Abstract
MicroRNAs (miRNAs) are small RNAs that modulate gene expression by binding target mRNAs. The hundreds of miRNAs expressed in the brain are critical for synaptic development and plasticity. Drugs of abuse cause lasting changes in the limbic regions of the brain that process reward, and addiction is viewed as a form of aberrant neuroplasticity. Using next-generation sequencing, we cataloged miRNA expression in the nucleus accumbens and at striatal synapses in control and chronically cocaine-treated mice. We identified cocaine-responsive miRNAs, synaptically enriched and depleted miRNA families, and confirmed cocaine-induced changes in protein expression for several predicted synaptic target genes. The miR-8 family, known for its roles in cancer, is highly enriched and cocaine regulated at striatal synapses, where its members may affect expression of cell adhesion molecules. Synaptically enriched cocaine-regulated miRNAs may contribute to long-lasting drug-induced plasticity through fine-tuning regulatory pathways that modulate the actin cytoskeleton, neurotransmitter metabolism, and peptide hormone processing.
Keywords: microRNA, RNA-Seq, cocaine, deep sequencing, bioinformatics
INTRODUCTION
Hundreds of microRNAs (miRNAs), a class of endogenous 21–25 nucleotide (nt) small RNAs that modulate gene expression by binding complementary sequences in the 3′-untranslated regions (3′ UTRs) of their target mRNA transcripts (Kim et al. 2009), are expressed in the mature mammalian brain (Lagos-Quintana et al. 2002; Krichevsky et al. 2003; Miska et al. 2004; Landgraf et al. 2007) and are thought to be involved in synapse development and plasticity (Schratt 2009). The biogenesis of mature miRNAs is characterized by successive RNA cleavage events in which long primary miRNA (pri-miRNA) transcripts are processed into shorter preliminary miRNA (pre-miRNA) hairpins, exported from the nucleus, and cleaved into ∼22 nt duplex RNAs prior to being loaded into RNA-induced silencing complexes (RISCs) (Kim et al. 2009). Each RISC contains an Argonaute protein (AGO1-4 in human and mouse) and a miRNA (Höck and Meister 2008). The RISC then interacts with the target mRNAs through base-pairing interactions specified by the miRNA, and either represses translation or induces mRNA degradation. AGO2, the only catalytically active AGO family member in humans and mice, has endonuclease (“slicer”) activity and the ability to cleave target mRNAs (Kim et al. 2009).
Exposure to drugs of abuse such as cocaine triggers persistent cellular and molecular changes in the limbic regions of the brain that process reward. The limbic system, in particular the striatum, is considered the epicenter of the long-term molecular and morphological changes that underlie addiction (Nestler 2001). The ventral striatum (nucleus accumbens) is considered the primary striatal subregion with limbic functions. Neurons can modulate gene expression with subcellular precision; polyribosomes and mRNAs are found at the base of dendritic spines and are involved in localized protein synthesis (Rao and Steward 1991; Kiebler and DesGroseillers 2000). Both AGO2 and DICER, an RNase III enzyme required for miRNA biogenesis, have been localized to dendritic spines (Lugli et al. 2005). Emerging evidence supports an autonomous role for individual synapses in the local control of protein translation coupled to neuronal excitation (Schratt 2009; Zukin et al. 2009).
Data from broad expression studies using microarray technology and from focused single-gene approaches indicate that cocaine exposure elicits widespread changes in the transcriptional landscape (Kauer and Malenka 2007). Several recent studies have begun to investigate the role of AGO2 and miRNAs in the biochemical, molecular, and behavioral response to cocaine (Chandrasekar and Dreyer 2009; Hollander et al. 2010; Im et al. 2010; Schaefer et al. 2010). For example, qPCR and in situ analysis of specific bioinformatically determined miRNAs identified let-7d, miR-124, and miR-181a as cocaine regulated in rats (Chandrasekar and Dreyer 2009). Two studies by the Kenny group identified induction of miR-132 and miR-212 in dorsal striatum after 7 d of cocaine self-administration in rats and implicate miR-212 in the behavioral and motivational response to cocaine through CREB, MeCP2, and brain-derived neurotrophic factor (BDNF) signaling (Hollander et al. 2010; Im et al. 2010). In addition, Schaefer et al. identified an overlapping subset of cocaine-induced and AGO2-knockdown-depleted miRNAs in D2 dopamine receptor (Drd2) expressing neurons of the NAc, and showed a reduction in cocaine self-administration when AGO2 is depleted from Drd2 neurons (Schaefer et al. 2010). Consistent with these findings, we observed cocaine-induced increases in Ago2 mRNA levels upon performing RNA-Seq (JE Eipper-Mains, DD Kiraly, MO Duff, MJ Horowitz, G May, CJ McManus, RE Mains, BA Eipper, and BR Graveley, in prep.). Together, these studies provide evidence that miRNA-mediated gene regulation plays an important role in cocaine-related changes in neurotransmission and behavior. However, all of these studies were targeted to a small number of candidate miRNAs or used microarrays, which suffer from issues of cross-hybridization, higher background signal, lower dynamic range, and lack the ability to identify unknown miRNAs (Metzker 2010), and therefore do not provide a complete picture of cocaine-induced changes in miRNA levels.
Here, we have more closely examined the role of AGO2 and miRNAs in response to chronic cocaine exposure. First, we confirmed synaptic Ago2 mRNA and protein expression in the striatum and increased expression upon chronic cocaine exposure. We also determined the repertoire of miRNAs expressed in total nucleus accumbens (NAc) tissue lysates and purified striatal post-synaptic densities (PSDs) harvested from saline- and cocaine-treated mice. This led to the identification of a subset of cocaine-regulated miRNAs in whole tissue and at the synapse, and the characterization of miRNAs by degrees of synaptic enrichment and depletion. We also identified mRNAs that are potentially targeted by the differentially expressed miRNAs, and validated cocaine-induced changes in protein levels for several of the identified targets. These data provide an overview of the global regulation and synaptic enrichment of miRNAs in response to cocaine.
RESULTS
Cocaine regulates striatal AGO2
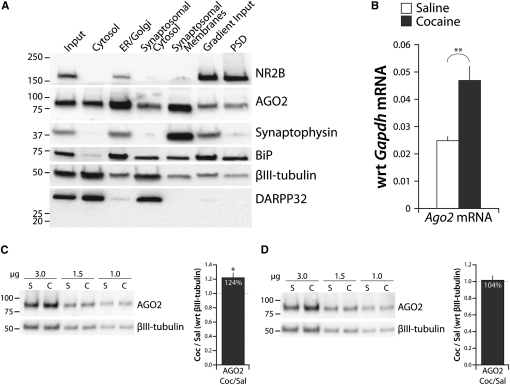
To assess whether AGO2 protein was present at striatal synapses, we subjected adult mouse striatum to subcellular fractionation (Fig. 1A). Strong enrichment of NR2B, an NMDA receptor subunit, in the PSD fraction, and depletion of Synaptophysin, a presynaptic protein, verified the success of the preparation (Petralia et al. 1994; Kornau et al. 1995; Ma et al. 2008). The ER lumenal protein BiP showed strong enrichment in the ER and Golgi fraction. βIII-tubulin was most abundant in the cytoplasmic fractions. Likewise, the striatally enriched cytoplasmic protein DARPP32 (Walaas and Greengard 1984) was highly enriched in soluble fractions and was absent from synaptic fractions. As expected, AGO2 was enriched in the ER/Golgi fraction and present in the cytoplasmic fractions. Consistent with prior studies demonstrating synaptic localization of AGO2 in hippocampus and frontal cortex (Lugli et al. 2005, 2008), our data indicate that AGO2 was present in striatal PSDs (Fig. 1A).
FIGURE 1.
Subcellular fractionation. (A) Wild-type mouse striata were subjected to subcellular fractionation. Aliquots containing equal amounts of protein (10 μg; 5 μg, PSD) were fractionated by SDS-PAGE, transferred, and analyzed by Western blot. NR2B, AGO2, and Synaptophysin were visualized from one gel; BiP, βIII-tubulin, and DARPP32 were visualized from an identical gel. PSD, post-synaptic density. (B) Total NAc RNA from adult male mice treated with saline or cocaine (four per group) was analyzed by qPCR; Ago2 mRNA expression was normalized to Gapdh. (C,D) PSDs purified from the striata (C) and prefrontal cortices (PFCs) (D) of saline (S)- and cocaine (C)-treated mice were fractionated by SDS-PAGE. AGO2 levels were normalized to βIII-tubulin by densitometry for the varying amounts of protein loaded; plot shows average C/S ratio for AGO2. Significance was tested by the two-tailed Student's t-test with unequal (B) or equal (C,D) variance. *P < 0.05; **P < 0.01.
We next used quantitative-PCR to examine whether chronic cocaine administration affected Ago2 mRNA expression in the ventral striatum and found a significant increase in the level of Ago2 mRNA following cocaine exposure (Fig. 1B). We then determined the impact of chronic cocaine on AGO2 protein levels at the PSD. Striata (dorsal and ventral) and medial prefrontal cortices (mPFCs) from 12 cocaine-treated and 12 saline-treated wild-type male C57BL/6 mice were pooled and subjected to subcellular fractionation. Levels of AGO2 and βIII-tubulin in PSDs purified from striata and mPFCs were assessed by quantitative Western blotting. We observed a 24% increase in synaptic AGO2 protein after cocaine treatment in the striatum, but not the mPFC (Fig. 1C,D). Together these results indicate that AGO2 is localized to the striatal PSD and increases after cocaine treatment. Because up-regulation of AGO2 was specific to the striatum, we chose to focus our subsequent analyses on this brain region.
The nucleus accumbens and purified striatal PSDs share abundant microRNAs
Given the synaptic localization and cocaine responsiveness of AGO2 protein, we hypothesized a role for AGO2 and miRNAs in regulating mRNA stability or translation at the striatal PSD in response to cocaine. To explore this idea, we used next-generation sequencing to analyze the global miRNA expression profiles in both NAc total lysates and purified striatal PSDs. For the NAc lysates, we sequenced 2.53 and 4.46 million reads from the saline and cocaine samples, respectively (Supplemental Table S1). We also generated 3.67 and 4.60 million reads from the striatal PSD saline and cocaine samples, respectively (Supplemental Table S1). After removing the 3′ linker sequence, reads ranging from 18 to 30 nt were aligned to the miRBase database (version 16) (Griffiths-Jones et al. 2006, 2008) using Bowtie (version 0.12.7) (Langmead et al. 2009) to identify and assess the abundance of known miRNAs.
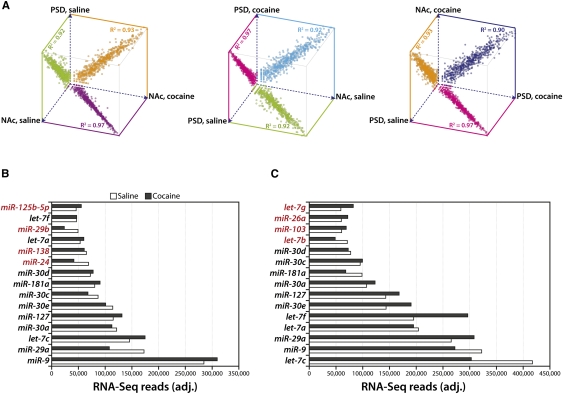
We generated pairwise scatter plots to compare expression of each mature miRNA (as a percentage of total reads) across all four samples (Fig. 2A). The two NAc total lysate samples correlated more closely with each other (R2 = 0.97) than with either striatal PSD sample (R2 = 0.90–0.93). The converse was also true, with striatal PSD miRNAs exhibiting a higher correlation to one another (R2 = 0.97) than to the NAc samples. For both the total tissue lysates and the PSDs, a handful of individual miRNAs appeared to be cocaine-regulated; cocaine treatment did not cause widespread alterations in the global miRNA profile.
FIGURE 2.
High-throughput sequence analysis of NAc and PSD miRNA. (A) Pairwise scatter plots of miRNA expression data for NAc lysate and striatal PSD libraries (saline and cocaine) prepared from an adult mouse; shown as log10(normalized miRNA frequency) per sample. R2 values were calculated by least squares best fit. Expression data for the 15 most abundant miRNAs in NAc lysates (B) and striatal PSDs (C) are shown as reads (adjusted for equal total reads per sample). The miRNAs in black italics are highly expressed in both NAc and PSD; miRNAs in red italics are highly expressed only in that sample.
While the total tissue lysate and purified PSD expression profiles differed, the most highly expressed miRNAs were largely identical; 11 of the 15 most abundantly sequenced miRNAs in both groups were the same (Fig. 2B,C). Of note, a number of miRNAs known to be brain-specific or neurally enriched, such as miR-9, miR-30, miR-127, miR-138, miR-181, and let-7 (Lagos-Quintana et al. 2002; Krichevsky et al. 2003; Miska et al. 2004; Landgraf et al. 2007), were very abundant in these samples.
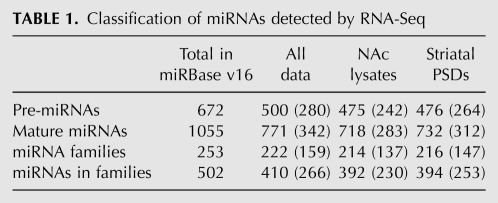
We detected one or more reads mapping to a total of 500 (74%) of the 672 mouse pre-miRNAs annotated in miRBase, of which 475 and 476 miRNAs were detected in the NAc and striatal PSD libraries, respectively (Table 1). We set a lower limit of 100 reads per miRNA for further analysis as we found that reliable qPCR validation of changes necessitated this level of expression. Although this read requirement decreased the diversity of miRNAs considered for further analysis, the miRNAs meeting the expression cutoff account for 99.7% and 99.8% of all reads in the NAc and striatal PSD libraries, respectively. A summary of the individual miRNAs and miRNA families identified in these experiments is presented in Table 1.
TABLE 1.
Classification of miRNAs detected by RNA-Seq
Cocaine modulates expression of ventral striatum microRNAs
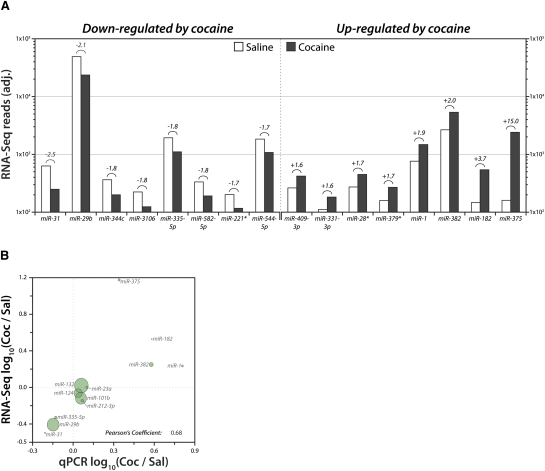
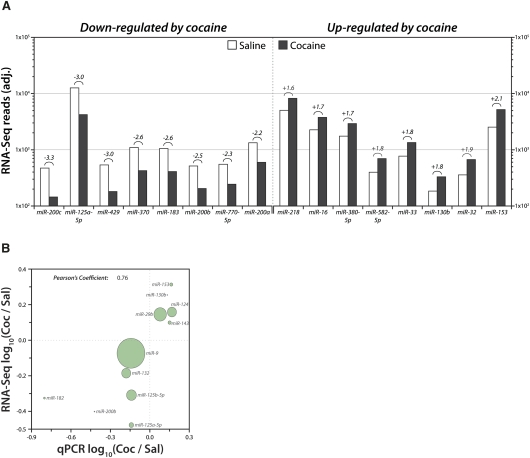
There were 283 mature miRNAs detected with at least 100 reads in both NAc lysate libraries (Table 1); 10% of these miRNAs exhibited at least a 1.5-fold change in expression, with 16 miRNAs up-regulated and 13 down-regulated after cocaine treatment. The NAc total lysate miRNAs that exhibit the largest changes in expression following cocaine are shown in Figure 3A. The regulated miRNAs were generally of low-to-moderate abundance, detected at a level between 100 and 1000 reads. One notable exception, miR-29b, was among the 15 most abundantly expressed miRNAs in the NAc (Fig. 2B).
FIGURE 3.
Cocaine-regulated miRNAs in NAc total lysates. (A) NAc lysate libraries (saline and cocaine) analyzed by high-throughput sequencing of miRNAs. The eight most cocaine down- and up-regulated miRNAs detected at >100 reads are shown as adjusted reads (equal total reads per sample) on log10 scale. The number above the bars indicates fold change. (B) Pairwise scatter plot comparing log10 ratio of Coc/Sal expression for selected miRNAs in NAc tissue lysate as computed from RNA-Seq data (vertical axis) and qPCR data (horizontal axis). Size of circle is proportional to average RNA-Seq expression value. Pearson's Coefficient represents the linear correlation coefficient, r.
We used qPCR to validate cocaine-induced changes in several of the most highly regulated miRNAs and to validate RNA-Seq data for miRNAs known to be brain-enriched or synaptically relevant (Supplemental Fig. S3). Commercially available probes were used for qPCR; these probes measure only one mature miRNA product, which did not always correspond to the most abundantly sequenced product due to 3′-end heterogeneity (Supplemental Fig. S4). Nevertheless, strong agreement across the two methods (r = 0.68) was observed when the cocaine/saline ratios obtained by qPCR and RNA-Seq were plotted (Fig. 3B). Importantly, interplatform agreement was consistent over a broad range of overall expression levels.
Cocaine regulates microRNAs in striatal PSDs
A specific subset of mRNAs is transported along dendrites and local protein synthesis occurs in stimulated dendritic spines, altering the components of the synapse (Rao and Steward 1991; Kiebler and DesGroseillers 2000). As AGO2 protein is localized to the PSD and its levels at the PSD increased following chronic cocaine exposure, we determined whether any PSD localized miRNAs were responsive to cocaine. There were 312 mature miRNAs detected at 100 reads or more in both striatal PSD libraries (Table 1); 11% of these miRNAs exhibited at least a 1.67-fold change in expression, with four times as many miRNAs down-regulated (28) as up-regulated (7) after cocaine treatment. Figure 4A shows those striatal PSD miRNAs with at least 100 reads that demonstrated the largest change in expression following cocaine. Similar to what was observed for the NAc miRNAs, the cocaine-regulated PSD miRNAs were generally of low-to-medium abundance, detected with 100 to 10,000 reads.
FIGURE 4.
Cocaine-regulated miRNAs in striatal PSDs. (A) Striatal PSD libraries (saline and cocaine) analyzed by high-throughput sequencing of miRNAs. The eight most cocaine down- and up-regulated miRNAs detected at >100 reads are shown as adjusted reads as described in Fig. 3. (B) Pairwise scatter plot comparing log10 ratio of Coc/Sal expression for selected miRNAs in striatal PSDs as computed from RNA-Seq data (vertical axis) and qPCR data (horizontal axis) as described in Fig. 3.
We again turned to qPCR to confirm responses observed with RNA-Seq. A comparison of the ratios of cocaine/saline measured by RNA-Seq and qPCR showed a high degree of concordance between the two techniques (r = 0.76) (Fig. 4B). Cross-platform agreement spanned a very broad range of expression levels, as seen in NAc lysates.
Synaptically enriched microRNA families
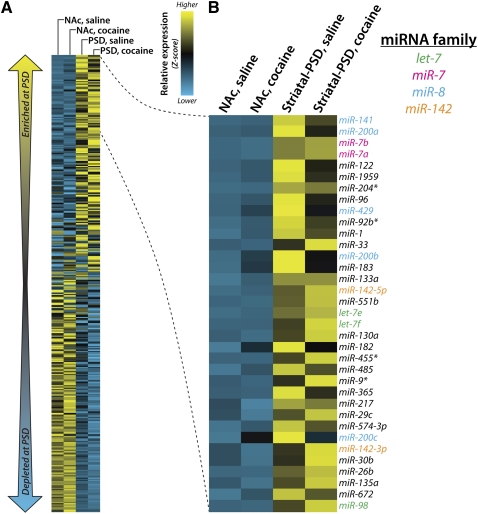
To determine the subset of miRNAs that are enriched or depleted at the synapse, we calculated the relative abundance of each miRNA in all four samples. We then computed the average abundance of each miRNA in the two NAc lysate and striatal PSD samples, and calculated the log10 ratio of the PSD average over the NAc average. The miRNAs with higher PSD-to-NAc lysate ratios are synaptically enriched (Fig. 5), whereas those with lower ratios are synaptically depleted (Supplemental Fig. S5). Z-scores of relative expression were calculated for each sample and the data are displayed as heat maps (Fig. 5A,B).
FIGURE 5.
Identification of miRNA families enriched at the PSD. (A) Normalized miRNA frequency data for saline- and cocaine-treated NAc lysate and striatal PSD libraries of wild-type mice were calculated; heat map is sorted by decreasing average PSD frequency/NAc frequency ratio. Z-score was computed on normalized miRNA frequency across all samples. Blue indicates low expression and yellow indicates high expression. (B) Expansion of heat map showing top PSD-enriched miRNAs. Text color indicates miRNA families with two or more family members appearing in the list.
Of the 672 annotated murine miRNAs, 75% (502) can be grouped into 253 families based on sequence homology in the “seed region” (nucleotides 2–8 at the 5′-end of the miRNAs) and the observation that these miRNAs are often functionally redundant (Kim et al. 2009). We detected expression of at least one member of 222 (88%) of the 253 annotated miRNA families, with 159 families expressed at 100 reads or more (Table 1). Strikingly, several of the synaptically enriched miRNAs we identified were members of the same family, as indicated by text color in Figure 5B. All five members of the miR-8 family (miR-141, miR-200a, miR-200b, miR-200c, and miR-429) were synaptically enriched. The same was true for the miR-7 and miR-142 families. The mouse genome contains 12 loci encoding members of the let-7 family, three of which were synaptically enriched (Fig. 5B). In contrast, we did not observe family enrichment in the subset of PSD-depleted miRNAs (Supplemental Fig. S5B).
PSD-localized, cocaine-regulated microRNAs target a functionally related subset of mRNAs
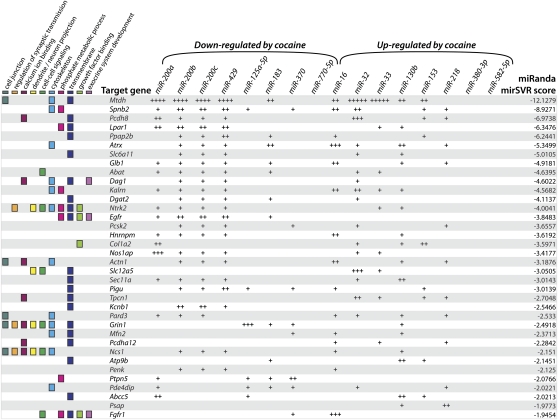
To provide insight into the role of the cocaine-regulated miRNAs, we performed a computational search for potential target mRNAs. First, we compiled a list of 1041 dendritically and synaptically localized mRNAs reported in the literature (Tian et al. 1999; Eberwine et al. 2002; Moccia et al. 2003; Sung et al. 2004; Poon et al. 2006; Suzuki et al. 2007; Supplemental Table S2). Of those mRNAs, 424 were PSD enriched in rat forebrain by microarray (Suzuki et al. 2007), and this subset of synaptically localized genes (Supplemental Table S2) was cross-referenced with the putative miRNA target genes identified by analysis using the miRanda microRNA target prediction algorithm (Betel et al. 2008, 2010). Briefly, each predicted miRNA binding site in the 3′ UTR of a given gene has a precomputed mirSVR score; this value directly correlates with the extent of mRNA down-regulation predicted by miRNA binding, and the composite score for each gene in Figure 6 represents the sum of all cataloged mirSVR scores across the queried miRNAs (Betel et al. 2010).
FIGURE 6.
Target prediction for cocaine-regulated miRNAs. The 16 most cocaine-regulated striatal PSD miRNAs from Fig. 4 were subjected to bioinformatic target prediction using miRanda. The list of potential target mRNAs was cross-referenced with the 424 PSD-enriched mRNAs identified by Suzuki et al. (2007) (Supplemental Table S2) and sorted by ascending mirSVR score. + symbols show presence of predicted miRNA binding site in 3′ UTR of target gene; number of + signs denotes number of predicted miRNA binding sites. The 36 target genes shown were run through gene ontology analysis using DAVID; colored boxes indicate enriched gene ontological functions with P < 0.05.
We performed target prediction on the 16 PSD-localized cocaine-regulated miRNAs depicted in Figure 4A. A subset of the list of synaptically localized predicted target genes for the cocaine-regulated striatal-PSD miRNAs sorted by mirSVR score is shown in Figure 6. A number of the identified target genes were predicted to have multiple binding sites for a given miRNA in their 3′ UTR (depicted by the number of + symbols) and the majority of the predicted target genes had putative binding sites for both up- and down-regulated miRNAs.
We carried out gene ontological analysis using DAVID (Dennis et al. 2003; Huang et al. 2009a,b) to investigate the functional properties of the identified target genes. The colored boxes next to the genes in Figure 6 show the enriched gene ontological categories (P < 0.05). Most of the enriched categories (cell junction, regulation of synaptic transmission, calcium ion binding, dendrite/neuron projection, cell–cell signaling, cytoskeleton, phosphate metabolic process, transmembrane, growth factor binding) are processes generally linked to synaptic plasticity, learning, and memory.
Identified target genes exhibit differential expression at striatal synapses after cocaine
Schematic drawings for Ntrk2, E-cadherin, Mtdh, and Pcsk2, potential target genes of cocaine-regulated synaptic miRNAs (Fig. 7; Supplemental Fig. S6), depict the sites within their annotated 3′ UTRs that are predicted to bind up- and down-regulated miRNAs. As these synaptically enriched transcripts may interact with multiple regulated miRNAs, the direction of cocaine-mediated changes in synaptic protein levels are difficult to predict.
FIGURE 7.
Validation of target predictions. PSDs purified from the striata of saline (S)- and cocaine (C)-treated mice were fractionated by SDS-PAGE; three different amounts of protein were analyzed to establish response linearity. Levels for pan-TRK (A); ECAD (➄) and Metadherin (MTDH) (B); and PC2 (➄), proPC2 (➄), and preproPC2 (←) (C) were normalized to βIII-tubulin by densitometry for the varying amounts of protein loaded; plot shows average C/S ratio. Based on published reports, the higher molecular weight band (∼120 kDa) above the major 105 kDa ECAD band corresponds to the precursor, proECAD (➄) (Miyashita and Ozawa 2007; van Roy and Berx 2008). For PC2, the smaller molecular weight band represents mature PC2 (➄), the middle band represents proPC2 (➄), and the upper band is the size of preproPC2 (←) (Bloomquist et al. 1991). Schematic drawings represent mRNAs of indicated genes; open reading frame (ORF) is gray, 3′ UTR is striped, arrowheads show miRanda predicted miRNA binding sites for miRNAs up-regulated (▴) or down-regulated (▾) after cocaine at PSD. Significance was tested by the two-tailed Student's t-test with equal variance. *P < 0.05; **P < 0.01. Error bars represent the standard deviation (SD).
We used quantitative Western blotting to investigate the effect of cocaine administration on the protein level of several potential target genes of cocaine-regulated synaptic miRNAs (Fig. 7). One identified target gene is Ntrk2 (also known as TrkB), a member of the neurotrophin receptor tyrosine kinase family, which binds BDNF and is critically involved in the process of activity-dependent synaptic plasticity and long-term potentiation (Minichiello 2009). The Ntrk2 3′ UTR is targeted by four miR-8 family members (miR-200a, miR-200b, miR-200c, and miR-429), which are down-regulated by cocaine, and by miR-33 and miR-130b, which are up-regulated by cocaine (Fig. 6; Supplemental Fig. S6). The level of synaptic TRK protein increased 18% in the striatum after chronic cocaine administration (Fig. 7A).
Similarly, synaptic expression of Metadherin (MTDH), encoded by Mtdh, the top synaptically regulated miRNA target gene (Fig. 6), increased to 142% of the saline level after chronic cocaine treatment (Fig. 7B). Mtdh is targeted by miR-183 and four miR-8 family members, which are down-regulated by cocaine, and by miR-16, miR-32, miR-33, miR-130b, and miR-153, which are up-regulated by cocaine (Fig. 6; Supplemental Fig. S6). Although not a direct target of the identified cocaine-regulated synaptically localized miRNAs, E-cadherin (also known as Cdh1 or ECAD) expression is controlled by transcriptional repressors (ZEB1 and ZEB2), which are directly targeted by members of the miR-8 family of miRNAs (Inui et al. 2010), and by several other miRNAs, which are down-regulated by cocaine and by a number of miRNAs, which are up-regulated by cocaine (Fig. 7B; Supplemental Fig. S6). Two forms of ECAD were detected at the synapse by Western blot (Fig. 7B). The 105 kDa major band is mature ECAD (black arrow) and the higher molecular weight band represents the precursor, proECAD (Miyashita and Ozawa 2007; van Roy and Berx 2008). The level of mature ECAD decreased to 68% of the saline level following chronic cocaine.
The prohormone convertase PC2 (also known as Pcsk2) was also identified in our bioinformatic analysis as a potential target of the cocaine-regulated miRNAs. Pcsk2 is targeted by three miR-8 family members and miR-370, which are down-regulated by cocaine, and by miR-32 and miR-218, which are up-regulated by cocaine (Fig. 6; Supplemental Fig. S6). Several forms of PC2 were detected at striatal PSDs by Western blot (Fig. 7C). The smaller molecular weight band represents mature PC2, the middle band represents proPC2, and the upper band is the size of preproPC2 (Bloomquist et al. 1991), indicative of dendritic synthesis and processing. When normalized to βIII-tubulin, mature PC2, and proPC2/preproPC2 increased significantly, 67% and 61%, respectively, after cocaine treatment.
DISCUSSION
The role of miRNAs in response to drugs of abuse is largely unknown. Here, we used deep sequencing to identify miRNAs that are differentially regulated in response to cocaine exposure, both in purified NAc tissue and at striatal synapses. This catalog of synaptically enriched and cocaine-regulated miRNAs provides new insight into the molecular mechanisms of the cocaine-response and serves as a resource for future studies.
Intriguingly, many (18) of the synaptically enriched miRNAs we identified are encoded in miRNA clusters—precursor transcripts that contain multiple miRNAs—or are members of the same miRNA families. Moreover, the expression patterns of the clustered miRNAs are similar to one another. Particularly strong examples include the two clusters encoding miR-8 family members (miR-429/200a/b and miR-141/200c), the cluster encoding let-7 family members (let-7f/miR-98), and clusters miR-1/133a, miR-182/96/183, and miR-216a/217 (Supplemental Fig. S7). This indicates that many of the miRNAs we identified are coordinately regulated by virtue of being encoded in clusters and potentially control the same set of mRNA targets as they share the same seed sequences.
It is well established that the growth and remodeling that occur in dendritic spines following synaptic activity require de novo protein synthesis, and that blocking protein synthesis attenuates formation of long-term memory (Schratt 2009). The ability to modulate the synthesis of a subset of synaptic proteins affords neurons the capacity to integrate and fine-tune extracellular cues with subcellular resolution, in individual dendrites or at specific synapses (Wang et al. 2009). MicroRNAs represent ideal candidates for regulating local translation of specific mRNAs at the synapse. It is therefore intriguing that we observed both increased levels of synaptically localized AGO2 protein and altered levels of specific miRNAs upon cocaine exposure. These observations indicate that modulations in miRNA-mediated gene regulation play an important role in the response to cocaine. Programmed changes in miRNA levels may result in specific changes in the stability and/or the translation rate of target mRNAs at the synapse.
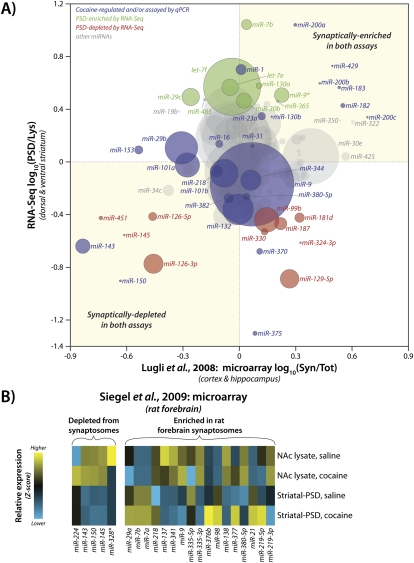
We identified several miRNAs that were synaptically enriched or depleted (Fig. 5B; Supplemental Fig. S5B). A comparison of our data to earlier microarray experiments performed in adult (2 mo old) mouse cortex and hippocampus (Fig. 8A; Lugli et al. 2008) revealed significant overlap. Notably, four of the five miR-8 family members (miR-200a/b/c/429) are highly enriched in both data sets. Additionally, miR-182 and miR-183 are highly PSD enriched in both data sets, whereas miR-126, miR-143, miR-145, miR-150, and miR-451 are synaptically depleted in both data sets (Fig. 8A). We also observed strong agreement between our data and a second microarray study that identified synaptosomally enriched and depleted miRNAs from rat P15 total forebrain samples (Siegel et al. 2009). In both data sets, miR-219-5p, miR-21, miR-377, miR-98, miR-376b, miR-218, miR-7a/b, and miR-29a are synaptosomally or PSD enriched, whereas miR-143, miR-145, and miR-150 are depleted (Fig. 8B). The high degree of concordance between these three data sets, which used different experimental platforms and strikingly different samples, may reflect the prevalence of glutamatergic synapses and other features common to these brain regions.
FIGURE 8.
Comparison of data for striatum and cortex/hippocampus. (A) Pairwise scatter plot comparing miRNA synaptic enrichment ratios from Lugli et al. 2008 (adult mouse cortex and hippocampus; measured by microarray) with RNA-Seq data (adult mouse striatum) from these experiments (Lugli et al. 2008). Horizontal axis, data from Lugli et al. 2008 computed as ratio of PSD (Syn)/total tissue (Tot) expression; vertical axis, data from RNA-Seq computed as PSD/NAc lysate ratio of average of saline and cocaine expression levels for adult mouse striatal PSD and NAc lysate. Blue circles were identified as cocaine-regulated and/or were assayed by qPCR, green circles indicate miRNAs enriched at striatal PSDs by RNA-Seq, red circles indicate miRNAs depleted at striatal PSDs by RNA-Seq, and gray circles represent all other miRNAs. Size of circle is proportional to average RNA-Seq expression value. (B) Synaptosomal enrichment and depletion categories for miRNAs from Siegel et al. 2009 as determined by microarray analysis of rat forebrain samples (Siegel et al. 2009). Heat map shows high-throughput sequencing data for saline- and cocaine-treated NAc lysates and striatal PSDs of wild-type mice; normalized miRNA frequency was calculated and Z-score was computed on normalized miRNA frequency across all samples. Blue indicates low expression and yellow indicates high expression.
The increased levels of AGO2 and altered levels of specific miRNAs at the synapse in response to cocaine strongly suggest that these factors play a role in controlling translation of localized mRNAs. To explore this, we identified potential mRNA targets from a set of PSD-enriched mRNAs (Fig. 6; Suzuki et al. 2007). Due to the fact that most mRNAs we identified are predicted to be targeted by many of the regulated miRNAs, it was difficult to predict whether translation of the targets would increase or decrease in response to cocaine. Nonetheless, we confirmed changes in protein levels for three predicted targets (TRKB, MTDH, and PC2) and one indirect target (ECAD) upon exposure to cocaine. One of the validated targets, TRKB (NTRK2), has also been shown to increase in the NAc of rats after chronic cocaine self-administration (Graham et al. 2009), and TRKB-BDNF signaling in the NAc may modulate behavioral responses to cocaine (Lobo et al. 2010). Importantly, several other predicted targets have been implicated in various aspects of the cocaine response (Przewłocka and Lasoń 1995; Hemby et al. 2005; Hubert et al. 2008).
Recent studies have demonstrated the importance of miRNAs in the pathophysiology of several neuropsychiatric disorders and mental retardation syndromes (Conrad et al. 2010; Sulzer 2011), including schizophrenia (Burmistrova et al. 2007; Beveridge et al. 2010; Moreau et al. 2011; Santarelli et al. 2011), bipolar disorder, Fragile X mental retardation (Edbauer et al. 2010; Krol et al. 2010; Wulff et al. 2011), Rett syndrome (Wu et al. 2010), Alzheimer's disease, Parkinson's disease (Junn et al. 2009), and Huntington's disease (Martí et al. 2010). A number of the cocaine-responsive miRNAs identified in this study are important in controlling dendritic morphology; these miRNAs, and many others, may contribute to long-lasting forms of drug-mediated synaptic plasticity through control of regulatory pathways that modulate changes to the actin cytoskeleton, neurotransmitter metabolism, and peptide hormone processing. It is likely that the miRNAs we identified are but one aspect of an exquisitely complex regulatory system that includes dendritic mRNA trafficking and excitation-coupled modulation of translation and protein phosphorylation to fine-tune translation of synaptically localized miRNAs. As the breadth of information about the molecular changes that occur in the brain in response to cocaine and other drugs of abuse increases, the importance of integrating these data in a meaningful and biologically informative way becomes increasingly clear.
MATERIALS AND METHODS
Cocaine treatment of mice
Adult wild-type C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME; 2–5 mo old) were used for these experiments. All mice were allowed to acclimate to the colony for at least 1 wk before handling or injections. Mice were kept in the University of Connecticut Health Center animal facility on a 12-h light/dark cycle (lights on from 7:00 am to 7:00 pm) and handled in accordance with University of Connecticut Health Center Institutional Animal Care and Use Committee guidelines. To reduce stress from handling during the injections, animals in all groups were handled for 1 min/animal/day for 2–3 d prior to the start of injections. During injections, animals were brought into the behavior room to acclimate to the new environment for 45–60 min each day before the beginning of experiments.
For these studies, we used two slightly different cocaine injection paradigms, both of which have been shown to produce robust locomotor sensitization. For all experiments, animals were given a saline or cocaine injection and had their locomotor activity monitored for the subsequent 45 min. In the experiments in which RNA from NAc lysates was used for miRNA-Seq library preparation, animals were given injections of 20 mg/kg cocaine or saline (10 mL/kg) intraperitoneally for 7 d. Our group and others have demonstrated the efficacy of this method at eliciting robust locomotor sensitization (Hiroi et al. 1999; Kiraly et al. 2010). For all other experiments (both RNA and protein), animals received a lower dose of cocaine (10 mg/kg) on the first and last (seventh) day, with the standard 20 mg/kg cocaine dose on days 2–6 (Kalivas and Duffy 1993; Pierce et al. 1996). The extent of locomotor sensitization was taken as the ratio of activity on day 7/day 1 (Pierce et al. 1996). We chose to switch to this method because it provided a clearer distinction between those animals that had sensitized and those that did not. Protein biochemistry from our laboratory has shown no biochemical differences between sensitized animals from the two paradigms (DD Kiraly, unpubl.). Neither tissue nor data from these different treatment paradigms were pooled. For all experiments, animals were sacrificed 24 h after the final injection.
Whole tissue lysate preparation and subcellular fractionation
Adult male mouse ventral striatum (nucleus accumbens, NAc) punches were harvested. For whole tissue protein studies, samples were sonicated and boiled in 1% SDS, 50 mM TrisHCl [pH 7.4], 5 mM EDTA, 50 mM NaF, 2 mM sodium orthovanadate, 1 mM PMSF, and protease inhibitor cocktail as described (Ma et al. 2008). Total RNA from tissue lysates for individual mice was prepared from freshly dissected tissue with TRIzol (Invitrogen) following the manufacturer's instructions, except that isopropanol precipitation was carried out overnight at −20°C with two ethanol washes instead of one.
For subcellular fractionation, whole striata (40 mg wet weight/mouse) or medial prefrontal cortices (mPFC) (30 mg/mouse) from 12 identically treated male mice were pooled, homogenized, and fractionated by differential centrifugation followed by sucrose gradient centrifugation. After extraction of the PSD-enriched fraction with 0.5% Triton X-100, PSDs were pelleted and subjected to Western blot analysis as described (Ma et al. 2008; Kiraly et al. 2010). The protocol is similar to that of Lugli et al. (2008) and was modified to protect against RNA degradation as follows: All rotors, homogenizers, pestles, and ultracentrifuge tubes were rinsed with RNaseZAP (Applied Biosystems) and DEPC water and air dried prior to use; fractionation buffers were prepared using DEPC water and were treated with RNASecure (1:25 dilution from stock; Applied Biosystems) at 60°C for 10 min; aliquots of different subcellular fractions were diluted into TRIzol for preparation of total RNA as described previously.
Small RNA cloning and sequencing
Small RNA sequencing libraries were prepared following the manufacturer's instructions for the Small RNA Sample Prep Kit (Illumina). Total RNA used for NAc tissue lysate library preparation was pooled in equal parts from RNA isolated from eight male mice from each treatment group. One saline and one cocaine NAc lysate library were prepared from the pooled RNA and these are referred to as NAc lysate saline and cocaine libraries. Total RNA used for striatal post-synaptic density (PSD) library preparation was isolated as outlined earlier from the pooled striata of 11 mice (7 male, 4 female) from each treatment group; these libraries are referred to as striatal PSD saline and cocaine libraries. Small RNAs of 18–35 nt were gel-purified from 1 μg of pooled total RNA. Eluted RNAs were ligated to 5′ and then to 3′ adaptors with RNA ligase, with gel purification following each ligation. RNAs were reverse transcribed and cDNAs were PCR-amplified for 16 cycles. PCR products were purified on a nondenaturing acrylamide gel and sequenced on the Illumina Genome Analyzer IIx for 50 (NAc lysate libraries) or 40 (striatal PSD libraries) cycles.
Small RNA genomic analysis
Sequences were extracted from image files using Firecrest and Bustard. The 3′ adaptor sequences were trimmed from the reads, and reads 18–30 nt in length were aligned to all annotated mouse microRNAs from miRBase (www.mirbase.org; version 16) (Griffiths-Jones et al. 2006, 2008) with Bowtie (bowtie-bio.sourceforge.net; version 0.12.7) (Langmead et al. 2009); only perfectly aligning reads were considered for miRNA expression level analysis. A custom Perl script was used to calculate the number of reads aligning to each specific miRNA gene and data were normalized by the total number of reads obtained in that sequencing run. A cutoff of 100 reads or more for an individual miRNA was required for it to be included in further expression analyses.
To identify reads mapping to a variety of noncoding genes and repetitive elements, the remaining non-miRNA sequences were aligned to the Rfam database (rfam.sanger.ac.uk; version 10.0), the Repbase database (www.girinst.org/repbase/index.html; version 16.01), and NONCODE (noncode.org; version 2.0). We subsequently mapped the remaining reads to the mouse transcriptome, including exons, introns, and exon–exon junctions annotated on the UCSC genome browser (genome.ucsc.edu; mm9; NCBI Build 37) as the following tracks: UCSC Genes, RefSeq Genes, Vega Protein Coding Annotations, and Ensembl Gene Predictions. Finally, the remaining reads were aligned to the mouse genomic sequence (genome.ucsc.edu; mm9; NCBI Build 37). All of these alignments were performed using Bowtie and allowing 1-mismatch. Alignment data are shown in Supplemental Figure S1.
Importantly, the trimmed reads were also aligned to miRBase allowing one or two mismatches. Neither the overall expression pattern nor the list of the most cocaine-responsive miRNAs changed significantly when allowing for more mismatches (JE Eipper-Mains, unpubl.). Read numbers presented in this paper were calculated from the 0-mismatch alignment.
The raw and processed sequence data have been deposited in GEO (GSE29261).
Generation of heat maps
Normalized miRNA frequency across all four sequenced libraries was calculated; expression in one sample of 100 reads or more was required. Z-scores were computed from frequency data using the following equation: (x − μ)/σ, where x is the sample frequency; μ is the mean across all samples; and σ is the standard deviation across all samples. Blue indicates low expression and yellow indicates high expression; heat maps were generated using Java Treeview software (sourceforge.net/projects/jtreeview/files).
Target gene prediction
MicroRNA target genes were identified using miRanda (www.microrna.org; August 2010 release) (Betel et al. 2008, 2010). Prediction scores were computed using the miRanda database of highly conserved target sites with good mirSVR scores (Betel et al. 2010). We chose this database because programs requiring target conservation across a broad range of species are more stringent than those with more relaxed definitions of conservation. For the miRanda-generated target gene list for PSD-localized cocaine-regulated miRNAs, we filtered the list to include only those target genes with supporting experimental data indicating dendritic or synaptic localization; data compiled from six studies (Tian et al. 1999; Eberwine et al. 2002; Moccia et al. 2003; Sung et al. 2004; Poon et al. 2006; Suzuki et al. 2007) yielded a list of 1041 target genes (Supplemental Table S2).
Functional analysis of target genes
We used the Database for Annotation, Visualization, and Integrated Discovery (DAVID; david.abcc.ncifcrf.gov) to interrogate multiple databases in parallel with a list of target genes (Dennis et al. 2003; Huang et al. 2009a,b). We used the GoCharts module to analyze a subset of the target mRNAs identified from miRanda analysis. GoCharts uses the classifications of the Gene Ontology Consortium (GO; www.geneontology.org), which assigns functional terms to annotated genes and proteins. Only those GO terms identified by DAVID with enrichment P < 0.05 were included in Figure 6.
Quantitative polymerase chain reaction (qPCR)
The cDNAs from tissue lysate total RNA were prepared using iScript (BioRad) with random primers; a 5-min 65°C step was added to the iScript protocol before chilling and adding the enzyme. Pairwise statistical comparisons used the t-test for two samples assuming unequal variance and the two-tailed value are reported. Real-time PCR for mRNAs was performed using an Eppendorf Realplex2 machine and Sybr-Green (BioRad), with the following parameters: 95°C 2 min; 95°C 15 sec, 55°C 15 sec, and 68°C 40 sec; repeat 40X. Maximal rates of amplification per cycle were calculated for all primer pairs and all samples in all assays and averaged 2.03 ± 0.06 (SD) in six runs of 96-well plates. Data are calculated with respect to GAPDH for each sample within an assay, and data across assays were then averaged because the values with respect to GAPDH for each transcript were very consistent. Primer pairs were chosen to keep the melt temperature (Tm) between 60°C and 62°C (calculated using www.basic.northwestern.edu/biotools/oligocalc.html) and products in the 120 ± 5-nt size range (Table 2). Differences between saline and cocaine samples were evaluated using the two-tailed Student's t-test with unequal variances.
TABLE 2.
qPCR primers
MicroRNA qPCR
Reverse transcription using mature miRNA-specific primers was performed on 1–10 ng total RNA from tissue lysates per the manufacturer's instructions for the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) using the suggested parameters (ice 5 min, 16°C 30 min, 42°C 30 min, and 85°C 5 min). Quantification was performed using the TaqMan MicroRNA Assay (Applied Biosystems) with the manufacturer's recommended amplification parameters (95°C 10 min, 95°C 15 sec, and 60°C 60 sec; repeat 40×). NAc lysate qPCR was performed on RNA isolated from individual animals, whereas striatal-PSD qPCR was carried out on the pooled preparations. Data are calculated with respect to an empirically determined endogenous control miRNA; total NAc lysate and striatal-PSD miRNA levels were calculated with respect to miR-9 and miR-101a, respectively.
Western blot analysis
Gels (4%–15% Mini-PROTEAN TGX precast gels, BioRad) of subcellular fractions were loaded with 1–10 μg protein, which was measured using the Bicinchoninic Acid Assay with bovine serum albumin as the standard (Pierce). Commercially available mouse monoclonal antibodies were used: βIII-tubulin (TUJ1; Covance; 1:1000), NR2B (clone N59/20; NeuroMab; 1:1000), Ago2 (2E12-1C9; Novus Biologicals; 1:1000), and Synaptophysin (SVP-38; Sigma; 1:10,000). Commercially available rabbit polyclonal antibodies were also used: BiP/GRP78 (Affinity BioReagents; 1:500), DARPP32 (2303; Cell Signaling Technology; 1:1000), pan-Trk (MCTrks, SC-414; Santa Cruz Biotechnology; 1:250), E-cadherin (ab53033; Abcam; 1:1000), and Metadherin (Ab2989; Millipore; 1:500). Rabbit polyclonal antibody JH1159 (1:250) (Zhou and Mains 1994) to the C-terminal 13 residues (amino acids 626-638) of rat PC2 was affinity-purified using the antigenic peptide linked to AffiGel 15 beads (Bio-Rad Laboratories).
For quantitative analysis of purified PSDs from saline- and cocaine-treated mice, three different amounts of sample were analyzed to verify the linearity of the response. After transfer to polyvinylidene difluoride membranes (Millipore), blots were stained with Coomassie Brilliant Blue R-250 and cut to separate the proteins of interest. Blots were incubated in primary antibody overnight at 4°C, washed, and incubated for 1 h at room temperature in the appropriate horseradish peroxidase-conjugated secondary antibody (Pierce). Washed blots were incubated in Super Signal West Pico chemiluminescence substrate (Pierce) for 5 min and multiple exposures were collected using a Syngene imaging system. Signals were quantified using GeneTools software; for each dilution of each PSD sample, the signal for each protein was normalized to βIII-tubulin and the cocaine-to-saline ratio was determined.
Statistical analyses
Data across qPCR assays were averaged because the values with respect to the endogenous control for each miRNA were very consistent. Outliers were determined with the GraphPad Grubbs' test outlier calculator (graphpad.com/quickcalcs/Grubbs1.cfm). Differences between saline and cocaine samples were evaluated using the two-tailed Student's t-test with unequal variances (NAc lysates) or equal variance (striatal-PSDs). For PSD Western blot quantification, cocaine-to-saline ratios for the three amounts of each sample were averaged and differences were evaluated using the two-tailed Student's t-test with equal variance. Pearson's coefficient represents the linear correlation coefficient between two samples, X and Y. Pearson's r = covariance(X,Y)/σXσy, where σ is the standard deviation.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Iris Lindberg (University of Maryland School of Medicine) for supplying PC2 peptide for antibody affinity purification and Dr. Victor May and Dr. Giordano Lippi for their comments on the manuscript. This work was supported by NIH grant nos. DA-15464, DA-23082, NS-41224 (to R.E.M. and B.A.E.), DA-26706 (to D.D.K.), and GM062516 (to B.R.G.).
Authors’ contributions: J.E.E.-M., B.R.G., B.A.E., R.E.M., and D.D.K. designed the experiments; D.P. prepared the sequencing libraries; D.D.K., R.E.M., and B.A.E. carried out the experiments; and J.E.E.-M. performed the bioinformatic analyses and generated the figures. The manuscript was written by J.E.E.-M. with input from all authors.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2775511.
REFERENCES
- Betel D, Wilson M, Gabow A, Marks DS, Sander C 2008. The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90 doi: 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ 2010. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry 15: 1176–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist BT, Eipper BA, Mains RE 1991. Prohormone-converting enzymes: regulation and evaluation of function using antisense RNA. Mol Endocrinol 5: 2014–2024 [DOI] [PubMed] [Google Scholar]
- Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI 2007. MicroRNA in schizophrenia: genetic and expression analysis of miR-130b (22q11). Biochemistry (Mosc) 72: 578–582 [DOI] [PubMed] [Google Scholar]
- Chandrasekar V, Dreyer J-L 2009. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci 42: 350–362 [DOI] [PubMed] [Google Scholar]
- Conrad KL, Ford K, Marinelli M, Wolf ME 2010. Dopamine receptor expression and distribution dynamically change in the rat nucleus accumbens after withdrawal from cocaine self-administration. Neuroscience 169: 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3 doi: 10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- Eberwine J, Belt B, Kacharmina JE, Miyashiro K 2002. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res 27: 1065–1077 [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang C-F, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M 2010. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DL, Krishnan V, Larson EB, Graham A, Edwards S, Bachtell RK, Simmons D, Gent LM, Berton O, Bolanos CA, et al. 2009. Tropomyosin-related kinase B in the mesolimbic dopamine system: region-specific effects on cocaine reward. Biol Psychiatry 65: 696–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemby SE, Tang W, Muly EC, Kuhar MJ, Howell L, Mash DC 2005. Cocaine-induced alterations in nucleus accumbens ionotropic glutamate receptor subunits in human and non-human primates. J Neurochem 95: 1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Fienberg AA, Haile CN, Alburges M, Hanson GR, Greengard P, Nestler EJ 1999. Neuronal and behavioural abnormalities in striatal function in DARPP-32-mutant mice. Eur J Neurosci 11: 1114–1118 [DOI] [PubMed] [Google Scholar]
- Höck J, Meister G 2008. The Argonaute protein family. Genome Biol 9: 210 doi: 10.1186/gb-2008-9-2-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im H-I, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ 2010. Striatal microRNA controls cocaine intake through CREB signalling. Nature 466: 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hubert GW, Jones DC, Moffett MC, Rogge G, Kuhar MJ 2008. CART peptides as modulators of dopamine and psychostimulants and interactions with the mesolimbic dopaminergic system. Biochem Pharmacol 75: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H-I, Hollander JA, Bali P, Kenny PJ 2010. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci 13: 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S 2010. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Junn E, Lee K-W, Jeong BS, Chan TW, Im J-Y, Mouradian MM 2009. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci 106: 13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Duffy P 1993. Time course of extracellular dopamine and behavioral sensitization to cocaine. I. Dopamine axon terminals. J Neurosci 13: 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer JA, Malenka RC 2007. Synaptic plasticity and addiction. Nat Rev Neurosci 8: 844–858 [DOI] [PubMed] [Google Scholar]
- Kiebler MA, DesGroseillers L 2000. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron 25: 19–28 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kiraly DD, Ma X-M, Mazzone CM, Xin X, Mains RE, Eipper BA 2010. Behavioral and morphological responses to cocaine require kalirin7. Biol Psychiatry 68: 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH 1995. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269: 1737–1740 [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T 2002. Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739 [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, Dietz DM, Zaman S, Koo JW, Kennedy PJ, et al. 2010. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science 330: 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94: 896–905 [DOI] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR 2008. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem 106: 650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X-M, Kiraly DD, Gaier ED, Wang Y, Kim E-J, Levine ES, Eipper BA, Mains RE 2008. Kalirin-7 is required for synaptic structure and function. J Neurosci 28: 12368–12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí E, Pantano L, Bañez-Coronel M, Llorens F, Miñones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X 2010. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 38: 7219–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML 2010. Sequencing technologies—the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Minichiello L 2009. TrkB signalling pathways in LTP and learning. Nat Rev Neurosci 10: 850–860 [DOI] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR 2004. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol 5: R68 doi: 10.1186/gb-2004-5-9-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Ozawa M 2007. A dileucine motif in its cytoplasmic domain directs beta-catenin-uncoupled E-cadherin to the lysosome. J Cell Sci 120: 4395–4406 [DOI] [PubMed] [Google Scholar]
- Moccia R, Chen D, Lyles V, Kapuya E, Yaping E, Kalachikov S, Spahn CMT, Frank J, Kandel ER, Barad M, et al. 2003. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci 23: 9409–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM 2011. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry 69: 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ 2001. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci 2: 119–128 [DOI] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Wenthold RJ 1994. The NMDA receptor subunits NR2A and NR2B show histological and ultrastructural localization patterns similar to those of NR1. J Neurosci 14: 6102–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce RC, Bell K, Duffy P, Kalivas PW 1996. Repeated cocaine augments excitatory amino acid transmission in the nucleus accumbens only in rats having developed behavioral sensitization. J Neurosci 16: 1550–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi S-H, Jamieson CAM, Geschwind DH, Martin KC 2006. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci 26: 13390–13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewłocka B, Lasoń W 1995. Adaptive changes in the proenkephalin and D2 dopamine receptor mRNA expression after chronic cocaine in the nucleus accumbens and striatum of the rat. Eur Neuropsychopharmacol 5: 465–469 [DOI] [PubMed] [Google Scholar]
- Rao A, Steward O 1991. Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized: analysis of proteins synthesized within synaptosomes. J Neurosci 11: 2881–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ 2011. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann Area 46 in schizophrenia. Biol Psychiatry 69: 180–187 [DOI] [PubMed] [Google Scholar]
- Schaefer A, Im H-I, Venø MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O'Carroll D, Greengard P 2010. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med 207: 1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G 2009. microRNAs at the synapse. Nat Rev Neurosci 10: 842–849 [DOI] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJL, Kane C, et al. 2009. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol 11: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D 2011. How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron 69: 628–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y-J, Weiler IJ, Greenough WT, Denman RB 2004. Selectively enriched mRNAs in rat synaptoneurosomes. Brain Res Mol Brain Res 126: 81–87 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Tian QB, Kuromitsu J, Kawai T, Endo S 2007. Characterization of mRNA species that are associated with postsynaptic density fraction by gene chip microarray analysis. Neurosci Res 57: 61–85 [DOI] [PubMed] [Google Scholar]
- Tian QB, Nakayama K, Okano A, Suzuki T 1999. Identification of mRNAs localizing in the postsynaptic region. Brain Res Mol Brain Res 72: 147–157 [DOI] [PubMed] [Google Scholar]
- van Roy F, Berx G 2008. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 65: 3756–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walaas SI, Greengard P 1984. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. I. Regional and cellular distribution in the rat brain. J Neurosci 4: 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC 2009. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 324: 1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, Ruan X, Ruan Y, Sun YE 2010. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci 107: 18161–18166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B-E, Sakurai M, Nishikura K 2011. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat Rev Genet 12: 81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Mains RE 1994. Endoproteolytic processing of proopiomelanocortin and prohormone convertases 1 and 2 in neuroendocrine cells overexpressing prohormone convertases 1 or 2. J Biol Chem 269: 17440–17447 [PubMed] [Google Scholar]
- Zukin RS, Richter JD, Bagni C 2009. Signals, synapses, and synthesis: how new proteins control plasticity. Front Neural Circuits 3: 14 doi: 10.3389/neuro.04014.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]