Abstract
Noncanonical microRNAs (miRNAs) and endogenous small interfering RNAs (endo-siRNAs) are distinct subclasses of small RNAs that bypass the DGCR8/DROSHA Microprocessor but still require DICER1 for their biogenesis. What role, if any, they have in mammals remains unknown. To identify potential functional properties for these subclasses, we compared the phenotypes resulting from conditional deletion of Dgcr8 versus Dicer1 in post-mitotic neurons. The loss of Dicer1 resulted in an earlier lethality, more severe structural abnormalities, and increased apoptosis relative to that from Dgcr8 loss. Deep sequencing of small RNAs from the hippocampus and cortex of the conditional knockouts and control littermates identified multiple noncanonical microRNAs that were expressed at high levels in the brain relative to other tissues, including mirtrons and H/ACA snoRNA-derived small RNAs. In contrast, we found no evidence for endo-siRNAs in the brain. Taken together, our findings provide evidence for a diverse population of highly expressed noncanonical miRNAs that together are likely to play important functional roles in post-mitotic neurons.
Keywords: Next Generation Sequencing, miRNA, neurons
INTRODUCTION
miRNAs are ∼22 nucleotide (nt) RNAs that simultaneously influence both the transcript stability and translation of mRNA targets (Bartel 2009). Canonical miRNAs are initially expressed as part of long primary transcripts (pri-miRNAs). The miRNAs themselves are contained within these transcripts as hairpin folds, which are recognized by the dsRNA-binding protein DGCR8 (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004; Landthaler et al. 2004; Han et al. 2006). DGCR8 then directs DROSHA, the RNase III–containing catalytic subunit of the Microprocessor complex (Lee et al. 2003), to cleave at the base of the hairpin, liberating it from the pri-miRNA. The resulting precursor miRNA (pre-miRNA) is then exported from the nucleus and further processed by the RNase III–containing protein DICER1 to generate the mature miRNA (Hammond 2005). The mature miRNA is transferred to the RNA-induced silencing complex (RISC) prior to binding sequences in the 3′ UTR and/or open reading frames of its mRNA targets (Tay et al. 2008; Bartel 2009; Melton and Blelloch 2010). Once bound, the miRNA/RISC mediates transcript destabilization and/or translational inhibition (Baek et al. 2008; Selbach et al. 2008; Guo et al. 2011).
In addition to the canonical miRNA biogenesis pathway, there exist classes of small RNAs that are DICER1 substrates but do not require processing by DGCR8 (for review, see Babiarz and Blelloch 2009). These include endogenous small interfering RNAs (siRNAs) and noncanonical miRNAs. siRNAs are generated by the successive cleavage of long, double-stranded RNAs (dsRNAs) by DICER1. The noncanonical miRNAs arise when pre-miRNA–like structures are generated in the absence of microprocessor activity and are cleaved directly by DICER1. This class of miRNAs is typified by the mirtron, which arises from the splicing of short introns. Following debranching, these short introns fold into pre-miRNAs and are cleaved by DICER1 (Okamura et al. 2007; Ruby et al. 2007; Babiarz et al. 2008). Additional noncanonical miRNAs include endogenous short hairpin RNAs (shRNAs), which are intergenic. There have been two proposed mechanisms for shRNA biogenesis: (1) they are potentially directly transcribed as pre-miRNA structures (Babiarz et al. 2008), or (2) they are processed into a pre-miRNA hairpin via an undetermined nuclease (Chong et al. 2010). Finally, noncanonical miRNAs derived from small nucleolar RNAs (snoRNAs) have been identified (Babiarz et al. 2008; Ender et al. 2008; Taft et al. 2009). Like the shRNAs, how the snoRNA-derived miRNAs are processed prior to DICER1 cleavage is unclear.
Recently, a central role for endo-siRNAs in mammalian oocytes has been shown using a combination of phenotypic analysis of Dgcr8 versus Dicer1 knockouts and deep sequencing (Tam et al. 2008; Watanabe et al. 2008; Ma et al. 2010; Suh et al. 2010). However, whether endo-siRNAs function or even exist in somatic tissues remains unknown. Furthermore, no developmental roles have been identified for noncanonical miRNAs. Because of the complex nature of neural development, we embarked on examining potential roles for endo-siRNAs and noncanonical miRNAs in the brain, specifically in post-mitotic neurons.
RESULTS
Phenotypic differences resulting from post-mitotic loss of Dgcr8 versus Dicer1 in neurons
To identify potential roles for endo-siRNAs and noncanonical miRNAs in post-mitotic neurons, we crossed conditional Dgcr8 (Rao et al. 2009) and Dicer1 (Harfe et al. 2005) mice to two distinct transgenic mouse lines with Cre expression driven from an α-calcium/calmodulin–dependent protein kinase II (CamK) promoter—the CamK-cre93 or CamK-cre159 lines (Rios et al. 2001). Both CamK-cre93 and CamK-cre159 transgenes are expressed in post-mitotic neurons in similar regions of the brain, including the hippocampus and cortex, starting at P0 for CamK-cre93 and P15 for CamK-cre159 (Rios et al. 2001). The cre lines were crossed to produce x-cre: Dgcr8 Δ/flox and x-cre:Dicer1 flox/flox mice (see Materials and Methods), referred to as dgcr8Δ/Δ and dicer1Δ/Δ for the remainder of the text.
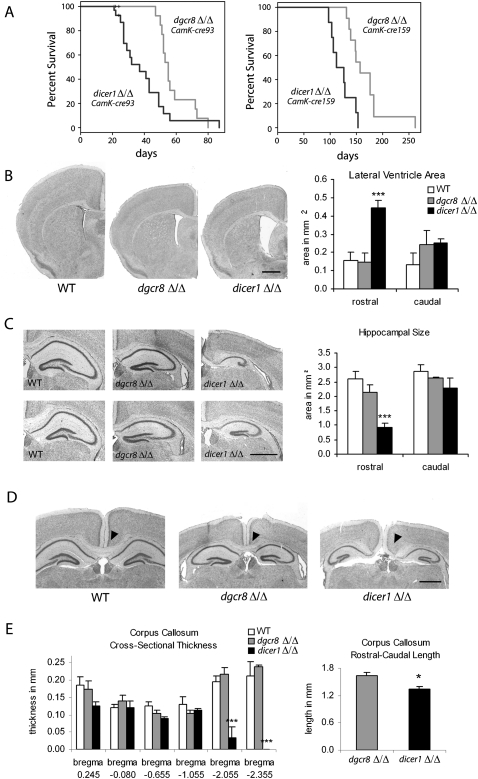
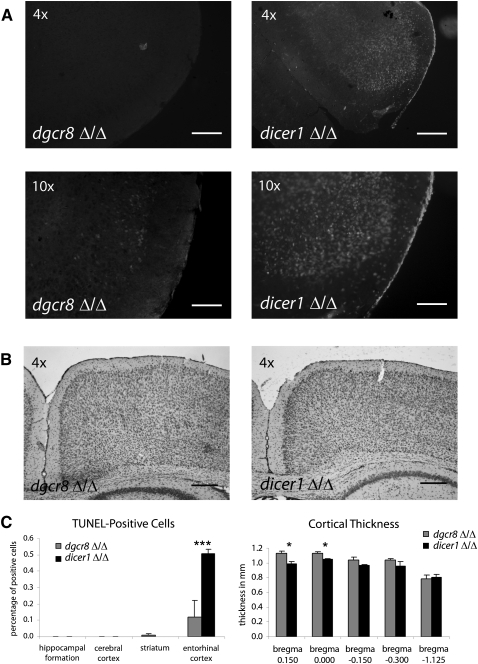
Deletion of either Dgcr8 or Dicer1 in post-mitotic neurons resulted in a fully penetrant lethal phenotype (Fig. 1A). However, the dicer1Δ/Δ mice consistently showed earlier lethality than did dgcr8Δ/Δ mice, independent of the cre line (Fig. 1A). To better understand the underlying causes for the differences in lethality, we further characterized the neuronal phenotypes at the neuroanatomical level, focusing on the CamK-cre93 mice. We performed this analysis on day 21–24 mice because it was the time point at which the dicer1Δ/Δ conditional mice began to die (Fig. 1A). dgcr8Δ/Δ and dicer1Δ/Δ mice were microcephalic with similar brain weights, which were greatly reduced compared with that of controls (Supplemental Fig. 1). However, Nissl labeling showed striking anatomical abnormalities in dicer1 compared with dgcr8 knockout mice, including enlarged lateral ventricles (P = 0.0009), smaller and malformed hippocampi (P = 0.008), as well as a truncated corpus callosum (CC; P = 0.027) (Fig. 1B–E). Terminal deoxynucleotidyl transferase–mediated biotinylated UTP nick end labeling (TUNEL) showed an increase in the number of cells undergoing apoptosis in both mutants relative to the wild type (WT); however, there were significantly more apoptotic cells in the dicer1Δ/Δ versus dgcr8Δ/Δ mice (P = 0.005) (Fig. 2A). The apoptotic population in both mutant lines was largely restricted to the lateral entorhinal cortex at this time point (Fig. 2B). Measurements of the cortical thickness throughout the brain was also reduced relative to the WT with a stronger phenotype in the dicer1Δ/Δ mice, particularly in the rostral brain corresponding to the prefrontal cortex (P = 0.030 and 0.038 at bregma 0.150 and 0.000, respectively) (Fig. 2C).
FIGURE 1.
Conditional loss of Dicer1 in post-mitotic neurons resulted in more severe phenotypes than loss of Dgcr8. (A) Survival analysis was performed on CamK-cre93 dgcr8Δ/Δ and dicer1Δ/Δ conditional mice. P-value was determined by the log-rank test. (B) dicer1Δ/Δ animals showed increases in lateral ventricle size at rostral (bregma −0.080, P = 0.0009) but not at caudal (bregma −1.255, P = 0.94) regions. (C) The rostral (bregma −2.055, P = 0.008), but not caudal (bregma −2.48, P = 0.371), hippocampal structure was malformed and reduced in size in the dicer1Δ/Δ brains compared with dgcr8Δ/Δ. (D,E) dicer1Δ/Δ animals exhibited agenesis of the caudal CC (note the lack of callosal tracts in the dicer1Δ/Δ animal and the intact callosum in the dgcr8Δ/Δ animal). Bregma values given for corresponding coronal sections from WT animals. Error bars, SEM; all scale bars, 3 mm.
FIGURE 2.
TUNEL labeling and cortical thickness measurements in dgcr8Δ/Δ and dicer1Δ/Δ brains. (A) Fluorescent images of TUNEL-positive cells in the entorhinal cortex taken at 4× and 10× magnification. (B) Representative images of cortical thickness at 4× magnification. (C) dicer1Δ/Δ animals showed a dramatic increase in apoptotic cells in the entorhinal cortex (P = 0.005). Cortical thinning was significantly higher in dicer1Δ/Δ animals in rostral (P < 0.038), but not in caudal, sections (P > 0.71). Bregma values given for corresponding sections from mutant animals. Error bars, SEM; scale bars, 0.25 mm (4×) and 1 mm (10×).
Identifying Dgcr8-independent, Dicer-dependent small RNAs in the mouse brain
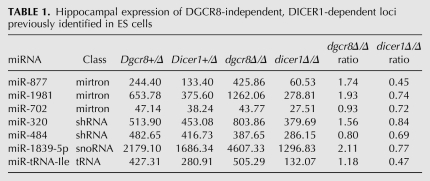
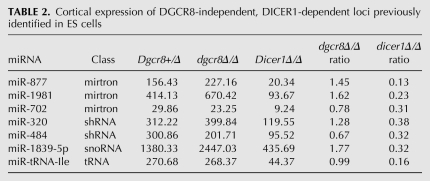
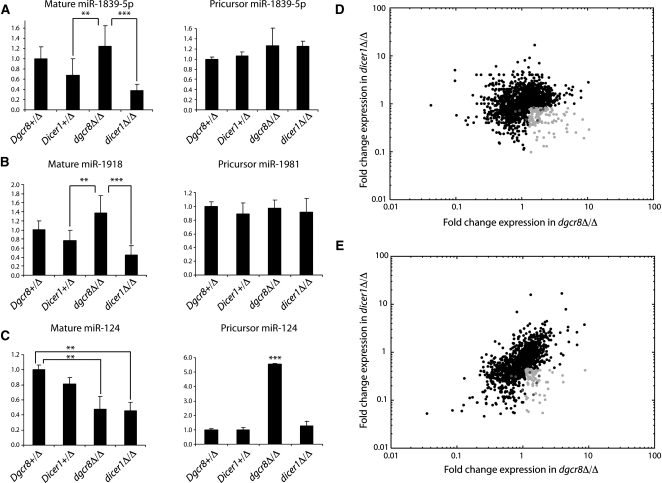
Given that the phenotypes we observed were stronger in the dicer1Δ/Δ mice compared with the dgcr8Δ/Δ mice, we sought to identify small RNAs that were selectively affected by DICER1, but not DGCR8 loss. To identify DGCR8-independent, DICER1-dependent small RNAs, we built small RNA sequencing libraries from the hippocampus and cortex of 21-d-old Dgcr8Δ/Δ, Dicer1Δ/Δ, and control mice. Libraries were sequenced using the Illumina deep sequencing platform, producing from 0.5–4 million genome-mapping reads (Gene Expression Omnibus [GEO] accession no. GSE21090) (Supplemental Table 1). The mapping of sequence reads to previously annotated pre-miRNA hairpins (miRBase 12.0) revealed that some but not all miRNAs changed their expression in both the mutants relative to controls (Supplemental Fig. 2). This result was expected since the cre transgene is expressed in a small fraction of cells in these regions (i.e., post-mitotic neurons), while we are isolating small RNAs from the entire tissue. We previously noted a pattern for DGCR8-independent, DICER1-dependent small RNAs in embryonic stem (ES) cells, where these small RNAs were enriched in dgcr8Δ/Δ and depleted in dicer1Δ/Δ (Babiarz et al. 2008). Therefore, we reasoned this pattern would allow us to identify noncanonical miRNAs expressed in the post-mitotic neurons. To test this hypothesis, we evaluated previously identified noncanonical miRNAs (Tables 1, 2; Babiarz et al. 2008). Four of seven of these known noncanonical miRNAs showed the expected pattern in both the hippocampus and cortex, suggesting that this approach should identify >50% of the DGCR8-independent, DICER1-dependent small RNAs in the brain. To confirm our results, two noncanonical miRNAs (miR-1981 and miR-1839-5p) were examined for their DGCR8-independent, DICER1-dependent nature by quantitative reverse transcriptase PCR (qRT-PCR). Both mature miR-1981 and miR-1839-5p were increased in dgcr8Δ/Δ and decreased in dicer1Δ/Δ, while the canonical neuron-specific miR-124 was decreased in both knockouts (Fig. 3A–C), in keeping with the expected pattern for DGCR8 independence and DICER1 dependence. Additionally, the pri-miRNA transcripts of both of these noncanonical miRNAs were unchanged in the dgcr8Δ/Δ, supporting the hypotheses that these RNAs are not processed by DGCR8. In contrast, the pri-miRNA transcript for canonical miR-124 accumulated in the dgcr8Δ/Δ (Fig. 3A–C). Given that these data from individual canonical and noncanonical miRNAs increased the confidence in our methodology, the analysis was expanded to the entire genome.
TABLE 1.
Hippocampal expression of DGCR8-independent, DICER1-dependent loci previously identified in ES cells
TABLE 2.
Cortical expression of DGCR8-independent, DICER1-dependent loci previously identified in ES cells
FIGURE 3.
DGCR8 and DICER1 dependence of small RNAs in the mouse brain. (A–C) qRT-PCR of mature miRNAs and pri-miRNAs in the hippocampus. Deep sequencing of the hippocampus (D) and cortex (E). Dark gray points are DGCR8-independent, DICER1-dependent loci identified in ES cells (Table 1). Light gray points are loci that were defined as DGCR8-independent, DICER1-dependent in the hippocampus (D) dgcr8Δ/Δ/Dgcr8+/Δ ≥ 1.3, dicer1Δ/Δ/Dicer1+/Δ ≤ 0.83 and cortex (E) dgcr8Δ/Δ/Dgcr8+/Δ ≥ 1.28, dicer1Δ/Δ/Dgcr8+/Δ ≤ 0.45.
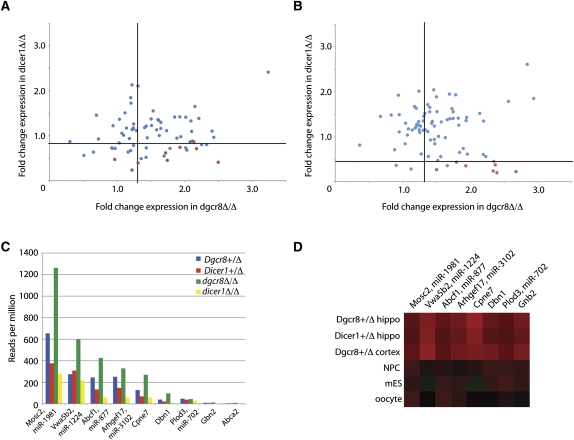
One hundred–base pair windows were slid across the entire genome at 10-bp intervals, and the number of reads mapping within each window were counted. These windows were then normalized to the total number of unique mapping genome reads, as previously described (Marson et al. 2008). Only windows with at least five reads per million in the Dgcr8+/Δ control were considered. When overlapping windows were collapsed, there were 1605 windows in the hippocampus and 1401 windows in the cortex (Supplemental Table 2). For each window, a ratio of the number of reads found in the conditional knockouts relative to controls was calculated. In both the hippocampus and the cortex, the majority of windows had ratios clustered around 1 as expected (Fig. 3D,E). However, a small but significant number of reads diverged from the larger cluster. Based on the read ratio of the known DGCR8-independent, DICER1-dependent small RNAs and known canonical miRNAs, criteria were developed to identify potential novel DGCR8-independent loci (Fig. 3, light gray points; Supplemental Table 2; see also Materials and Methods). While these criteria capture 4/7 (57%) of known DGCR8-independent, DICER1-dependent loci (Fig. 3D,E, dark gray points; Tables 1, 2), 10.4% of the windows overlapped with known canonical miRNAs, suggesting a small but significant false-positive rate. Therefore, each locus was examined carefully based on known mechanisms of DGCR8 independence as described below. Additionally, it should be noted that the conservative criteria and false-negative rate with known noncanonical miRNAs suggest the loci found with this analysis are likely an underestimate of the total noncanonical miRNAs in the mammalian brain.
Taking a similar approach, we established equivalent cutoffs for the cortex. However, 35.9% of those reads were from previously annotated miRNAs. Therefore, it was more difficult to identify novel loci in the cortex, likely due to the greater heterogeneity of the conditional deletion even within post-mitotic neurons (Rios et al. 2001). Because of the lack of sensitivity and specificity in the cortex, we largely used the cortex data to confirm and expand on DGCR8-independent, DICER1-dependent loci identified in the hippocampus.
Endogenous shRNAs are expressed at low levels in the brain
Our previous studies have determined that the dominant class of DGCR8-independent, DICER1-dependent small RNAs in ES cells are endogenous shRNAs, consisting largely of miR-320 and miR-484 (Babiarz et al. 2008). In addition to being enriched in dgcr8Δ/Δ, these miRNAs were intergenic and folded into hairpins, and the reads were predominantly localized to the 3′ end of the hairpin. Examination of miR-320 and miR-484 in the hippocampus and cortex revealed that miR-320 met the DGCR8-independent, DICER1-dependent criteria. However, miR-484 did not meet these strict criteria (Tables 1, 2). We sought to identify additional shRNA loci expressed in the brain. One locus that met these criteria was present in an intergenic region on chromosome 7 (Supplemental Fig. 3); however, it was only expressed at 131 reads per million in the hippocampus. Surprisingly, all the shRNAs were present at low levels, making up only a small percentage of the total noncanonical miRNA complement in the post-mitotic neurons. Therefore, relative to the other noncanonical miRNAs, the shRNAs are less likely to have a critical role in neurons, although knockout studies will be required to confirm these findings.
Identification of novel mirtrons in the mouse brain
Mirtrons are a subclass of miRNAs that are DGCR8-independent and DICER1-dependent and derive from the folding of spliced and debranched short introns into pre-miRNA structures. The small RNA reads derive from two extreme ends of the introns (i.e., at the exon/intron boundaries). To identify mirtrons within our hippocampal data set, we analyzed the enzymatic dependencies as described above for all small RNAs mapping to introns ≤500 bp. Seventy-four introns contained at least five reads per million in the Dgcr8+/Δ control (Fig. 4A; Supplemental Table 4). Fifteen of these introns met our criteria for DGCR8-independent, DICER1-dependent loci. Four of these were previously annotated mirtrons: miR-3102 (Arhgef17), miR-1981, miR-1224, and miR-877 (Supplemental Fig. 4). As described previously (Chiang et al. 2010), miR-3102 derives from the intron of the Arhgef17 gene, which forms a longer hairpin structure than that typically seen for mirtrons and appears to be sequentially cleaved to produce two sets of mirtrons, although the internal mirtron pair was present at much reduced levels (24 internal reads vs. 214 from the intron/exon boundary in the Dgcr8+/Δ hippocampus) (Supplemental Fig. 4). Closer examination of the remaining 11 intronic loci, including the fold structure and positioning of reads, identified an additional mirtron derived from an intron within the Dbn1 gene (Supplemental Fig. 4). Our findings confirm the DICER1 dependence and DGCR8 independence of this novel miRNA.
FIGURE 4.
Mirtrons in the mouse brain. (A) The DGCR8 and DICER1 dependencies of small RNAs derived from introns ≤500 bp in the hippocampus. Red points represent previously identified mirtrons. The lines show the cutoffs for DGCR8-independent, DICER1-dependent reads. (B) Small RNA dependencies in the cortex as in A. (C) Read counts for the nine mirtrons identified in A (red points) in each of the libraries from the hippocampus. (D) Expression of mirtrons in the mouse hippocampus, cortex, NPCs (Marson et al. 2008), ES cells (Babiarz et al. 2008), and oocytes (Tam et al. 2008). Reads from each tissue were log transformed and median centered.
As mammalian mirtrons are typically derived from introns <110 bp (Babiarz et al. 2008), we more closely evaluated all introns <110 bp long that had at least five reads in the control regardless of the enzymatic dependencies (Fig. 4A, red points). This careful analysis identified an additional previously annotated mirtron, miR-702 (Babiarz et al. 2008), as well as three more novel mirtrons derived from introns within the Cpne7, Gnb2, and Syvn1 genes. Similar analysis in the cortex (Fig. 4B; Supplemental Table 4) confirmed four of the mirtrons identified in the hippocampus (Abcf1/mir-877, Mosc2/mir-1981, Dbn1, and Cpne7) as well as two additional mirtrons that derived from introns in the Fbxw9 and Fam116b genes (Supplemental Fig. 4). Therefore, all together, our analysis of the hippocampal and cortex data revealed six novel mirtrons.
Mirtron expression levels are low in ES cells (fewer than 20 reads per million) (Babiarz et al. 2008), making their functional relevance in that setting questionable. Similarly, mirtrons are of extremely low abundance in the embryonic skin, where miR-877 and miR-1224 were present at fewer than 10 reads per million (Yi et al. 2009). In contrast, we observed that the expression levels of the uncovered mirtrons in hippocampus and cortex were relatively high (Fig. 4C), with the most highly expressed mirtron, Mosc2/miR-1981, present at greater than 600 reads per million. A direct comparison of expression levels for the different mirtrons found in the hippocampus and cortex to other tissues, including oocytes (Tam et al. 2008), ES cells (Babiarz et al. 2008), and neural precursor cells (NPCs) (Marson et al. 2008), revealed consistently higher levels (Fig. 4D). The differences in expression are even more impressive when considering the heterogeneity of cells in the hippocampus compared with the relative homogeneity of the oocytes, NPCs, and ES cells. That is, if only a subset of the cells in the hippocampus is expressing any individual mirtron, the ratio of mirtron to total small RNAs within those cells should be much higher. These relatively high levels of expression of mirtrons in the brain suggest they may have an important functional role in the adult nervous system.
snoRNAs predominate the Dgcr8-independent, Dicer-dependent small RNAs in the hippocampus and cortex
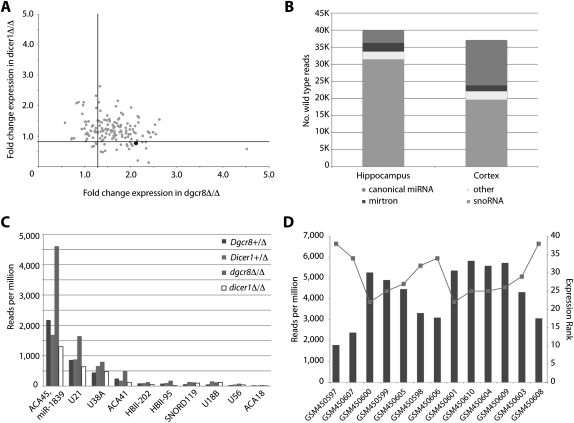
Mirtrons only explained six of the 15 introns ≤500bp that produced DGCR8-independent, DICER1-dependent small RNAs in the hippocampus. Examination of the remaining introns revealed that all nine contained highly conserved snoRNAs from which the small RNAs were derived. snoRNAs have recently been shown to produce DGCR8-independent, DICER1-dependent small RNAs that behave like miRNAs (Babiarz et al. 2008; Ender et al. 2008; Taft et al. 2009). Therefore, we expanded our analysis to include all snoRNAs. However, snoRNAs are not well annotated in the mouse genome, so a previously published strategy was employed to analyze snoRNAs on a genome-wide scale (Taft et al. 2009), taking advantage of the high degree of conservation of snoRNAs and the well-curated human snoRNA database. By use of this approach, 149 snoRNA loci contained at least five reads per million in the control hippocampus (Fig. 5A; Supplemental Table 5). Nineteen of the 149 met the DGCR8-independent, DICER1-dependent criteria (Fig. 5A). Unexpectedly, 74% (14 of the 19) of the snoRNAs meeting these criteria were of the C/D box family. As an alternative approach, we utilized the snoRNA annotations of the mm9 mouse genome assembly generated using the snoseeker tool (Yang et al. 2006, 2010). From the snoseeker-derived snoRNA loci, 147 loci were identified with at least five reads per million, and 24 of the 147 were DGCR8-independent, DICER1-dependent (Supplemental Table 6). Previously described mammalian snoRNA-derived small RNAs were exclusively from H/ACA box family snoRNAs (Babiarz et al. 2008; Ender et al. 2008; Taft et al. 2009). However, a recently published study has suggested that C/D box-derived small RNAs can be incorporated into RISC in human CD4+ T cells and silence mRNAs (Brameier et al. 2010).
FIGURE 5.
Small RNAs derived from snoRNAs in the hippocampus and cortex. (A) DGCR8 and DICER1 dependencies of small RNAs derived from conserved snoRNAs in the hippocampus. The black point represents the previously identified DGCR8-independent, DICER1-dependent, snoRNA-derived miRNA, miR-1839-5p. (B) DGCR8-independent, DICER1-dependent loci from the hippocampus and cortex were classified according to the UCSC genome browser annotations (mm8). (C) Read counts for the snoRNAs identified in B in each of the libraries from the hippocampus. (D) Read counts of miR-1839-5p from prefrontal cortex small RNA libraries described by Somel et al. (2010) in dark gray. Rank order of expression relative to other miRNAs in light gray.
Analysis of the DGCR8 dependence and DICER1 dependence of the snoRNAs was also examined in published data sets from other mouse tissues. In both mES cells (Babiarz et al. 2008) and CD4+ T cells (Chong et al. 2010), only the previously described H/ACA-derived small RNA, miR-1839-5p, was identified as DGCR8-independent, DICER1-dependent (Supplemental Table 6). Furthermore, analysis of neural Ago IP deep sequencing data (Chi et al. 2009) showed loading of the expected 21-nt RNA species from H/ACA-box snoRNA miR-1839-5p (Supplemental Fig. 5). In contrast, a 28-nt species representing the opposite end of the hairpin from the expected small RNA of the C/D-box snoRNA was enriched in the Ago pull-down. Therefore, the exact mechanism of the biogenesis and function for the C/D box–derived small RNAs remains unclear, and further studies will be required to identify the precise nature of these small RNAs in the brain.
A majority of the DGCR8-independent, DICER1-dependent small RNA reads in the hippocampus and cortex were derived from snoRNAs, 78.7% and 52.9%, respectively (Fig. 5B). Furthermore, a number of the individual snoRNA-derived small RNAs were present at high levels (Fig. 5C). For example, miR-1839-5p (derived from a H/ACA box snoRNA) produced over 2000 reads per million in the control hippocampus. This level of expression placed miR-1839-5p into the top 100 most highly expressed miRNAs in the hippocampus, suggesting it is likely to play an important functional role in this tissue. To identify if the high level of expression of miR-1839-5p is conserved in humans, we examined a recently published human prefrontal cortex deep sequencing data set (Somel et al. 2010). Indeed, miR-1839-5p expression was in the top 50 miRNAs in all 12 individuals, ranking between the 22nd and 38th most highly expressed (Fig. 5D; Supplemental Table 7).
Novel mirtrons and snoRNA-derived small RNAs share seed sequence homology and conserved predicted targets with highly conserved canonical miRNAs
To gain insight into the broad functional roles of mirtrons and snoRNA-derived small RNAs in the mouse brain, we sought to identify their predicted targets. To this end, we used Targetscan (Friedman et al. 2009) to generate predicted targets with a user-specified seed sequence. By using this approach, we generated lists of conserved predicted targets for miR-1981 and miR-1839-5p, which are the most highly expressed noncanonical miRNAs in the mouse brain. The miR-1981 mature sequence is uniquely annotated in mouse, and the miR-1839-5p sequence is uniquely annotated in only five mammalian species, although the parent snoRNA, ACA45, is highly conserved among mammals (Griffiths-Jones et al. 2008). Despite the lack of conservation of these small RNAs themselves, both miR-1981 and miR-1839-5p have a large number of predicted targets with highly conserved target sites in their 3′ UTRs. The diversity and high degree of conservation in the target sites suggest that the target sites may have evolved prior to the evolution of these particular small RNAs. We reasoned that this could occur if these target sites are also targeted by other small RNAs with highly similar seed sequences and that these evolutionarily newer small RNAs evolved to co-opt these pre-existing target sites.
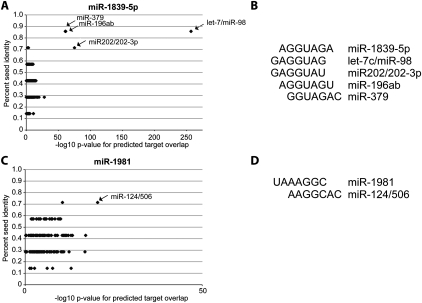
To test this hypothesis, we computationally measured the similarity of the seed matches of miR-1981 and miR-1839-5p to the seed matches of 152 miRNA families. We simultaneously measured the overlap in the predicted targets of miR-1981 and miR-1839-5p to the predicted targets of each set of miRNA families. The results of these analyses show a trend whereby miRNAs that have a higher degree of seed sequence share a higher number of predicted targets. Of note, miR-1981 shares 71% seed identity to miR-124, and the two miRNAs share 52 predicted targets with a –log10 P-value of 20. miR-1839-5p shares 86% seed identity to the let-7/miR-98 family, and they share 139 predicted targets with a –log10 P-value of 256 (Fig. 6A,C). In the case of let-7 and miR-1839-5p, the seed sequences are shifted by a single base; as a result, these two small RNAs share not only an overlapping pool of mRNA targets but also target sites within these mRNAs (Fig. 6B). In the case of miR-124 and miR-1981, the seed sequences are shifted by two bases (Fig. 6D). These results suggest a mechanism whereby the evolution and function of new mirtrons and snoRNA-derived small RNAs are supported by a pre-existing repertoire of mRNAs with conserved target sites. In this way newly evolved small RNAs like miR-1981 and mir-1839-5p can immediately have a significant molecular impact on a large set of pre-existing targets and the corresponding molecular pathways within which they function.
FIGURE 6.
miR-1839-5p and miR-1981 share extensive seed sequence and predicted targets with conserved canonical miRNA families. (A) The percentage of seed identity (fraction of nucleotides overlapping in seed sequence after ClustalW alignment) of miR-1839-5p compared with each of 152 conserved miRNA families and the –log10 P-value for the overlap in predicted targets of miR-1839-5p and each of the conserved miRNA families are plotted. Names and arrows are displayed for highly significant miRNA families. (B) The alignment of the seed of miR-1839-5p with the seeds of highly significant miRNA families from A. (C) Same analysis as in A for miR-1981. (D) Same analysis as in B for miR-1981. P-values in A and C are calculated by Fisher's exact test.
Novel mirtrons and snoRNA-derived small RNAs are predicted to target cellular pathways important for neuronal survival and function
To follow up on the predicted miRNA targets for the two most highly expressed noncanonical miRNAs, miR-1981 and miR-1839-5p, we sought to identify pathways potentially regulated by these miRNAs. We reasoned such an analysis might shed insight into the molecular basis of the observed differences between dicer1Δ/Δ and dgcr8Δ/Δ knockout brains with respect to cell survival and gross morphology. To this end, we performed an unbiased pathway analysis of the predicted targets of miR-1981 and miR-1839-5p using MSigDB (Subramanian et al. 2005). This analysis predicts that miR-1981 modulates PI3-kinase receptor signaling by targeting PIK3R1, EGFR, PTPN11, and SOS1 (Supplemental Table 8). Likewise, the program predicts that miR-1839-5p targets the IGF1 pathway (with the predicted targets in IGF1 and IGFR1) as well as amine ligand binding receptors (with the predicted targets ADRB1, ADRB2, ADRB3, and HTR1D) (Supplemental Table 8). These pathways are known to be critical regulators of nervous system function, particularly in cell survival and proliferation (Rodgers and Theibert 2002; Ye and D'Ercole 2006), suggesting that the downstream effects of miR-1839-5p and miR-1981 on their mRNA targets could contribute to the phenotypic differences between Dicer1 and Dgcr8 conditional knockout brains.
DISCUSSION
In this study, we provide evidence for potential functional roles of noncanonical miRNAs in adult mouse brain. Specifically, we show that the loss of DICER1 in post-mitotic neurons confers a much more severe phenotype than seen with the equivalent loss of DGCR8. Furthermore, we uncovered a large number of novel noncanonical miRNAs by deep sequencing, including six novel mirtrons and 23 potential snoRNA-derived small RNAs that are expressed at relatively high levels in post-mitotic neurons.
A primary goal in the current study was to identify DGCR8-independent, DICER1-dependent small RNAs within the post-mitotic neurons of the adult mouse. We were motivated to search for such small RNAs because of the differences in phenotypes following post-natal neuron-specific deletion of DGCR8 versus DICER1. In particular, the Dicer1Δ/Δ mice died earlier and their brains showed an increase in apoptotic cells together with cortical thinning, enlarged ventricles, and smaller hippocampi, suggesting potential roles for DGCR8-independent, DICER1-dependent small RNAs in cell survival and/or proliferation. Some of these DICER1-dependent phenotypes are reminiscent of those seen in human neurological disorders. For example, in the dicer1Δ/Δ brain, the rostral CC developed normally, while there is a near complete loss of the caudal CC. A similar agenesis of the caudal CC, along with smaller hippocampi, has been shown in a mouse model of Alzheimer's disease (Gonzalez-Lima et al. 2001), and structural abnormalities, including cortical thinning along with enlarged ventricles, has been reported in the brains of schizophrenic and autistic patients (Shenton et al. 2001; Brambilla et al. 2003). Therefore, it is intriguing to consider roles for noncanonical miRNAs underlying these structural abnormalities in human psychiatric diseases.
One of the challenges of this study was determining the enzymatic dependencies of small RNA-generating loci in a complex tissue. The CamK-cre transgenes are active only in post-mitotic neurons (Rios et al. 2001), which make up only a fraction of the total brain tissue. Additionally, CamK-cre activity is unlikely to be in all post-mitotic neurons of the hippocampus or cortex. We typically observed a 10%–20% decrease in canonical miRNAs in the conditional knockouts (Supplemental Fig. 2). However, the most abundant miRNAs, such as members of the let-7 family, were largely unchanged in both dgcr8Δ/Δ and dicer1Δ/Δ (Supplemental Table 2). This lack of change is likely due to the broader expression of let-7 in the different cell types of the hippocampus and cortex, especially the glia, which make up the majority of cells in the brain. Other miRNAs like miR-24, miR-127, and miR-137 (Supplemental Table 3) were dramatically reduced in both dgcr8Δ/Δ and dicer1Δ/Δ, which could be explained by higher levels of expression of these miRNAs, specifically in the cre-expressing cells. Indeed, our analysis demonstrates that it is possible to determine expression of cell type–specific miRNAs within a complex tissue by comparing the expression patterns of miRNAs between normal tissue and tissue deleted for Dgcr8 or Dicer1 in specific subsets of cells within the tissue.
Previous analysis of DGCR8-independent, DICER1-dependent small RNAs in ES cells revealed the presence of mirtrons, shRNAs, endogenous siRNAs, and a single snoRNA-derived small RNA (Babiarz et al. 2008). In ES cells, the most highly expressed DGCR8-independent, DICER1-dependent small RNAs were derived from endogenous shRNAs (miR-320 and miR-484) and endogenous siRNAs (two loci containing tandem, inverted SINEs), while mirtrons and snoRNA-derived small RNAs were expressed at very low levels. In contrast, small RNAs derived from mirtrons and snoRNAs appear to be expressed at high levels in the adult brain and, therefore, may underlie the significant differences in phenotype. DGCR8-independent, DICER-dependent phenotypes have also been recently ascribed to the retina, where loss of Dicer, but not Dgcr8, Drosha, or Ago2, resulted in macular degeneration (Kaneko et al. 2011). Interestingly, the retinal phenotype was associated with an increase in B1/Alu SINE RNAs, and B1/Alu SINE-derived endo-siRNAs have been described in mouse ES cells (Babiarz et al. 2008).
While several of the mirtrons we identified appear to be expressed at high enough levels to be functionally relevant in the brain, the DGCR8-independent small RNAs that predominated were derived from snoRNAs. This finding differs from ES cells, where only a small number of small RNAs were from snoRNAs (Babiarz et al. 2008; Taft et al. 2009). Furthermore, the ES cell small RNAs were derived from H/ACA box snoRNAs (miR-1839-5p), although at low levels. Another group, using HEK 293 cells (Ender et al. 2008), showed the H/ACA box–derived small RNAs can indeed function to suppress a target. The work presented here indicates that the H/ACA-derived small RNAs are highly expressed in the murine hippocampus and cortex. Additionally, our analysis of published human prefrontal cortex small RNA sequencing (Somel et al. 2010) shows these miRNAs are highly expressed in the human brain. Finally, putative target mRNA identification suggests that these miRNAs play important roles in neural development. In addition to the highly expressed H/ACA-derived miR-1839-5p, we identified a number of small RNAs derived from C/D box snoRNAs. C/D box–derived small RNAs have also been recently uncovered in immortalized human cell lines, where they displayed a miRNA-like function as assayed by luciferase reporters (Brameier et al. 2010). However, their biogenesis is currently unclear. For instance, in the mouse hippocampus and cortex, the secondary structure prediction algorithms do not predict a pre-miRNA–like structure and several of the identified small RNAs are 28 nt in length, rather than the expected 21 nt of a DICER1 cleavage.
Our data in the context of previous analyses of small RNA composition in diverse tissues suggest that the classes and expression levels of DGCR8-independent, DICER1-dependent small RNAs vary widely across cell types. In the three cases where dicer1Δ/Δ cells had a more severe phenotype than dgcr8Δ/Δ (ES cells [Wang et al. 2007], oocytes [Suh et al. 2010; and this study]), there has been an abundant, but different, cohort of small RNAs that may be responsible for the phenotypic differences. In contrast, in the embryonic skin, where dgcr8Δ/Δ and dicer1Δ/Δ have identical phenotypes, few DGCR8-independent, DICER1-dependent small RNAs were identified, and those that were identified were present in low levels (Yi et al. 2009). It remains an open question whether other tissues may produce an entirely different cohort of DGCR8-independent, DICER1-dependent small RNAs. An especially interesting question is whether endogenous siRNAs are found in any somatic tissues or are limited to the oocytes and early embryos where there is no interferon-based response to long dsRNAs (Wianny and Zernicka-Goetz 2000; Yang et al. 2001).
Our finding that DGCR8-independent small RNAs share seed sequences and targets with more highly conserved miRNA families suggests a mechanism for the rapid evolution of noncanonical small RNAs, likely with important functional roles. The co-evolution of a small RNA and a large repertoire of targets de novo would require more evolutionary time than the evolution of just the small RNA itself. Our data suggest that newly evolved small RNAs can co-opt mRNA targets and target-sites with shared seeds that have already been evolved to be effective in miRNA mediated suppression. Small RNAs that evolve in such a way would be predicted to not only share mRNA targets but also function with the miRNA families with which they share seed sequence. In particular, our findings suggest that the mirtron miR-1981 and the snoRNA-derived small RNA miR-1839-5p share targets and function with the miR-124 and let-7 families of miRNAs, respectively.
MATERIALS AND METHODS
Animals
All animal experiments described in this article have been approved by UCSF's Institutional Animal Care and Use Committee. CamK-cre93 and 159 transgenic mouse lines were bred to Dgcr8+/Δ mice, and the subsequent offspring, which were CamK-cre, Dgcr8+/Δ were bred to Dgcr8 flox/flox mice to generate conditional CamK-cre, Dgcr8 flox/Δ mice. Dicer1 conditional mice were generated similarly by crossing CamK-cre to Dicer1 flox/flox to generate CamK-cre Dicer1+/ flox. These mice were mated with Dicer1 flox/flox to generate conditional CamK-cre Dicer1 flox/flox mice. Mice were aged until death or sacrificed between 21 and 25 d of age for molecular analysis.
Neuroanatomical analysis
dgcr8Δ/Δ and dicer1Δ/Δ animals were sacrificed at post-natal day 22–24, and brains were fixed in 4% PFA, cryoprotected in 30% sucrose in PBS, and embedded in Shandon cryochrome compound (Thermo no. 9990426). Serial cryosections of 25 μm were collected and used for both Nissl staining and the TUNEL assay. For Nissl staining, coronal serial sections were stained in a gallocyanine-chrome alum solution (0.15% gallocyanine, 5% chrome alum in water) for 2 h, dehydrated, and mounted using Permount (Fisher Scientific). The TUNEL assay was performed on serial sections using the FragEL DNA Detection Kit (Calbiochem) according to the manufacturer's instructions, and slides were mounted using Vectashield with DAPI (Vector Laboratories). Images were collected on a Nikon Eclipse 80i microscope, and all anatomical measurements were performed using ImageJ (National Institutes of Health). Ventricle and hippocampal areas were measured by tracing the structural boundaries and calculating the area based on the number of pixels. The dorsal-ventral cross sectional thickness of the CC was measured at the midline from coronal sections, and the anterior-posterior length of the CC was calculated based on the number of serial sections spanned by the structure. Quantification of apoptotic cells was evaluated by identifying fluorescent cell bodies within a defined area and normalizing to total cell bodies stained with DAPI. Cortical thickness measurements were taken in the primary somatosensory cortex, measuring across cortical layers I–VI. Structural features and morphometric calculations were identified and based on the Allen Mouse Brain Reference Atlas (Allen Institute for Brain Science).
Statistical analysis
Survival analysis was performed using survdiff function in R (http://www.r-project.org/), and P-values were calculated using the log-rank test.
Small RNA cloning and sequencing
The hippocampus and cortex were manually dissected from Dgcr8+/Δ, dgcr8Δ/Δ, Dicer1+/Δ, and dicer1Δ/Δ brains. Total RNA was extracted using Trizol Reagent (Invitrogen). Small RNAs were cloned using the small RNA cloning kit v1.5 (Illumina). Briefly, the 3′ adapter was ligated with truncated Rnl2 (NEB) for 3 h at room temperature. The 5′ adapter was then added with ATP and Rnl1 (NEB) for an additional 15 h at room temperature. Following 3′ and 5′ adapter ligation, reverse transcription was performed with Super Script III (Invitrogen). Fifteen cycles of PCR were performed with Phusion Polymerase (NEB). The resulting product was purified on a 6% acrylamide TBE gel and stained with SYBR Gold (Invitrogen), and the products from ∼95–110 nt, representing 15- to 30-nt small RNAs, were purified. The library was analyzed with a DNA 1000 Chip on the Bioanalyzer 2100 (Agilent) for the appropriate size distribution. Libraries were quantified using Qubit (Invitrogen). Libraries were diluted to 10 nM and sequenced on a Genome Analyzer 2 with a 36 cycle kit (Illumina).
Bioinformatic analysis of small RNAs
Sequences were trimmed of the 3′ adapter by requiring 6 nt of perfect match to the adapter sequence. Sequences not containing a 6-nt perfect match were discarded. Sequences from 15–30 nt were mapped to the mm8 assembly of the mouse genome (Church et al. 2009) using Eland (Pipeline version 0.2.2.3). All uniquely mapping reads were considered for analysis of DGCR8 and DICER1 dependence. To determine miRNA-derived reads, trimmed sequences of 15–30 nt were mapped to the annotated pre-miRNA hairpins, miRBase 12.0 (Griffiths-Jones et al. 2008) using ELAND (Pipeline version 1.0). Small RNAs were mapped to RepBase Mouse Repeats (Jurka et al. 2005) using ELAND (Pipeline version 1.0).
Small RNA dependencies were determined by sliding 100-bp windows across the genome at 10-bp increments. Overlapping windows were collapsed by determining if the start of a window was contained within a previous window and combining the window coordinates. The number of small RNAs mapping within each window were counted for each library. Data were normalized to the total number of unique genome mapping reads, as previously described (Marson et al. 2008). Windows containing five or more reads in Dgcr8+/Δ were considered. DGCR8-independent, DICER1-dependent cutoffs were determined empirically to be the most inclusive of loci previously identified in ES cells (Babiarz et al. 2008) and exclude as many known canonical miRNAs as possible. Hippocampus cutoffs were dgcr8Δ/Δ / Dgcr8+/Δ ≥ 1.3, dicer1Δ/Δ / Dicer1+/Δ ≤ 0.83; 134 of the 1601 windows met these criteria and were classified as DGCR8-independent, DICER1-dependent. Cortex cutoffs were dgcr8Δ/Δ / Dgcr8+/Δ ≥ 1.28, dicer1Δ/Δ / Dgcr8+/Δ ≤ 0.45; 61 of the 1401 windows met these criteria and were classified as DGCR8-independent, DICER1-dependent. For all windows and DGCR8-independent, DICER1-dependent for the hippocampus and cortex data sets, see Supplemental Table 2. Loci that met the DGCR8-independent, DICER1-dependent criteria were classified according to overlapping features in the UCSC genome browser (Karolchik et al. 2008). For the short intron analysis, intron coordinates were determined from the RefSeq database at the UCSC Genome Browser. For the snoRNA analysis, coordinates for the snoRNAs were extracted from the sno/miRNA track in the hg18 human assembly and converted to the mouse mm8 coordinates using the liftover tool, requiring 95% sequence identity, as previously described (Taft et al. 2009). For additional snoRNA analysis, the snoseeker mm9 snoRNA coordinates were converted to mm8 using the liftover tool.
Human brain small RNA sequences (Somel et al. 2010) were downloaded from GEO (accession no. GSE18012) and processed as described above, except the small RNAs were mapped to the hg18 human genome assembly. The number of sequences mapping overlapping with snoRNAs and miRNAs described in the sno/miRNA track were determined.
miRNA qRT-PCR
The hippocampus and cortex were dissected out of knockout and littermate control mice brains, and total RNA was isolated using Trizol. To evaluate miRNA expression, 250 ng of total RNA was polyA-tailed with polyA polymerase (NEB) and reverse transcribed using Superscript II (Invitrogen) with a modified oligo dT adaptor, as described previously (Hurteau et al. 2006). qPCR was performed using primers against the adaptor and the mature miRNAs. To evaluate pri-miRNA expression, 200 ng of total RNA was used to generate cDNA using the Taqman Reverse Transcription Kit (Applied Biosystems). Gene-specific primers were designed against the precursor sequences and spanning introns. All qPCRs were performed using Bio-Rad iQ Supermix on a CFX96 Real-Time System and a C1000 Thermal Cycler (Bio-Rad). The U6 snRNA and GAPDH were used as internal controls. All qPCR reactions were performed in triplicate and relative quantitations were calculated using the Pfaffl method (Pfaffl 2001).
Accession numbers
Deep sequencing data can be found at the GEO database, accession no. GSE21090.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Robert Judson, Ron Parchem, and members of the Blelloch laboratory for critical reading of the manuscript. We also thank Michael McManus for providing the Dicer1 conditional mice, and Jon Woo and the UCSF Genome Core Facility for deep sequencing support. We thank Leonid Tetylman for helpful discussions about perl and sql. R.B. is supported by National Institute of Health (K08 NS48118 and R01 NS057221) and California Institute of Regenerative Medicine (New Faculty Award RN2-00906). E.M.U. is supported by the NIH (R21-MH083090), the Research to Prevent Blindness Career Development Award, Autism Speaks, the March of Dimes, That Man May See, the Sandler Foundation, and the Alfred P. Sloan Foundation. C.M. is supported by an NSF graduate research fellowship.
Author contributions: J.E.B. generated small RNA libraries and performed bioinformatic analyses of the data. R.H. analyzed the neuroanatomical features of mutant brain samples and performed the qRT-PCR analysis. C.M. performed the mouse genetics, analyzed the mortality, brain weights, prepared brain tissue and RNA samples, and performed miRNA predicted target, miRNA seed homology, and pathway enrichment analysis. M.T. provided important insights into the bioinformatic analysis of snoRNAs. J.B., R.H., C.M., and R.B. wrote the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2442211.
REFERENCES
- Babiarz JE, Blelloch R 2009. Small RNAs: their biogenesis, regulation and function in embryonic stem cells. In StemBook, ed. The Stem Cell Research Community. StemBook, Cambridge, MA: [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla P, Hardan A, di Nemi SU, Perez J, Soares JC, Barale F 2003. Brain anatomy and development in autism: review of structural MRI studies. Brain Res Bull 61: 557–569 [DOI] [PubMed] [Google Scholar]
- Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J 2010. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res 39: 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR 2010. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev 24: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi: 10.1371/journal.pbio.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lima F, Berndt JD, Valla JE, Games D, Reiman EM 2001. Reduced corpus callosum, fornix and hippocampus in PDAPP transgenic mouse model of Alzheimer's disease. Neuroreport 12: 2375–2379 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2011. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM 2005. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett 579: 5822–5829 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ 2005. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci 102: 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurteau GJ, Spivack SD, Brock GJ 2006. Potential mRNA degradation targets of hsa-miR-200c, identified using informatics and qRT-PCR. Cell Cycle 5: 1951–1956 [DOI] [PubMed] [Google Scholar]
- Jurka J, Kohany O, Pavlicek A, Kapitonov VV, Jurka MV 2005. Clustering, duplication and chromosomal distribution of mouse SINE retrotransposons. Cytogenet Genome Res 110: 117–123 [DOI] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471: 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. 2008. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res 36: D773–D779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T 2004. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM 2010. microRNA activity is suppressed in mouse oocytes. Curr Biol 20: 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Blelloch R 2010. MicroRNA regulation of embryonic stem cell self-renewal and differentiation. Adv Exp Med Biol 695: 105–117 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45 doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, et al. 2009. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 105: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios M, Fan G, Fekete C, Kelly J, Bates B, Kuehn R, Lechan RM, Jaenisch R 2001. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol 15: 1748–1757 [DOI] [PubMed] [Google Scholar]
- Rodgers EE, Theibert AB 2002. Functions of PI 3-kinase in development of the nervous system. Int J Dev Neurosci 20: 187–197 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shenton ME, Dickey CC, Frumin M, McCarley RW 2001. A review of MRI findings in schizophrenia. Schizophr Res 49: 1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, et al. 2010. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res 20: 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R 2010. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 20: 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Wang Y, Melton C, Medvid R, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2: 70–75 [DOI] [PubMed] [Google Scholar]
- Yang S, Tutton S, Pierce E, Yoon K 2001. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol 21: 7807–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH 2006. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res 34: 5112–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Shao P, Zhou H, Chen YQ, Qu LH 2010. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res 38: D123–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, D'Ercole AJ 2006. Insulin-like growth factor actions during development of neural stem cells and progenitors in the central nervous system. J Neurosci Res 83: 1–6 [DOI] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, et al. 2009. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci 106: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]