean distance, and clustering was performed among centroids. Clustering heatmaps were drawn using the R standard library “heatmap” function without scaling and with color saturation values at fold changes of 2(−2.5) shown as blue and 2(2.5) shown as yellow. Row and column annotations, including RPG transcript location and k-means clustering classes, were also drawn using this package. K-means clustering was performed with the C Clustering Library (v. 1.49) implementation (de Hoon et al. 2004) using Euclidean distance to classify transcript behaviors (Fig. 8A) and Kendall's tau to classify environmental treatments (Fig. 1). For k-means clustering, solutions were calculated for all possible response class numbers [k = 2−(n−1)], and models failing to show robust solutions over 103 independent runs were discarded. For pairwise transcript distance matrix calculations, experimental time points were chosen from each stress data set representing the largest average absolute transcript level change between experimental and control samples. Similarity in transcript behaviors across all stresses was calculated using Euclidean distance. This similarity matrix was then clustered using hierarchical clustering and k-means clustering (both as described above).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article and may be viewed online at http://biochemistry.ucsf.edu/labs/guthrie/bergkesselWhitworth2011.html.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM21119. M.B. was supported by an HHMI predoctoral fellowship. G.B.W. received funding from the Committee for the Support of Faculty Scholarship at Grinnell College. C.G. is an American Cancer Society Research Professor of Molecular Genetics. We thank Quinn Mitrovitch, Alex Plocik, Rebecca Holmes, Jaclyn Greimann, and Phil Esra for helpful suggestions and critical reading of this manuscript. We also thank Joe DeRisi and Jeff Pleiss for technical assistance and members of the Guthrie lab for useful discussion.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2754011.
REFERENCES
- Alarcon CM, Heitman J, Cardenas ME 1999. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol Biol Cell 10: 2531–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JJ 2008. Development and application of technologies to study individual kinase substrate relationships. PhD thesis, University of California, San Francisco [Google Scholar]
- Ashe MP, De Long SK, Sachs AB 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell 11: 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker RA, Chambers JM, Wilks AR 1988. The new S language. Chapman & Hall, New York [Google Scholar]
- Beilharz TH, Preiss T 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby AC, Litchfield DW 2005. The multiple personalities of the regulatory subunit of proteína kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2β. Int J Biol Sci 1: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwai AP, Reed JC, Glover CV 1994. Casein kinase II of Saccharomyces cerevisiae contains two distinct regulatory subunits, β and β′. Arch Biochem Biophys 309: 348–355 [DOI] [PubMed] [Google Scholar]
- Boer VM, Amini S, Botstein D 2008. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci 105: 6930–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg I, Groenen P 1997. Modern multidimensonal scaling. Theory and applications. Springer, New York [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Bond U 1999. Reassembly and protection of small nuclear ribonucleoprotein particles by heat shock proteins in yeast cells. RNA 5: 1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581 [DOI] [PubMed] [Google Scholar]
- Cherkasova VA, Hinnebusch AG 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev 17: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM 2006. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W 2008. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 22: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME 2008. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE 3: e3273 doi: 10.1371/journal.pone.0003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. 2009. Functional organization of the S. cerevisiae phosphorylation network. Cell 136: 952–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM 2006. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MJ, Rios G, Ali R, Belles JM, Serrano R 1997. Comparative physiology of salt tolerante in Candida tropicalis and Saccharomyces cerevisiae. Microbiology 143: 1125–1131 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.116/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Gresham D, Boer V, Caudy A, Ziv N, Brandt N, Storey J, Botstein D 2011. System-level analysis of genes and functions affecting survival during nutrient starvation in Saccharomyces cerevisiae. Genetics 187: 299–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B, Viggiano S, Hibbs MA, Troyanskaya OG, Amberg DC 2007. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev 21: 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Hohmann S 2009. Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett 583: 4025–4029 [DOI] [PubMed] [Google Scholar]
- Honorine R, Mosrin-Huaman C, Hervouet-Coste N, Libri D, Rahmouni A 2010. Nuclear mRNA quality control in yeast is mediated by Nrd1 co-transcriptional recruitment, as revealed by the targeting of Rho-induced aberrant transcripts. Nucleic Acids Res 39: 2809–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia MH, Larossa RA, Lee JM, Rafalski A, Derose E, Gonye G, Xue Z 2000. Global expresión profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics 3: 83–92 [DOI] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Nierras CR, Warner JR 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol 19: 5393–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A 1992. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132: 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci 101: 14315–14322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979 [DOI] [PubMed] [Google Scholar]
- Moehle CM, Hinnebusch AG 1991. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 11: 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin C, Jauhiainen A, Warringer J, Nerman O, Sunnerhagen P 2009. mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA 15: 600–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munding EM, Igel AH, Shiue L, Dorighi KM, Trevino LR, Ares MJ 2010. Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev 24: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21: 4347–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsten MEK, Litchfield DW 2004. Order or chaos? An evaluation of the regulation of protein kinase CK2. Biochem Cell Biol 82: 681–693 [DOI] [PubMed] [Google Scholar]
- Pagano MA, Bain J, Kazimierczuk Z, Sarno S, Ruzzene M, Di Maira G, Elliott M, Orzeszko A, Cozza G, Meggio F, et al. 2008. The selectivity of inhibitors of protein kinase CK2: an update. Biochem J 415: 353–365 [DOI] [PubMed] [Google Scholar]
- Patil C, Walter P 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 13: 349–355 [DOI] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell 12: 1607–1613 [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C 2007a. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell 27: 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C 2007b. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol 5: e90 doi: 10.1371/journal.pbio.0050090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Guthrie C 2006. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA 12: 994–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Kim KS, Guthrie C 2002. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent R, Sautel CF, Moucadel V, Laudet B, Filhol O, Cochet C 2010. In vitro and in vivo assays of protein kinase CK2 activity. Methods Enzymol 485: 597–610 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Strasser K, Hurt E 2002. An intron in the YRA1 gene is required to control Yra1 protein expression and mRNA export in yeast. EMBO Rep 3: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Cardenas ME 2003. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol Cell Biol 23: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Santacreu L, Moreno J, Perez-Ortin JE, Alepuz P 2009. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15: 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Mallick J, Zhao Y, Warner JR 2007. Potential interface between ribosomal protein production and pre-rRNA processing. Mol Cell Biol 27: 4815–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa C, Nedea E, Krogan N, Wada T, Handa H, Greenblatt J, Buratowski S 2004. Bromodomain factor 1 (Bdf1) is phosphorylated by protein kinase CK2. Mol Cell Biol 24: 4734–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH 2008. Quality control of mRNP in the nucleus. Chromosoma 117: 419–429 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Clark R, Liu CY, Kaufman RJ 2004. The unfolded protein response represses differentiation through the RPD3-SIN3 histone deacetylase. EMBO J 23: 2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Dietrich FS, Roy SW 2007. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol 8: R223 doi: 10.1186/gb-2007-8-10-r223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC 2010. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem 285: 16893–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen E, Wanke V, Roosen J, Smets B, Dubouloz F, Pedruzzi I, Cameroni E, De Virgilio C, Winderickx J 2006. Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae. Cell Div 1: 3 doi: 10.1186/1747-1028-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarek N, Cameroni E, Jaquenoud M, Luo X, Bontron S, Lippman S, Devgan G, Snyder M, Broach JR, De Virgilio C 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol Cell 38: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674 [DOI] [PubMed] [Google Scholar]
- Vogel JL, Parsell DA, Lindquist S 1995. Heat-shock proteins Hsp104 and Hsp70 reactivate mRNA splicing after heat inactivation. Curr Biol 5: 306–317 [DOI] [PubMed] [Google Scholar]
- Wang J, Nygaard V, Smith-Sørensen B, Hovig E, Myklebost O 2002. MArray: analysing single, replicated or reversed microarray experiments. Bioinformatics 18: 1139–1140 [DOI] [PubMed] [Google Scholar]
- Warner JR 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. 2008. A genetic interaction map of RNA processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell 32: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WA, Roach PJ 2002. Nutrient-regulated protein kinases in budding yeast. Cell 111: 155–158 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN 2006. TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, O'Shea EK 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost HJ, Lindquist S 1991. Heat shock proteins affect RNA processing during the heat shock response of Saccharomyces cerevisiae. Mol Cell Biol 11: 1062–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR 2008. How Saccharomyces responds to nutrients. Annu Rev Genet 42: 27–81 [DOI] [PubMed] [Google Scholar]
- Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, Warner JR 2006. Fine-structure analysis of ribosomal protein gene transcription. Mol Cell Biol 26: 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]