Abstract
Biogenesis of the vast majority of plant siRNAs depends on the activity of the plant-specific RNA polymerase IV (PolIV) enzyme. As part of the RNA-dependent DNA methylation (RdDM) process, PolIV-dependent siRNAs (p4-siRNAs) are loaded onto an ARGONAUTE4-containing complex and guide de novo DNA methyltransferases to target loci. Here we show that the double-stranded RNA binding proteins DRB2 and DRB4 are required for proper accumulation of p4-siRNAs. In flowers, loss of DRB2 results in increased accumulation of p4-siRNAs but not ta-siRNAs, inverted repeat (IR)-derived siRNAs, or miRNA. Loss of DRB2 does not impair uniparental expression of p4-dependent siRNAs in developing endosperm, indicating that p4-siRNA increased accumulation is not the result of the activation of the polIV pathway in the male gametophyte. In contrast to drb2, drb4 mutants exhibit reduced p4-siRNA levels, but the extent of this reduction is variable, according to the nature and size of the p4-siRNAs. Loss of DRB4 also leads to a spectacular increase of p4-independent IR-derived 24-nt siRNAs, suggesting a reallocation of factors from p4-dependent to p4-independent siRNA pathways in drb4. Opposite effects of drb2 and drb4 mutations on the accumulation of p4-siRNAs were also observed in vegetative tissues. Moreover, transgenic plants overexpressing DRB2 mimicked drb4 mutants at the morphological and molecular levels, confirming the antagonistic roles of DRB2 and DRB4.
Keywords: RNAi, siRNA, double-stranded RNA binding protein, non-coding RNA, Dicer
INTRODUCTION
Plants contain complex populations of small RNAs involved in different RNA-based silencing pathways that function at the transcriptional and/or post-transcriptional level to control genes, viruses, and transposable elements (Jamalkandi and Masoudi-Nejad 2009; Ruiz-Ferrer and Voinnet 2009; Voinnet 2009; Law and Jacobsen 2010). Arabidopsis small RNAs are produced by the activity of four Dicer-like ribonucleases (DCLs) on double-stranded RNA precursors of different origins and structures (Bouche et al. 2006; Henderson et al. 2006; Mlotshwa et al. 2008; Liu et al. 2009). The vast majority of Arabidopsis small RNAs (>90% of the plant global small RNA mass) consists of siRNAs that depend for their biogenesis on the capacity of RNA polymerase IV (PolIV), a homolog of DNA-dependent RNA polymerase II (PolII), to transcribe thousands of intergenic loci (Rajagopalan et al. 2006; Kasschau et al. 2007; Zhang et al. 2007). PolIV-derived single-stranded RNA precursors are converted to long double-stranded molecules by the action of the RNA-dependent RNA polymerase 2 (RDR2) and are mainly cleaved in small 24-nt dimers by the action of DCL3 (Lahmy et al. 2010). The accumulation of some, but not all, PolIV-dependent small RNAs (p4-siRNAs) depends on the activity of RNA polymerase V (PolV), an enzyme related to PolIV but presenting a specific carboxy-terminal domain containing evolutionarily conserved GW/WG repeats (El-Shami et al. 2007). P4-siRNAs have been classified in two categories named type I and type II (Mosher et al. 2009). Type I p4-siRNAs are produced exclusively in flowers and siliques, while type II p4-siRNAs are produced in almost all plant tissues with, again, a maximum of expression in flowers and siliques (Kasschau et al. 2007; Mosher et al. 2009). P4-siRNAs are completely absent from the paternal lineage, and all p4-siRNAs accumulating in the developing seeds are inherited maternally (Mosher et al. 2009). P4-siRNAs associate with ARGONAUTE 4 (AGO4), AGO6, or AGO9 (Havecker et al. 2010) and play a critical role in the RNA-directed DNA methylation (RdDM) process, as they can guide a complex set of proteins leading to DNA methylation and chromatin structure modifications at RdDM loci (Chinnusamy and Zhu 2009).
MicroRNAs (miRNAs) represent the second largest population of plant small RNAs. MiRNAs are produced from PolII (and not polIV) transcripts that can adopt a foldback structure (Rajagopalan et al. 2006; Kasschau et al. 2007). Most miRNAs result from the cleavage of this precursor by a complex composed of DCL1, the double-stranded RNA binding protein (dsRBP) DRB1, and the zinc-finger protein SERRATE (Voinnet 2009). A few miRNA precursors escape this rule, as they are processed by DCL4 instead of DCL1 (Rajagopalan et al. 2006). Although miRNAs represent only ∼5% of the plant global small RNA mass, they are important regulators of mRNA translation and degradation, and their contribution is critical for normal plant development.
In addition to p4-siRNAs and miRNAs, three minor populations of small RNAs are present in plants. Trans-acting siRNAs (ta-siRNAs) originate from the targeting of long PolII precursors by specific miRNAs (Allen and Howell 2010). The biogenesis of ta-siRNAs requires the RNA-dependent RNA polymerase 6 (RDR6), DCL4, and the dsRBP DRB4. Like miRNAs, ta-siRNAs target mRNA for degradation and are critical regulators of plant development (Allen and Howell 2010). Inverted repeat (IR)-derived siRNAs are transcribed from loci that generate long RNA foldback structures that are mainly cleaved by DCL2 and DCL3 to generate 22-nt and 24-nt siRNAs, respectively (Dunoyer et al. 2010). The biogenesis of these endogenous IR-derived siRNAs is completely independent of PolIV, yet these small RNAs can participate in RdDM as well as post-transcriptional gene silencing (PTGS) (Dunoyer et al. 2010). Finally, overlapping natural cis-antisense transcripts can generate double-stranded RNA molecules that can be cleaved by DCL1 or DCL2 in a two-step process to generate small natural antisense RNAs (nat-siRNAs) (Borsani et al. 2005; Jen et al. 2005; Jin et al. 2008). Curiously, despite the production of double-stranded molecules by the pairing of the two antisense transcripts, the biogenesis of nat-siRNAs requires PolIV and RDR6 (Borsani et al. 2005). In most cases, the overlapping natural cis-antisense transcripts are only simultaneously produced under specific environmental conditions (Jin et al. 2008), so that nat-siRNAs are not found in plants grown in standard conditions. Nat-siRNAs usually target one of the two generating transcripts for degradation (Borsani et al. 2005; Jen et al. 2005; Jin et al. 2008).
Arabidopsis possesses a small family of five closely related dsRBP (DRB1 to DRB5) (Hiraguri et al. 2005). DRB1, also known as HYL1, is required for DCL1-mediated processing of most miRNA precursors (Kurihara et al. 2006). DRB1 probably binds as a dimer to the miRNA/miRNA* duplex and recruits DCL1, in cooperation with SERRATE (Yang et al. 2010). It is likely that DRB1 guides the selection of the miRNA strand (Eamens et al. 2009). DRB4 is a partner of DCL4 and is involved in the biogenesis of ta-siRNAs and DCL4-dependent miRNAs (Adenot et al. 2006; Nakazawa et al. 2007; Pouch-Pélissier et al. 2008; Eamens et al. 2009). The DRB4/DCL4 complex is also required to produce siRNAs from exogenous viral dsRNAs. Recently, DRB4 was found to be essential in vitro for DCL4 activity (Qu et al. 2008; Fukudome et al. 2011). DRB2, DRB3, and DRB5 were reported not to be involved in small RNA biogenesis (Curtin et al. 2008), and their functions remained elusive. Here, we show that DRB2 and DRB4 are both involved in the production of p4-siRNAs and have antagonistic effects on this pathway.
RESULTS
DRB2 and DRB4 are required for proper accumulation of p4-siRNAs
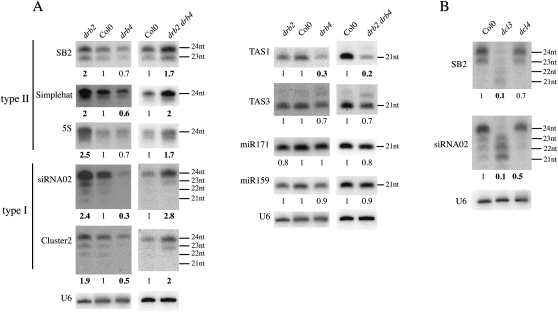
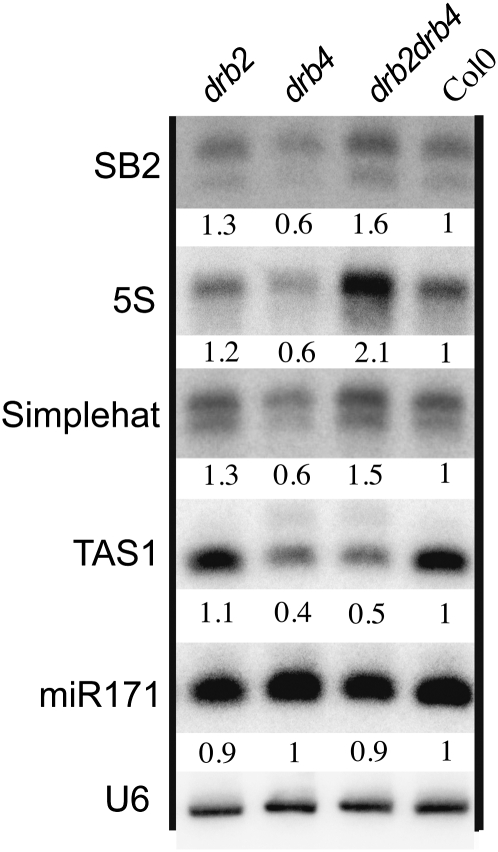
Accumulation of p4-siRNAs in immature flowers was analyzed in a large number of Arabidopsis thaliana mutants to identify new actors involved in their biogenesis. Immature flowers were chosen because both type I and type II p4-siRNAs accumulate in floral tissues (Mosher et al. 2009). In addition to PolIV, PolV, RDR2, DCL3, and AGO4, this screen identified DRB2 and DRB4 as important players in this pathway (Fig. 1). drb2 mutants accumulated two- to 2.5-fold more p4-siRNAs (type I and II) than wild-type plants (Fig. 1A), whereas drb4 mutants exhibited reduced accumulation of p4-siRNAs. This reduction was more pronounced for type I p4-siRNAs (0.3- to 0.5-fold) than type II p4-siRNAs (0.6- to 0.7-fold). drb2drb4 double mutants exhibited a pattern similar to drb2 single mutants, indicating that drb2 is epistatic to drb4 for p4-siRNAs accumulation. miRNA accumulation was largely unaffected by the loss of DRB2 and/or DRB4 as reported previously (Curtin et al. 2008). Also, as previously found (Adenot et al. 2006; Nakazawa et al. 2007; Curtin et al. 2008), accumulation of ta-siRNAs was unaffected by the loss of DRB2 but was reduced in drb4 and in the drb2drb4 double mutant.
FIGURE 1.
DRB2, DRB4, DCL3, and DCL4 are required for proper accumulation of polymerase IV-dependent (p4)-siRNAs. (A) Impact of the absence of DRB2, DRB4 or DRB2, and DRB4 on accumulation of different small RNA species. RNA extracts were obtained from immature flowers, and three independent biological replicates were performed for each sample. Values are normalized to U6 RNA and are expressed as a ratio relative to the wild-type Col0. For p4-siRNAs, only the 24-nt species was used for normalization. Probe sequences are given in Supplemental Table S1. The SB2 probe targets one of the six Arabidopsis thaliana SINE dispersed repeat families (Deragon and Zhang 2006). (B) Impact of DCL3 and DCL4 on p4-siRNA accumulation.
Western blot hybridization using specific antibodies raised against DCL3 and AGO4 revealed that these two major actors of the polIV pathway accumulated in similar amounts in drb2 and drb4 compared to wild type (data not shown), indicating that the changes in p4-siRNA accumulation observed in drb2 and drb4 did not result from changes in DCL3 or AGO4 levels. Consistently, 24-nt but not 21-, 22-, or 23-nt p4-siRNAs were lost in dcl3 (Fig. 1B), whereas siRNAs of the four sizes were simultaneously affected in drb2 and drb4. Interestingly, the accumulation of both type I and type II p4-siRNAs was reduced to the same extent in dcl4 and drb4 (Fig. 1B), suggesting a coordinated action of DRB4 and DCL4 in the p4-siRNA pathway.
Sequencing of small RNA populations from the drb2 and drb4 mutants
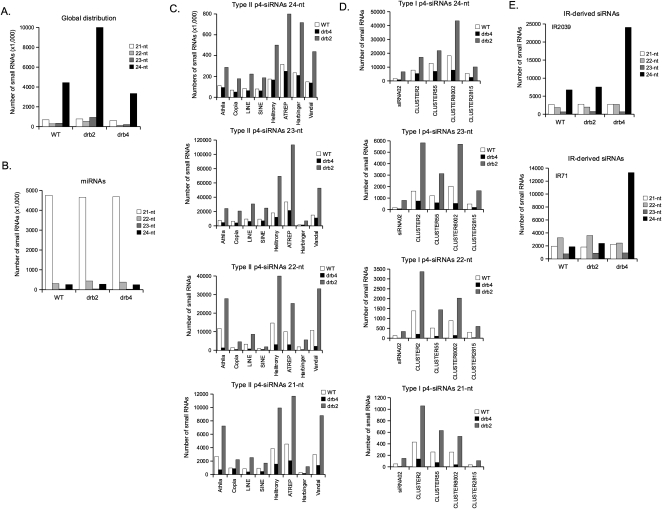
To extend our analysis of the role of DRB2 and DRB4 in the p4-siRNA pathway, low molecular weight RNAs from wild type, drb2, and drb4 immature flower extracts were reverse-transcribed and sequenced using the Illumina HiSeq 2000 system. After elimination of background RNA and normalization (see Materials and Methods), the global distribution of 21- to 24-nt RNAs was analyzed (Fig. 2A). As expected, the 24-nt population (mainly composed of p4-siRNAs (Rajagopalan et al. 2006; Kasschau et al. 2007; Zhang et al. 2007) was by far the major component. Compared to wild type, the 24-nt population clearly was increased (2.3-fold) in drb2 and reduced (0.75-fold) in drb4. The 21-nt population (mainly composed of miRNAs) (Rajagopalan et al. 2006; Kasschau et al. 2007) represented the second largest population of small RNAs. In our global analysis, the 21-nt population showed a modest increase in drb2 (1.2-fold) and a small decrease in drb4 (0.87-fold). To test if these small variations were the result of a change in the miRNA population, we analyzed miRNAs independently in the two mutants (Fig. 2B). We observed that 21-nt miRNA populations were unchanged in drb2 and drb4 (0.98- and 0.99-fold, respectively) compared to wild type. This observation is in agreement with our Northern hybridization results (Fig. 1) and suggests that the small variations of the 21-nt population observed in the global analysis were not the result of changes in miRNA populations. Finally, the 22- and 23-nt small RNA populations behaved similarly to the 24-nt population in the two mutants, although the 22-nt population shows a stronger reduction in drb4 (0.48-fold) compared to wild type.
FIGURE 2.
Genome-wide profiling of the different populations of small RNAs in the drb2 and drb4 mutants compared to wild-type (Col0). (A) Global distribution of 21–24-nt small RNAs. (B) General distribution of miRNAs. (C) Distribution of 21–24-nt type II p4-siRNAs issued from different dispersed repeat families. (D) Distribution of 21–24-nt type I p4-siRNAs issued from five different genomic loci. (E) Distribution of 21–24-nt small RNAs from two representative endogenous inverted repeats (IR) loci.
To test if global variations of 21- to 24-nt RNAs in drb2 and drb4 result mainly from changes in type I and II p4-siRNAs, we analyzed the distribution of small RNAs originating from various genomic loci. First, populations of type II p4-siRNAs from eight transposable element (TE) families were studied (Fig. 2C). All TE-related type II siRNAs (21- to 24-nt) were increased two- to threefold in the drb2 mutant. In contrast, the TE-related type II siRNAs of different sizes were affected differently by the loss of DRB4. Indeed, the 23- and 24-nt populations were less affected by the loss of DRB4 (0.7-fold reduction) compared to the 21-nt (0.4-fold) and the 22-nt (0.2-fold). Next, populations of type I p4-siRNAs from five genomic loci (Mosher et al. 2009) were analyzed (Fig. 2D). Here again, all type I siRNAs (21- to 24-nt) were increased two- to threefold in the drb2 mutant. Type I siRNAs were more affected by the loss of DRB4 than type II, with, again, strong differences depending on RNA sizes. Indeed, 23- and 24-nt type I siRNAs showed a 0.4-fold reduction, while 21-nt exhibited a 0.2-fold reduction and 22-nt accumulated to less than 0.1-fold compared to the wild type.
To evaluate the impact of DRB2 and DRB4 on the accumulation of p4-independent IR-derived siRNAs, populations of siRNAs from two IR loci were analyzed in wild type and in the drb2 and drb4 mutants (Fig. 2E). In sharp contrast to p4-dependant siRNAs, loss of DRB2 did not affect significantly IR-derived siRNAs, suggesting that DRB2 is not involved in the accumulation of p4-independent siRNAs. In contrast, loss of DRB4 resulted in increased levels (five- to sixfold) of 24-nt, but not 21- to 23-nt IR-derived siRNA (Fig. 2E), similar to a previous report (Dunoyer et al. 2010).
Both DRB2 and DRB4 are involved in the accumulation of DCL4-dependent miRNAs
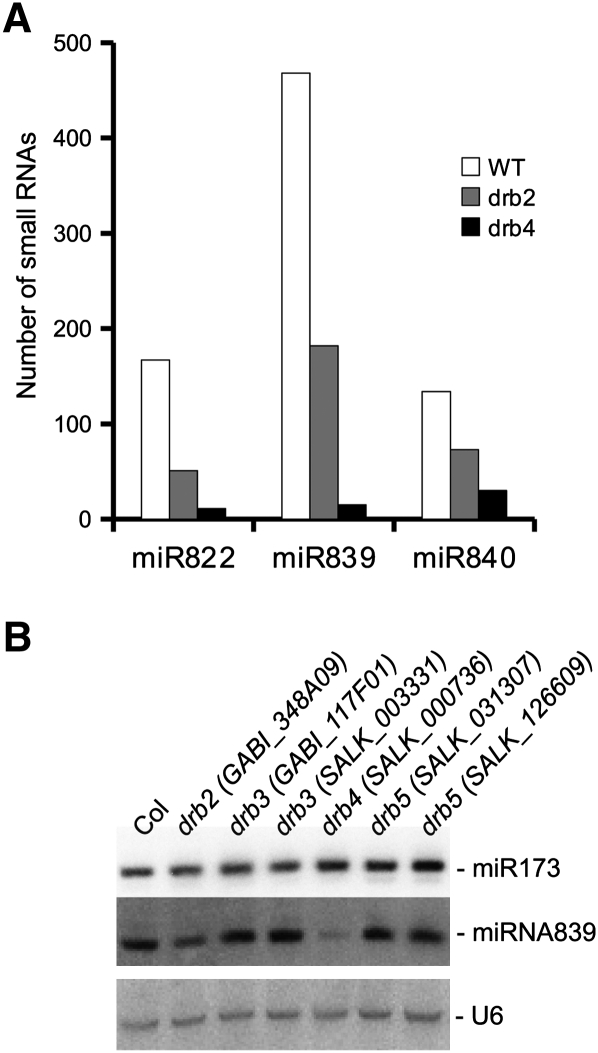
Young miRNAs such as miR822, miR839, or miR840 are generated by DCL4 instead of DCL1 (Rajagopalan et al. 2006; for miR840, see the ASRP database http://asrp.cgrb.oregonstate.edu/). Two of these RNAs (miR822 and miR839) accumulate at lower levels in drb4 (Pouch-Pélissier et al. 2008; Eamens et al. 2009), suggesting that their biogenesis requires the coordinate action of DCL4 and DRB4. Our sequencing data confirmed that the accumulation of these three DCL4-dependent miRNAs was strongly reduced in the absence of DRB4 (Fig. 3A). A two- to threefold reduced accumulation of these miRNAs was also observed in drb2, suggesting that DRB2 is also required for the proper biogenesis of DCL4-dependent miRNAs (Fig. 3A). To confirm the specific action of DRB2 and DRB4 on DCL4-dependent miRNAs, Northern blot hybridization was performed using drb2, drb3, drb4, and drb5 mutants. Accumulation of miR839 was unchanged in drb3 and drb5 but was reduced in drb2 and drb4, consistent with sequencing data. In contrast, the accumulation of miR173, a DCL1-dependent miRNA, was unchanged in drb2, drb3, drb4, and drb5 (Fig. 3B).
FIGURE 3.
Impact of DRB2 and DRB4 on DCL4-dependent miRNAs. (A) The three miRNAs are strongly reduced in the small RNA population of drb4 and are also significantly affected in the drb2 background. (B) Northern hybridization using RNA extracts from immature flowers, confirming a reduction of miRNA839 accumulation in drb2 and drb4 but not in two independent drb3 and drb5 mutant lines. References for the different mutant lines are indicated in brackets. The accumulation of DCL1-dependent miRNA173 was unaffected in these different conditions.
DRB2 does not repress p4-siRNA expression in the paternal lineage
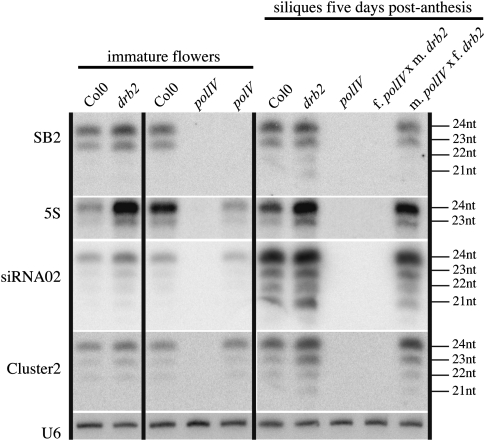
The paternal lineage does not contribute to p4-siRNA populations in developing seeds of Arabidopsis (Mosher et al. 2009). This uniparental expression of p4-siRNAs could be explained if an activating factor is produced in the maternal lineage and/or if a repressive factor is expressed in the paternal lineage (Mosher et al. 2009). Because it is highly abundant in anthers and pollen (Curtin et al. 2008), DRB2 is a good candidate for the shutdown of the p4-siRNA pathway in the paternal lineage, potentially explaining the twofold increased accumulation of p4-siRNAs observed in drb2. To test this hypothesis, we performed reciprocal crosses between drb2 and polIV mutants and analyzed the accumulation of type I and type II p4-siRNAs five days post-anthesis (Fig. 4). If DRB2 represses p4-siRNAs in the male lineage, crossing drb2 as male with polIV as female should result in the accumulation of paternally inherited p4-siRNAs in the resulting siliques. However, siliques resulting from crosses between polIV as female and drb2 as male did not accumulate p4-siRNAs, suggesting that DRB2 is not acting as a repressor in the paternal lineage. The reciprocal cross (polIV as male and drb2 as female) resulted in a higher level of p4-siRNAs in siliques compared to wild type but in a level similar to the one found in drb2 siliques, suggesting that the antagonistic effect of DRB2 on p4-siRNAs levels occurs in the maternal lineage.
FIGURE 4.
DRB2 does not repress p4-siRNA expression in the paternal lineage. For reciprocal crosses, emasculated flowers were pollinated manually, and RNA was extracted from the developing cross at five days after fertilization. Loss of DRB2 function does not induce paternal-specific expression of p4-siRNAs in the developing seeds. As reported previously, type II p4-siRNAs (SB2, 5S) are sensitive to PolV action, while type I p4-siRNAs (siRNA02 and Cluster2) accumulation is affected weakly, or not at all, in the polV mutant (Pontier et al. 2005; Zheng et al. 2009).
DRB2 and DRB4 are required for proper accumulation of p4-siRNAs in vegetative tissues
Whereas type I p4-siRNAs accumulate only in flowers and siliques, type II p4-siRNAs are additionally produced in vegetative tissues, allowing analysis of the impact of the loss of DRB2 and DRB4 in these later tissues. Changes in type II p4-siRNA, ta-siRNA, and miRNA accumulation in drb2, drb4, and drb2drb4 17-day-old seedlings were similar to those observed in flowers (cf. Figs. 1 and 5). Although reproducible, the small increase in type II p4-siRNAs in drb2 was much less pronounced in seedlings than in flowers. Therefore, the impact of the loss of DRB2 is lower in vegetative tissues compared to flowers. Apart from this difference, our results suggest that DRB2 and DRB4 modulate similarly the biogenesis of p4-siRNAs in flowers and vegetative tissues.
FIGURE 5.
DRB2 and DRB4 also impact p4-siRNA expression in vegetative tissues. Seventeen-day-old plantlets showed accumulation defects of type II p4-siRNAs similar to that observed with flowers but with a lower impact in the drb2 background. Hybridization signals are normalized to U6 RNA and expressed as a ratio relative to the wild-type Col0.
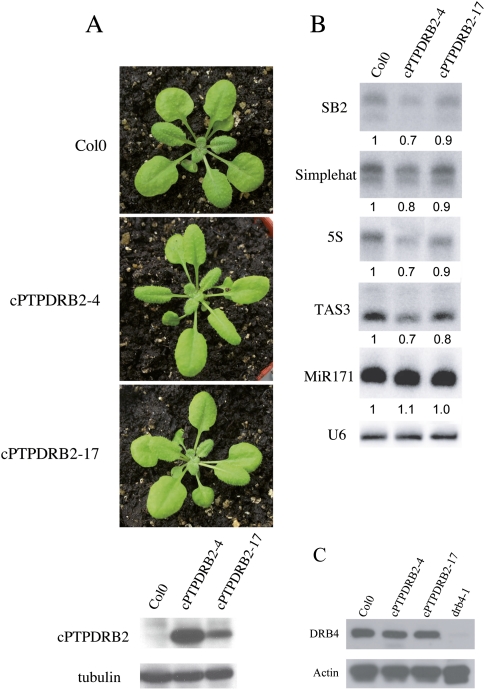
DRB2 overexpression in seedlings mimics a drb4 mutation at the morphological and molecular levels
To determine how DRB2 limits the production/accumulation of p4-siRNAs in flowers and vegetative tissues, we constitutively expressed a tagged-DRB2 protein under the control of the 35S promoter and measured levels of p4-siRNAs. Seventeen-day-old seedlings accumulating low levels of tagged-DRB2 (cPTPDRB2-17) looked similar to wild-type plants (Fig. 6A). In contrast, plants accumulating high levels of tagged-DRB2 (cPTPDRB2-4) and unchanged levels of DRB4 (Fig. 6C) exhibited a zippy phenotype (Fig. 6A), similar to drb4 and dcl4 mutants that have a zippy phenotype due to reduced accumulation of TAS3 ta-siRNAs (Adenot et al. 2006). Consistently, the level of TAS3 ta-siRNAs was reduced in plants accumulating tagged-DRB2 (Fig. 6B). The accumulation of tagged-DRB2 also correlated with a slight reduction of p4-siRNA accumulation (Fig. 6B), similar to that observed in drb4 (Fig. 5), indicating that DRB2 over-accumulation functionally antagonizes DRB4.
FIGURE 6.
Constitutive expression of DRB2 is associated with a zippy-like phenotype and a reduction of p4-siRNAs. (A) A downward-curled leaf margins phenotype was clearly associated with plant lines that express elevated levels of PTP-tagged DRB2 protein under the control of the 35S promoter. (B) Plants accumulating tagged-DRB2 present a slight reduction of p4-siRNAs, a situation similar to that observed in drb4 (Fig. 5). The level of TAS3 ta-siRNAs is also reduced in these plants. (C) Western blot probed with an antibody raised against DRB4. DRB4 accumulates to a similar level in Col0 and in the two DRB2 overexpressing lines, ruling out the possibility that the effects observed result from a reduction of DRB4 levels when DRB2 levels are increased. As expected, no specific signal was detected when a protein extract from the drb4 mutant line was used.
DISCUSSION
Our study reveals that a correct balance between DRB2 and DRB4 is necessary for proper accumulation of p4-siRNAs. Loss of DRB2 results in a general two- to threefold increase in p4-siRNA accumulation (Figs. 1, 2), whereas over-accumulation of DRB2 results in a reduction of p4-siRNA accumulation (Fig. 6B), similar to that observed in drb4 (Figs. 1, 2). Uniparental expression of p4-dependent siRNAs in developing endosperm is not affected by the drb2 mutation (Fig. 4), indicating that p4-siRNA increased accumulation does not result from the reactivation of the polIV pathway in the male gametophyte. Interestingly, all sizes of p4-siRNAs over-accumulate in drb2. By far, DCL3-dependent 24-nt siRNAs represent the most abundant form of p4-siRNAs (Fig. 1B). However, 21-, 22-, and 23-nt p4-siRNAs are also found. The enzymatic machinery responsible for their production in wild-type plants remains unclear. Analysis of double, triple, and quadruple dcl mutants revealed that, in the absence of DCL3, DCL2 produces 22- and 23-nt p4-siRNAs, while DCL4 produces 21-nt p4-siRNAs. Moreover, DCL1 appears able to produce 21-nt p4-siRNAs in the absence of DCL2, DCL3, and DCL4 (Bouche et al. 2006; Henderson et al. 2006; Liu et al. 2009). Although these results suggest that 21-, 22-, and 23-nt p4-siRNAs are produced by DCL2 and DCL4, it remains possible that DCL3 produces all sizes of p4-siRNAs in wild-type plants. Over-accumulation of all sizes of p4-siRNAs in drb2 mutants could be explained by an antagonistic effect of DRB2 on DCL3 and other DCL(s) involved in p4-siRNA biogenesis. However, drb2 mutants exhibit unchanged accumulation of polymerase IV-independent miRNAs, ta-siRNAs, and endogenous IR-derived siRNAs, which are processed by DCL1, DCL2, DCL3, and DCL4, indicating that the antagonistic effect of DRB2 is strictly limited to the p4-siRNA pathway. Therefore, the antagonistic effect of DRB2 on the p4-siRNA pathway likely occurs upstream of the DCL(s). One possibility could be that DRB2 interacts with RDR2 and binds to RDR2-derived dsRNA, thus limiting their processing by DCL(s). Alternatively, PolIV primary transcripts or RDR2-derived dsRNA bound to DRB2 could enter a degradation pathway involving RNases that remain to be identified. Under any of these two scenarios, loss of DRB2 would allow more RDR2-derived dsRNA to be processed into all sizes of p4-siRNAs by DCL(s).
Loss of DRB4 results in reduced accumulation of DCL4-dependent miRNAs and ta-siRNAs because DCL4 functions with DRB4 for their processing (Adenot et al. 2006; Pouch-Pélissier et al. 2008). Loss of DRB4 also results in reduced accumulation of all sizes of p4-siRNAs. Type I p4-siRNAs are more affected than type II p4-siRNAs, and 22-nt are more affected than 21-nt, which themselves are more affected than 23- and 24-nt (Figs. 1, 2). One possibility to explain these results is that, in addition to DCL4, DCL2 and DCL3 could also require DRB4 for efficient dsRNA processing. However, we failed to immunoprecipitate DCL3 from a transgenic line expressing a tagged-DRB4 fusion protein in conditions where we can readily immunoprecipitate DCL4 (see Supplemental Fig. S1) suggesting that DRB4 is not associated with DCL3 in vivo. Because type I and type II p4-siRNAs accumulate at reduced levels in both dcl4 and drb4 mutants, it is more likely DRB4 controls p4-siRNA levels through DCL4. How the DRB4/DCL4 complex regulates the accumulation of all sizes of p4-siRNAs remains to be determined. A spectacular increase in the accumulation of 24-nt p4-independent IR-derived siRNAs is also observed in drb4 (Fig. 2E), similar to a previous report (Dunoyer et al. 2010). In this case, DRB4 acts independently of DCL4 because IR-derived siRNA levels and patterns are unchanged in the dcl4 mutant (Dunoyer et al. 2010). How DRB4 limits the production of IR-derived siRNAs is unknown. One possibility is that lowering the global efficiency of the p4-dependent siRNA pathway in the drb4 mutant could lead to the reallocation of factors from the p4-dependent to the p4-independent siRNA pathway. Alternatively, DRB4 could directly bind IR precursor RNAs and limit their cleavage by DCL3.
Overall, our analyses reveal specialized, redundant, and antagonistic functions for DRB2 and DRB4. Analysis of drb2 and drb4 mutants points to an antagonistic effect in the DCL3-dependent p4-siRNA pathway. In addition, plants over-expressing DRB2 mimic the zippy phenotype of drb4 mutants and display reduced levels of p4-siRNAs (Fig. 6), supporting an antagonistic effect of DRB2 on DRB4 and pointing to a potential interference of ectopically expressed DRB2 in the DRB4/DCL4-dependent ta-siRNA pathway. In contrast, both drb2 and drb4 mutations cause a reduction in the accumulation of young DCL4-dependent miRNAs (Fig. 3), suggesting partial redundancy in this pathway. Lastly, loss of DRB2 does not affect IR-derived siRNA accumulation, whereas loss of DRB4 results in increased levels of DCL3-dependent 24-nt but not DCL2/4-dependent 21- to 23-nt IR-derived siRNA, indicating specialized functions for DRB4. Together, these results suggest that DRB proteins are part of multiple complexes, allowing independent actions in different pathways. Elucidation of the DRB interaction network will shed light on the functioning of this complex family of proteins.
MATERIALS AND METHODS
Genetic stocks and plant growth conditions
The stocks of drb2-1, drb4-1, drb2-1drb4-1, dcl4-2, dcl3-1, nrpd1a-4 (polIV), and nrpe1-11 (polV) used in this study are in a Columbia (Col0) genetic background and were described previously (Xie et al. 2004; Pontier et al. 2005; Xie et al. 2005; Curtin et al. 2008). For ectopic DRB2 expression, the DRB2 coding sequence was associated to a C-terminal PTP (protC-TEV-protA) tag (Schimanski et al. 2005) and placed under the 35S promoter expression. Wild-type Arabidopsis Col0 plants were transformed as previously described (Clough and Bent 1998), and transformed lines displaying different levels of DRB2 expression were identified using PAP antibody. Plant seeds were stratified for two days at 4°C before growth in chambers on soil at 23°C under a 16-h-light/8-h-dark cycle. For in vitro analyses, seeds were sterilized and sowed on solid Murashige and Skoog (MS) medium containing 1% sucrose (w/v) and grown under continuous light at 20°C.
RNA isolation and hybridization
Total RNA was extracted as described elsewhere (Pélissier et al. 2004), using immature inflorescences (stages 1–12), siliques five days post-anthesis, or rosette from in vitro growing 17-day-old plantlets. For the detection of small RNAs, 12–18μg of total RNA samples were heat-treated in 1.5 volume of standard formamide buffer and loaded on 15% polyacrylamide (19:1 acrylamide:bis-acrylamide)–8.3 M urea–0.5X TBE gel and separated by electrophoresis. The samples were electroblotted to hybond-NX membranes (GE Healthcare) and fixed following a carbodiimide-mediated cross-linking procedure (Pall et al. 2007). Pre-hybridization and hybridization were carried out in 5X SSC, 20 mM Na2HPO4 pH7.2, 7% SDS, 2X Denhardt solution, 50 mg/mL herring DNA at 50°C. Sequences of the different probes used are given in Supplemental Table S1. Filters were washed twice with 3X SSC, 25 mM NaH2PO4 pH7.5, 5% SDS at 50°C for 10 min, followed by one to two washes with 1X SSC, 1% SDS at 50°C for 10 min. Signals were visualized using a phosphorimager (Molecular Imager FX ; Bio-Rad) for quantification.
Sequencing of small RNAs and data analysis
Total RNAs were separated on polyacrylamide gel electrophoresis as above and small RNAs (18–36 nucleotides) were cut out of the gels, used to construct libraries according to the manufacturer's protocol (Illumina Small RNA Sample Prep Kits), and sequenced on a HiSeq 2000 system (Illumina) on the MGX platform (http://www.mgx.cnrs.fr/). Fourteen and 0.6 million small RNA reads were obtained for Col0, 16.1 million for drb2, and 10.4 million for drb4. Small read counts were normalized by taking highly expressed conserved miRNAs that were shown by northern hybridization to be invariant in Col0 (Fig. 1) and calculating normalization factors for each condition. The normalized data was cross-confirmed by calculating the normalization factor for all significantly expressed 36-nt (that represent general breakdown RNA products), which yielded similar values. Only small RNAs having 10 or more read counts in at least one condition were considered in Figures 2 and 3.
Immunoblot analysis
Protein extracts were obtained by grinding frozen tissues in liquid nitrogen; After resuspension in 4X Laemli Buffer, the extracts were treated for 5 min at 95°C and centrifuged before loading on SDS/PAGE gels. Proteins were transferred onto PVDF membrane (Immobilon-P; Millipore), and protA fusion proteins were visualized using the PAP (Peroxidase-Anti-Peroxidase) soluble complex (Sigma) diluted at 1/10,000. The level of DRB4 was evaluated using a custom made specific antibody (Eurogentec) diluted at 1/500. Equal protein loading was assessed using anti-tubulin or anti-actin specific antibody.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Cecile Bousquet-Antonelli and Thierry Lagrange for discussions. This work was supported by l'Agence Nationale de la Recherche (ANR-06-BLAN-0203-02), by the CNRS, and by the Université de Perpignan (UPVD).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2680711.
REFERENCES
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H 2006. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E, Howell MD 2010. miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin Cell Dev Biol 21: 798–804 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK 2009. RNA-directed DNA methylation and demethylation in plants. Sci China C Life Sci 52: 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM 2008. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett 582: 2753–2760 [DOI] [PubMed] [Google Scholar]
- Deragon JM, Zhang X 2006. Short interspersed elements (SINEs) in plants: Origin, classification, and use as phylogenetic markers. Syst Biol 55: 949–956 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, Alioua A, Himber C, Voinnet O 2010. An endogenous, systemic RNAi pathway in plants. EMBO J 29: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM 2009. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T 2007. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev 21: 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome A, Kanaya A, Egami M, Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T 2011. Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17: 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC 2010. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing, and DNA methylation patterning. Nat Genet 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T 2005. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57: 173–188 [DOI] [PubMed] [Google Scholar]
- Jamalkandi SA, Masoudi-Nejad A 2009. Reconstruction of Arabidopsis thaliana fully integrated small RNA pathway. Funct Integr Genomics 9: 419–432 [DOI] [PubMed] [Google Scholar]
- Jen CH, Michalopoulos I, Westhead DR, Meyer P 2005. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol 6: R51 doi: 10.1186/gb-2005-6-6-r51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Vacic V, Girke T, Lonardi S, Zhu JK 2008. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol 9: 6 doi: 10.1186/1471-2199-9-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC 2007. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57 doi: 10.1371/journal.pbio.0050057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Takashi Y, Watanabe Y 2006. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S, Bies-Etheve N, Lagrange T 2010. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics 5: 4–8 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE 2010. Establishing, maintaining, and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Feng Y, Zhu Z 2009. Dicer-like (DCL) proteins in plants. Funct Integr Genomics 9: 277–286 [DOI] [PubMed] [Google Scholar]
- Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V 2008. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One 3: e1755 doi: 10.1371/journal.pone.0001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC 2009. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286 [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T 2007. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol Biol 63: 777–785 [DOI] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A 2007. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA, and piRNA by northern blot. Nucleic Acids Res 35: e60 doi: 10.1093/nar/gkm112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélissier T, Bousquet-Antonelli C, Lavie L, Deragon JM 2004. Synthesis and processing of tRNA-related SINE transcripts in Arabidopsis thaliana. Nucleic Acids Res 32: 3957–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T 2005. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouch-Pélissier MN, Pélissier T, Elmayan T, Vaucheret H, Boko D, Jantsch MF, Deragon JM 2008. SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLoS Genet 4: e1000096 doi: 10.1371/journal.pgen.1000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O 2009. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60: 485–510 [DOI] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Gunzl A 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell 4: 1942–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: E104 doi: 10.1371/journal.pbio.0020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC 2005. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA 2010. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE 2007. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci 104: 4536–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X 2009. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]