Abstract
The eukaryotic RNA exosome is a highly conserved multi-subunit complex that catalyzes degradation and processing of coding and noncoding RNA. A noncatalytic nine-subunit exosome core interacts with Rrp44 and Rrp6, two subunits that possess processive and distributive 3′-to-5′ exoribonuclease activity, respectively. While both Rrp6 and Rrp44 are responsible for RNA processing in budding yeast, Rrp6 may play a more prominent role in processing, as it has been demonstrated to be inhibited by stable RNA secondary structure in vitro and because the null allele in budding yeast leads to the buildup of specific structured RNA substrates. Human RRP6, otherwise known as PM/SCL-100 or EXOSC10, shares sequence similarity to budding yeast Rrp6 and is proposed to catalyze 3′-to-5′ exoribonuclease activity on a variety of nuclear transcripts including ribosomal RNA subunits, RNA that has been poly-adenylated by TRAMP, as well as other nuclear RNA transcripts destined for processing and/or destruction. To characterize human RRP6, we expressed the full-length enzyme as well as truncation mutants that retain catalytic activity, compared their activities to analogous constructs for Saccharomyces cerevisiae Rrp6, and determined the X-ray structure of a human construct containing the exoribonuclease and HRDC domains that retains catalytic activity. Structural data show that the human active site is more exposed when compared to the yeast structure, and biochemical data suggest that this feature may play a role in the ability of human RRP6 to productively engage and degrade structured RNA substrates more effectively than the analogous budding yeast enzyme.
Keywords: 3′-to-5′ decay, EXOSC10, PM/SCL-100, exosome, structure
INTRODUCTION
The RNA exosome is an essential multiprotein complex that catalyzes RNA processing and decay (Lorentzen et al. 2008a; Schmid and Jensen 2008; Houseley and Tollervey 2009; Lykke-Andersen et al. 2009, 2011). The exosome is highly conserved in all eukaryotes and has been studied in many organisms that include Arabidopsis thaliana, Saccharomyces cerevisiae, Drosophila melanogaster, Trypanosoma brucei, and Homo sapiens (Mitchell et al. 1997; Allmang et al. 1999b; Chekanova et al. 2000; Estevez et al. 2001; Andrulis et al. 2002; Evguenieva-Hackenberg et al. 2003; Lebreton and Seraphin 2008). Depending on the organism, the exosome can degrade RNA substrates that include rRNA intermediates, specific mRNAs, intergenic transcripts, and hypermodified tRNAs (Allmang et al. 1999a; Kadaba et al. 2004). In addition to its role in RNA degradation, the exosome is required for efficient processing and maturation of ribosomal RNA, stable small nucleolar RNA, and small nuclear RNA. Evidence also supports a role for the exosome in controlling the levels of histone mRNAs (Chen et al. 2001; Canavan and Bond 2007; Reis and Campbell 2007), cryptic unstable transcripts (CUTs), and antisense transcripts through an association with the TRAMP complex in processes that contribute to post-transcriptional gene silencing and heterochromatin formation (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Davis and Ares 2006; Camblong et al. 2007; Houseley and Tollervey 2008; Wang et al. 2008a).
Human and budding yeast exosomes share similar subunit composition (Januszyk and Lima 2010). Structural studies on the human exosome determined that the nine-subunit core is composed of a six-member ring of RNase PH-domain proteins (Rrp41, Rrp42, Rrp43, Mtr3, Rrp46, and PM/Scl-75 [Rrp45]) that is capped by two proteins that contain S1 and KH domains (Rrp4 and Rrp40), and one protein that contains an S1 domain, Csl4 (Liu et al. 2006). While the ring-like architecture of the human exosome core is structurally similar to 3′-to-5′ phosphorolytic exoribonucleaseses such as bacterial polynucleotide phosphorylase and archaeal exosomes (Symmons et al. 2000; Buttner et al. 2005; Lorentzen et al. 2005), biochemical and genetic analysis revealed that budding yeast and human exosome cores were devoid of phosphorolytic or hydrolytic exoribonuclease activity (Liu et al. 2006; Dziembowski et al. 2007). Instead, the 3′-to-5′ exoribonuclease activity of eukaryotic exosomes is achieved by exosome core association with two hydrolytic enzymes: the tenth subunit, Rrp44 or Dis3, and the eleventh subunit, Rrp6 (Briggs et al. 1998; Phillips and Butler 2003; Liu et al. 2006; Midtgaard et al. 2006; Dziembowski et al. 2007; Schneider et al. 2007; Wang et al. 2007; Lorentzen et al. 2008b; Bonneau et al. 2009). While the activities of budding yeast Rrp44 and Rrp6 have been characterized (Liu et al. 2006; Dziembowski et al. 2007; Assenholt et al. 2008; Callahan and Butler 2008; Lebreton et al. 2008; Bonneau et al. 2009; Schaeffer et al. 2009; Schneider et al. 2009; Callahan and Butler 2010), much less is known about the intrinsic activities of the human paralogs of Rrp44, DIS3 and DIS3L (Staals et al. 2010; Tomecki et al. 2010), or human RRP6.
Human RRP6 and budding yeast Rrp6 are members of the DEDD nuclease superfamily that encompasses ribonucleases and deoxyribonucleases (Steitz and Steitz 1993), and are characterized by an EXO domain that shares sequence similarity to EXO I, II, and III motifs of the exonuclease domains of DNA polymerases that contain the four conserved acidic residues, DEDD (Ollis et al. 1985a,b). Some family members include an additional conserved histidine or tyrosine residue and are designated DEDD-H or DEDD-Y, respectively. Human RRP6, yeast Rrp6, and the bacterial RNA processing enzyme RNase D are members of the DEDD-Y family (Bluthner and Bautz 1992; Phillips and Butler 2003; Zuo et al. 2005; Midtgaard et al. 2006).
The crystal structure of the RNase D enzyme revealed that the EXO domain interacts with two conserved HRDC (helicase and RNase D C-terminal) domains (Zuo et al. 2005), first identified as nucleic acid–binding domains within the RecQ helicase protein family (Morozov et al. 1997). The only available crystal structure of Rrp6 was derived from the yeast enzyme, revealing that the EXO domain is flanked by a single C-terminal HRDC domain (Midtgaard et al. 2006). While HRDC domains are observed in budding yeast Rrp6 and RNase D, their relative positions differ. In addition, the EXO and single HRDC domain of budding yeast Rrp6 are sufficient to retain catalytic activity (Assenholt et al. 2008), but yeast Rrp6 possesses additional domains that are required for proper function in vivo including an N-terminal PMC2NT domain that is proposed to interact with the double-stranded RNA- and DNA-binding protein Rrp47 (also called C1D in human) (Mitchell et al. 2003; Schilders et al. 2007; Stead et al. 2007; Costello et al. 2011), a region C-terminal to the HRDC domain that is necessary for interaction with the core exosome (Callahan and Butler 2008), and a putative NLS domain at the C terminus that is nonessential insofar as a mutant Rrp6 that lacks the NLS grows without a noticeable phenotype in budding yeast (Phillips and Butler 2003).
RRP6 is the only exosome gene within the 11-subunit complex that is not essential for budding yeast cell viability, although deletion of RRP6 confers a slow growth phenotype and high temperature sensitivity, resulting in accumulation of an external transcribed spacer, an extended form of 5.8S rRNA (5.8S + 30 species) (Briggs et al. 1998), polyadenylated rRNA (Kuai et al. 2004), and oligoadenylated box C/D snoRNAs (Allmang et al. 1999a; van Hoof et al. 2000; Mitchell et al. 2003; Grzechnik and Kufel 2008). Furthermore, functional analysis of budding yeast rrp6-3 (D238A), rrp6-4 (E240A), rrp6-5 (D296A), rrp6-6 (Y361A), rrp6-7 (Y361F), and rrp6-8 (D365A) mutants in the EXO domain confirm that the phenotypes observed in budding yeast are attributed to the absence of exoribonucleolytic activity in these strains (Phillips and Butler 2003).
Human RRP6 is localized to nucleoli and nucleoplasm, which is in agreement with its major function in snoRNA and rRNA processing (Bluthner and Bautz 1992; Fomproix and Hernandez-Verdun 1999; Bluthner et al. 2000; Chen et al. 2001; Brouwer et al. 2002; Mahler and Raijmakers 2007; Staals et al. 2010; Tomecki and Dziembowski 2010); however, little else is known about its intrinsic catalytic activities. To characterize human RRP6, we isolated full-length human RRP6 and constructs that include EXO and/or HRDC domains, analyzed their activities, and compared these activities to budding yeast Rrp6. The crystal structure of a minimal catalytic domain of human RRP6 includes the EXO and HRDC domains, and subsequent structure-based mutational analysis confirms that human RRP6 is a bona fide member of the DEDD-Y family; although, human RRP6 differs to some extent from its budding yeast counterpart with respect to degradation of RNA substrates containing secondary structure.
RESULTS AND DISCUSSION
Domain boundaries and purification of human RRP6
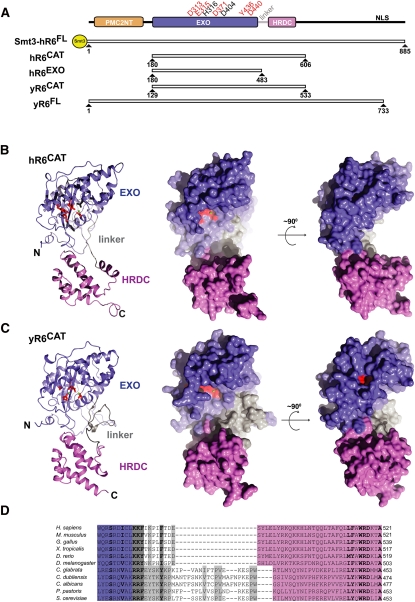
Constructs were designed to express and purify different regions of human RRP6 for structural and biochemical analysis. However, only three of 11 proteins produced were stable enough to facilitate purification. We named these constructs hR6CAT, hR6EXO, and full-length hR6 (Fig. 1A). To produce a stable full-length human RRP6, an Smt3 solubility tag (Mossessova and Lima 2000) was fused to the N terminus of hR6FL (referred to as Smt3-hR6FL). hR6CAT spans residues 180 to 606 and contains the EXO and HRDC domains. hR6EXO comprises residues 180 to 483 and encompasses the EXO domain. For comparative analyses, budding yeast constructs were also produced. Full-length yR6 was purified as described previously (Liu et al. 2006; Greimann and Lima 2008). Lastly, the yR6CAT construct was prepared based on sequence comparison to hR6CAT; it corresponds to residues 129 to 533, which also include the EXO and HRDC domains (Midtgaard et al. 2006; Assenholt et al. 2008).
FIGURE 1.
Structure of human RRP6. (A) Domain structure and construct description. Domain structure is depicted with a PMC2NT domain (orange), an EXOribonucleolytic domain (EXO, purple), an HRDC (helicase and RNase D C-terminal) domain (pink), and a nuclear localization signal (NLS). Residues that were mutated and tested for their effect on exoribonucleolytic decay activity are depicted above the domain structure. Active-site residues (red); additional residues D404 and H316 used in exoribonucleolytic decay activity studies (black). Constructs used for structural and biochemical studies are a Smt3 tagged full length human RRP6 (Smt3-hR6FL), the human exoribonucleolytic catalytic region (hR6CAT, residues 180 to 606), a human construct that contains only the EXO domain (hR6EXO, residues 180 to 483), a budding yeast full-length Rrp6 (yR6FL) construct, and the budding yeast Rrp6 truncation construct that contains the exoribonucleolytic catalytic region (yR6CAT, residues 129 to 533). (B) Overall structural comparison of human hR6CAT in cartoon (left panel) and orthogonal views in the space filling representation (middle and right panels). (C) Overall structure of yR6CAT (PDB ID = 2HBK) in cartoon (left panel) and orthogonal views in the space filling representation (middle and right panels). The EXO (purple), linker (gray), and HRDC (pink) domains. (D) Alignment of the linker with reference to secondary structure for higher eukaryotes (H. sapiens, M. musculus, G. gallus, X. tropicalis, D. rerio, and D. melanogaster) and lower eukaryotes (S. cerevisiae, C. glabrata, C. dubliensis, C. albicans, and P. pastoris). Within the linker, similar residues that are conserved across all species within higher or lower eukaryotes are shown in bold. Alignments were produced using ClustalW (Thompson et al. 1994).
Structure of a catalytically active fragment of human RRP6
We were unable to obtain diffracting crystals of wild-type hR6CAT but were able to crystallize and determine structures for two mutant forms of hR6CAT that contained either D313N or Y436A substitutions in the active site, each with two protomers in the asymmetric unit (Table 1) (see Materials and Methods). The overall architecture of hR6CAT resembles that of other DEDD-H and DEDD-Y exoribonucleases, especially yR6CAT (Fig. 1B,C), not surprising given that yR6CAT and hR6CAT share 37% identity and 59% similarity at the level of primary sequence over 426 amino acids. The structures of the six hR6CAT protomers can be aligned to the manganese-bound yR6CAT structure with an RMSD of 2.1 Å for 360 of the 427 Cα atoms. Similar to yeast Rrp6, the EXO domain of hR6CAT forms a classical α/β-fold, comprising a six-stranded, mixed β-sheet flanked by multiple α-helices. In addition, side chains that constitute the hR6CAT DEDD-Y active site are oriented in similar positions to yR6CAT when analogous secondary structure motifs are aligned (Figs. 1B,C, 2). The hR6CAT structure also resembles yeast Rrp6 with respect to the relative orientations of the EXO domain and the HRDC domain, a facet of Rrp6 that differs from Escherichia coli RNase D (Zuo et al. 2005; Midtgaard et al. 2006).
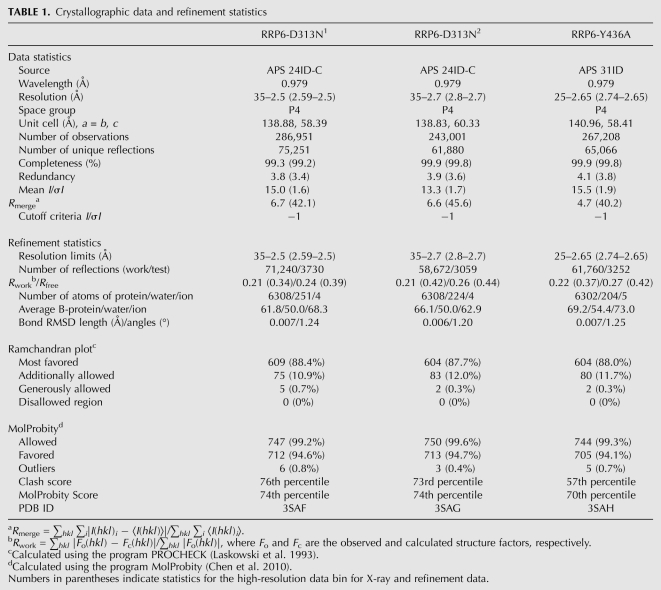
TABLE 1.
Crystallographic data and refinement statistics
Human RRP6CAT and yeast Rrp6CAT structures share similarity throughout the EXO and HRDC domains, but some important differences exist. An approximately 26-amino-acid linker between the EXO and HRDC domains (residues R399 to W425) is conserved at the level of 58% sequence similarity within the fungi S. cerevisiae, Pichia pastoris, Candida glabrata, Candida dubliniensis, and Candida albicans (Fig. 1D). This linker forms a wall on one side of the active site in the yeast Rrp6 structure that appears to deepen the active-site cleft, presumably to allow ingress of single-stranded RNA substrates to the active site while precluding larger substrates such as double-stranded RNA (Fig. 1B,C). In higher eukaryotes, this element is replaced by a shorter 10-amino-acid linker (hR6CAT residues R479 to E489) that possesses a 67% sequence similarity among the higher eukaryotes: H. sapiens, Mus musculus, Gallus gallus, Xenopus tropicalis, Danio rerio, and Drosophila melanogaster (Fig. 1D). This shorter linker in the human RRP6CAT structure and other higher eukaryotes exposes the active site to solvent, making it appear more accessible to both single- and double-stranded RNA substrates when compared to the yeast enzyme (Fig. 1B,C).
The active-site residues D313, E315, D371, D440, and Y436 exhibit similar conformations in five of the six hR6CAT protomers solved in our three crystal structures, conformations similar to that observed in the yR6CAT structure (Fig. 2). The DEDD-Y active sites of both hR6CAT and yR6CAT are similar to the canonical DEDD-Y DNA polymerase I exonuclease. The four acidic residues are predicted to coordinate two divalent cations that interact with the negatively charged RNA, activate a water molecule for hydrolysis by nucleophilic attack of the phosphodiester backbone of RNA, and stabilize the transition state and products of the reaction (Beese and Steitz 1991). The tyrosine is believed to work in concert with the glutamate residue to orient the nucleophilic water.
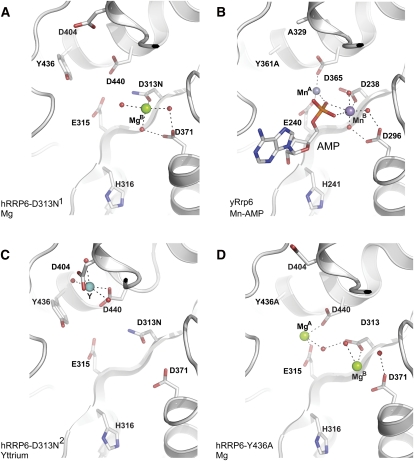
FIGURE 2.
The DEDD-Y active site of RRP6. (A) hR6CAT D313N1 mutant with Mg. Active-site residues D313N, E315, D371, D440, and Y436 are shown coordinating one of the magnesium ions MgB, depicted as a green sphere. Also shown are residues D404 and H316. (B) yR6CAT Y361A-AMP bound structure. D238, E240, D296, D365, and Y361A are shown coordinating two manganese atoms MnA and MnB (purple spheres) and the end product AMP. Also shown are residues A329 and H241. (C) hR6CAT D313N2 mutant with Y. Active-site residues D313N, E315, D371, D440, and Y436 are shown. Residue D440 is depicted coordinating an yttrium (shown as a cyan sphere) with D404 and D440. (D) hR6CAT Y436A mutant with two magnesiums. Active-site residues D313, E315, D371, D440, and Y436A are shown coordinating the two magnesium ions: MgA and MgB (green spheres). All waters are shown as red spheres.
Electron density revealed positions for one or two divalent ions in the active sites of the D313N and Y436A hR6CAT protomers, respectively (Fig. 2A,D). Manganese was used to identify the position of the divalent ions, as determined for the yeast Rrp6 structure (Midtgaard et al. 2006); but these attempts failed for hR6CAT. However, soaking magnesium into the crystal facilitated detection of electron density that was consistent with magnesium insofar as (1) the ions were coordinated with octahedral geometry; (2) the distances between the presumed magnesium ion and corresponding ligands were shorter than expected for typical hydrogen bonding interactions; and (3) no anomalous electron density could be detected at these positions, which eliminated these ions as yttrium, an ion required for crystallization (Supplemental Fig. S1A,C). In addition, the presumed magnesium ions occupy positions consistent with those observed in other canonical DEDD-Y active sites and to the manganese ions that were definitively identified in the structure of the yR6CAT Y361A mutant (Fig. 2B); magnesium A is coordinated by D313, E315, and D440; and magnesium B is coordinated by D313 and D371 (Fig. 2A,D). Attempts to soak in the products of hydrolysis (AMP or GMP) failed in the presence of magnesium, perhaps consistent with results obtained from the yeast system, where it was determined that the divalent cations (Mn2+ and Zn2+) were necessary for stabilizing the yeast Rrp6 product complex (Midtgaard et al. 2006).
One major difference was observed with respect to the conformation of active-site residues for one protomer of the hR6CAT D313N2 mutant, which diffracted X-rays to 2.7 Å. Instead of D440 pointing toward the other active-site residues to facilitate coordination of a magnesium ion, the D440 side chain was turned away from the active site and instead was jointly coordinating an yttrium ion with D404 (Fig. 2C). We assigned this ion as yttrium due to the presence of an anomalous signal, which was detected by Fourier analysis (Supplemental Fig. S1B). While this conformation was likely induced by cocrystallization with yttrium, we noted that D404 is conserved as an acidic residue in vertebrates (Fig. 6A, below) but is conserved as alanine in budding yeast, other lower eukaryotic organisms, and E. coli RNase D. The functional relevance of this finding is probed and discussed below.
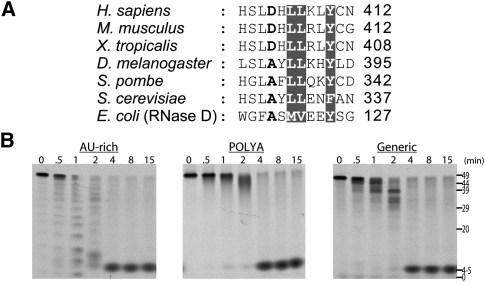
FIGURE 6.
Activities of hRRP6 mutant D404A. (A) Sequence comparison of RRP6. (Gray) Residues that are highly conserved; the D404 residue is shown in bold. Accession numbers are included in the legend to Figure 3. (B) Activity of hR6CAT, D404A on AU-rich, poly(A), and generic RNA 49-mers. Time points (in minutes) and apparent size (in nucleotides) are indicated at the top and right.
Activities and mutagenesis of human RRP6
Budding yeast Rrp6 has been extensively characterized, but exoribonucleolytic activity has not yet been demonstrated for Rrp6 from higher eukaryotes. We tested hR6CAT protein at high and low concentrations (1000 and 10 nM) in single-time-point RNA degradation assays, using two 5′-fluorescein-labeled RNA substrates (at 10 nM) containing either AU-rich or a generic sequence that were each incubated at 90 min (Fig. 3C; Greimann and Lima 2008). Exoribonucleolytic activity was observed for hR6CAT, as evidenced by the detection of a distribution of intermediates at 10 nM protein and a final product with a length of 4 to 5 nt at 1000 nM protein.
FIGURE 3.
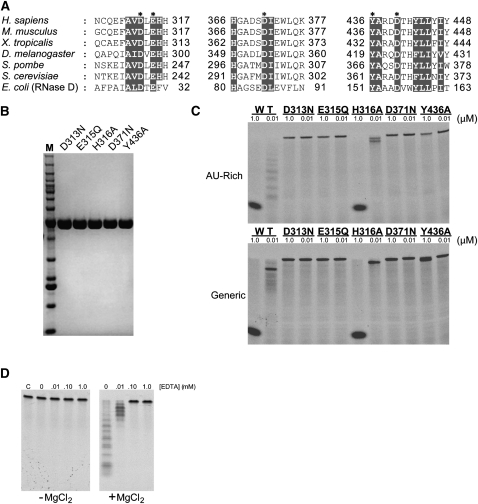
Mutagenesis of the DEDD-Y RRP6 active site. (A) Sequence comparison of residues within the RRP6 active site. Alignment was created using ClustalW (Thompson et al. 1994) with the following accession numbers: H. sapiens NP_001001998, M. musculus NP_057908, X. tropicalis NP_998833, D. melanogaster NP_001097795, Schizosaccharomyces pombe NP_593004, S. cerevisiae NP_014643, and E. coli NP_310540. (Gray) Residues that are highly conserved; (asterisks) residues that comprise the DEDD-Y active site. (B) SDS-PAGE analysis of hR6CAT active-site mutants. Equal amounts of mutant proteins were loaded into each lane; the gel was stained with Coomassie blue for detection. The molecular weight standard is the Invitrogen BenchMark Molecular Weight ladder with sizes: 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 90, 100, 120, 160, and 220 kDa. (C) Exoribonucleolytic activity of hR6CAT mutants. Wild-type and mutant versions of hR6CAT were each assayed at high (1 μM) and low (0.01 μM) concentrations of protein on 10 nM AU-rich RNA 49-mer substrate (top panel) and generic RNA 49-mer substrate (lower panel) for 90 min at 37°C. (D) Disruption of exoribonucleolytic activity of wild-type hR6CAT by EDTA. Assays were conducted with increasing amounts of EDTA for 45 min at 37°C with 10 nM AU-rich RNA 49-mer substrate in the presence (right panel) and absence (left panel) of 5 mM MgCl2.
To probe the importance of side chains that constitute the highly conserved DEDD-Y active site, the single-amino-acid substitutions D313N, E315Q, D371N, and Y436A were constructed, and proteins containing these mutations were purified and assayed for activity (Fig. 3A,B). As expected, exoribonuclease activity was not observed using any concentration of these mutant hR6CAT isoforms (Fig. 3C), and similar results were obtained for wild-type hR6CAT in the presence of EDTA (Fig. 3D). While the hR6CAT Y436A mutant exhibited little activity for the AU-rich substrate, some activity was detected at the highest concentration of mutant protein on the generic RNA, suggesting that the tyrosine side chain is critical but not essential for positioning the nucleophilic water during hydrolysis.
The structure of yeast Rrp6 in complex with an AMP product revealed contacts between a conserved histidine side chain (H241) and the AMP 2′ hydroxyl (Midtgaard et al. 2006). We tested the importance of the analogous residue in the human enzyme (Fig. 3A) by substituting H316 with alanine; however, the resulting mutant hR6CAT protein retained some exoribonuclease activity in decay assays using AU-rich or generic RNA substrates (Fig. 3C), indicating that the H316 side chain is not essential for hR6CAT exoribonuclease activity. Previous work on the yeast enzyme also revealed that removal of the HRDC domain from the full-length protein in budding yeast disrupted the processing of certain RNA substrates in vivo (Callahan and Butler 2008). Interestingly, the hR6EXO protein, which lacks the HRDC domain, exhibited a >100-fold reduction in exoribonuclease activity on all of the RNA substrates tested (Supplemental Fig. S2A). However, hR6EXO retains the ability to interact with RNA substrates with affinities estimated in the nanomolar range, as assessed by electrophoretic mobility shift assays (Supplemental Fig. S2B).
Substrate specificity
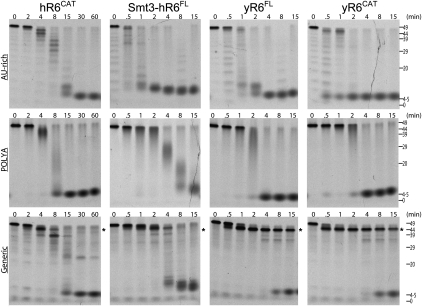
The catalytic activities of full-length human and yeast Rrp6 as well as the isolated catalytic regions from both enzymes (hR6CAT and yR6CAT) were assessed in time-course assays to compare their activities and substrate specificities in the degradation of three 49-mer RNA substrates with distinct sequence and structure [AU-rich, generic, or poly (A) sequences]. The yeast proteins and hR6CAT degraded poly(A) and AU-rich RNA substrates to final products that ranged between 4 and 5 nt, while the Smt3-hR6FL produced a final product with a slightly larger size (Fig. 4). The detection of a decreasingly sized distribution of intermediates with increasing time prior to accumulation of the final product is indicative of the distributive, exoribonucleolytic activity in which the enzyme cleaves, releases, and engages another RNA substrate. For comparison, a processive activity would contain both full-length RNA substrates as well as the cleaved end product during the course of the time-course assay, as a processive enzyme engages each RNA substrate to completion. The intermediates and final products of the assayed reactions were similar when results are compared for Smt3-hR6FL, yR6FL, and yR6CAT, although it is notable that the degradation was ∼10-fold slower for hR6CAT. In contrast, some interesting differences were observed when decay assays were performed using the generic substrate, an RNA molecule predicted to have significant secondary structure, a 2-nt 5′ overhang, and an 8-nt 3′ overhang (Liu et al. 2006). In this case, both yR6FL and yR6CAT quickly process this RNA as evidenced by removal of the 3′ overhang as early as 1–2 min, leaving a stable RNA intermediate (marked by asterisks) that is apparent even after 15 min of incubation. The size differences between the substrate and intermediate product suggest that yeast Rrp6 removes ∼5 nt but leaves the presumed stem–loop intact. This result is consistent with the structure of the yeast catalytic domain because the active site is located in a deep cleft that presumably restricts access to single-stranded RNA. In contrast, both hR6FL and hR6CAT appeared more adept at degrading the generic RNA substrate, and the pattern of intermediates was distinct; perhaps most notable was the absence of the intermediate, which is more stable in assays with the yeast enzymes. Again, this result is consistent with the structure of the human catalytic domain because it shows the active site to be more exposed, presumably to allow access to the 3′ end of the structured RNA.
FIGURE 4.
Comparison of exoribonucleolytic activities and substrate preferences for human RRP6 and yeast Rrp6 proteins. Demonstration of exoribonucleolytic activities over time for different Rrp6 constructs that include (column 1) the exoribonucleolytic catalytic region (hR6CAT, residues 180 to 606), (column 2) Smt3 tagged full-length human RRP6 (Smt3-hR6FL), (column 3) budding yeast full-length Rrp6 (yR6FL), and (column 4) the budding yeast Rrp6 truncation construct that contains the exoribonucleolytic catalytic region (yR6CAT, residues 129 to 533). Constructs were incubated with RNA substrates that are 49 nt in length with an AU-rich RNA sequence (row 1), poly(A) sequence (row 2), and a generic RNA sequence (row 3). Time points (in minutes) and apparent size (in nucleotides) are indicated at the top and right, respectively. For generic RNA exoribonucleolytic decay assays, asterisks indicate the major intermediate detected for the yeast enzymes and the relative position if the intermediate was detected in assays of the human enzymes. The RNA sequences are included in Materials and Methods.
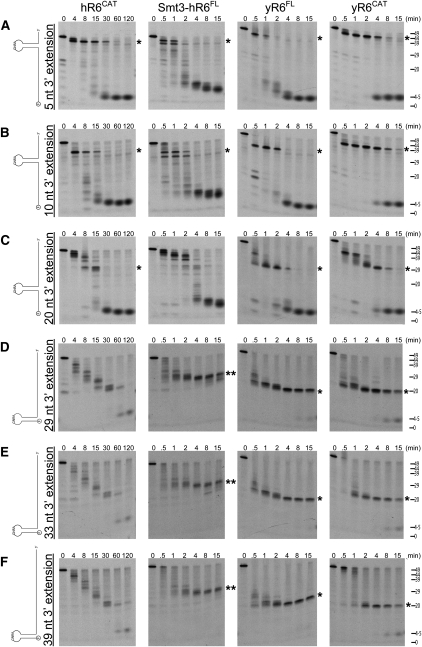
To further explore dependencies on secondary structure, we used substrates that contained a 20-nt GC-rich GNRA stem–loop flanked by AU-rich RNA sequences in RNA degradation assays using human (Smt3-hR6FL, hR6CAT) and budding yeast (yR6FL and yR6CAT) proteins (Fig. 5A–C). These three substrates contained the stem–loop in different positions within the body of the RNA flanked by 5′ and 3′ AU-rich extensions, and each was a substrate for hR6CAT and Smt3-hR6FL as well as the yeast enzymes, as judged by their ability to degrade these RNA substrates to a final product. While yR6CAT, Smt3- hR6FL, hR6CAT, and yR6FL degraded these substrates to completion, a primary intermediate (highlighted with a single asterisk) was observed in the degradation of each of these substrates that corresponded to a degradation product up to the base of the stem–loop. The degradation intermediates were detected in reactions containing yeast proteins and the hR6CAT protein; however, the Smt3-hR6FL protein only produced a significant build-up of the corresponding intermediate present in reactions with the other proteins for substrates containing 3′-single-stranded extensions of 5 and 10 nt.
FIGURE 5.
Comparison of exoribonucleolytic activities on structured GNRA stem–loop containing RNAs for human RRP6 and yeast Rrp6 proteins. (A–F) Time course and activities for the human RRP6 catalytic region (hR6CAT, column 1), Smt3 tagged full-length human RRP6 (Smt3-hR6FL, column 2), budding yeast full-length Rrp6 (yR6FL, column 3), and the budding yeast Rrp6 exoribonucleolytic catalytic region (yR6CAT, column 4). Protein constructs were each incubated with (A) a 49-mer RNA containing AU-rich RNA, a GNRA stem–loop, and a 5-nt extension; (B) a 49-mer RNA containing AU-rich RNA, a GNRA stem–loop, and a 10-nt extension; (C) a 49-mer RNA containing AU-rich RNA, a GNRA stem–loop, and a 20-nt extension; (D) a 49-mer RNA containing AU-rich RNA, a GNRA stem–loop, and a 29-nt extension; (E) a 53-mer RNA containing AU-rich RNA, a GNRA stem–loop, and a 33-nt extension; and (F) a 59-mer RNA containing AU-rich RNA, a GNRA stem–loop, and a 39-nt extension. Time points (in minutes) and apparent size (in nucleotides) are indicated at the top and right, respectively. A single asterisk indicates the major intermediates, consistent with sizes corresponding to a degradation product up to the base of the stem–loop. (Double asterisk in D–F) The major intermediate detected in the presence of Smt3-hR6FL. The RNA sequences are included in Materials and Methods.
Three additional substrates were also assessed for their exoribonucleolytic activity that included the GC-rich GNRA stem–loop at the 5′ end of the substrate with 29-, 33-, or 39-nt 3′-AU-rich extensions (Fig. 5D–F). In these assays, both human and yeast enzymes appeared to have difficulty degrading these substrates in comparison to substrates containing single-stranded AU-rich sequences located 5′ of the stem–loop. In addition, the full-length human RRP6 protein did not accumulate intermediates (double asterisks) similar to those observed in the exoribonucleolytic assays of both yeast proteins (single asterisks), which correspond to a degradation product up to the base of the stem–loop. Moreover, hR6CAT degraded these RNA substrates with no apparent accumulation of any major degradation intermediate. In summary, all of the versions of the human and yeast Rrp6 proteins used in this study appear able to degrade through secondary structure more readily when a 5′-single-stranded region is present in the substrate (Fig. 5, cf. A–C to D–F). The mechanistic basis for this phenomenon remains unclear, although it is attractive to hypothesize that the HRDC domain interacts with the 5′-single-stranded sequence, thus maintaining interactions with the substrate for long enough periods of time to wait out fraying of the RNA duplex so that the active site can reengage the 3′ end. Evidence in support of this role for the HRDC is based on the observation that removal of a salt bridge that bridges the EXO–HRDC interface (by mutation of D457 to alanine) or the entire deletion of the HRDC domain results in a loss of the processing activity but preservation of decay activity in vivo (Assenholt et al. 2008; Callahan and Butler 2008).
Gain of function hR6CAT mutant
As mentioned previously, residue D404 is conserved in higher eukaryotes but is present as an alanine in protozoans (Fig. 6A). In our crystallography studies, we detected an yttrium that was coordinated by residue D404 and active-site residue D440. Since yttrium can serve as a mimetic of divalent cations, we hypothesized that this structure might actually represent a nonproductive state of the enzyme. To test this hypothesis, we changed hR6CAT D404 to alanine to alter the human enzyme to appear more like the yeast enzyme, and assayed this mutant in comparison to its yeast and human counterparts. Consistent with our hypothesis, the D404A substitution resulted in a fourfold to eightfold increase in catalytic activity as assessed by the time it takes to completely degrade three different RNA substrates [AU-rich, poly(A), and generic] by comparing wild-type hR6CAT in Figure 4 and hR6CAT, D404A in Figure 6B. Moreover, a similar increase in catalytic activity was also detected on the aforementioned stem–loop-containing RNA substrates (Supplemental Fig. S3). While it is impossible to ascertain at this time if a nonproductive state of the full-length human enzyme exists in vivo, the gain-of-function mutation in the human active site raises the possibility that human RRP6 exoribonucleolytic activity could be modulated by changing the properties of this residue or the regions surrounding it.
CONCLUSIONS
The biochemical and structural studies presented here confirm that human RRP6 is a member of the DEDD-Y family of exoribonucleases. Although structurally similar to other family members, some unique differences were observed both structurally and biochemically for the human enzyme in comparison to yeast Rrp6. These include an active site that appears more exposed to solvent and a potential inhibited or nonproductive state of the enzyme, which was alleviated by mutating an acidic residue conserved in higher eukaryotes to an alanine present in lower eukaryotes. Further structural characterization of human and yeast Rrp6 in complex with specific RNA substrates will be required to fully understand how these differences affect the ability of Rrp6 to interact, process, and degrade RNA.
Biochemical analysis of human RRP6 in RNA decay assays and comparison to the corresponding yeast Rrp6 proteins revealed some interesting differences, namely, that full-length human RRP6 is better able to degrade substrates containing secondary structure in comparison to its yeast counterparts, but only when these substrates contained single-stranded elements 5′ to the structured domain. This may be a relevant observation because Rrp6 has been implicated in processing of many structured RNAs that include the 7S pre-rRNA, pre-tRNAiMet, and small nuclear and small nucleolar RNAs (Allmang et al. 1999a; van Hoof et al. 2000; Mitchell and Tollervey 2003; Kadaba et al. 2004; Callahan and Butler 2008, 2010; Grzechnik and Kufel 2008; Wang et al. 2008b). While the in vitro activities observed for budding yeast Rrp6 are consistent with this role, the activities of the human enzyme are less so. In other words, yeast Rrp6 appears to be an intrinsically good processing enzyme, whereas human RRP6 seems to be less inhibited by secondary structure. This suggests that human RRP6 may require a different mode of regulation than budding yeast Rrp6 to facilitate decisions that govern processing versus complete decay of structured RNA substrates. This hypothesis will require further analysis, including assays on larger and more structured RNAs, and reconstitution of human RRP6 into RNA exosomes to observe if the exosome core alters RRP6 activity.
MATERIALS AND METHODS
Protein expression and purification
The H. sapiens RRP6 (PM/SCL-100) gene encoding residues for hR6FL, hR6CAT, and hR6EXO regions were PCR-amplified from placenta cDNA pool (Ambion) using Pfu Turbo DNA polymerase (Stratagene). The DNA encoding budding yeast yRrp6CAT was PCR-amplified from a full-length budding yeast RRP6 construct (Greimann and Lima 2008). Digested PCR products were inserted into pSMT3-TOPO (hR6CAT, hR6EXO, yR6CAT) or a pCDF-Smt3 (hR6FL) vector. Point mutations (D313N, E315Q, H316A, D371N, Y436A, and D404A) of hR6CAT were introduced by a site-directed mutagenesis kit (QuikChange; Stratagene). All constructs were confirmed by DNA sequencing. To produce protein, constructs were transformed into BL21 (DE3) CodonPlus-RIL (Stratagene), grown in shaker flasks to ∼0.6 OD600, and induced overnight at 18°C with 0.25 mM IPTG in the presence of 2% ethanol. After induction, cells were harvested and flash-frozen. To prepare cell lysate, sonication was performed using a cell disrupter in 50 mM Tris-HCl (pH 8.0), 20% (w/v) sucrose, 350 mM NaCl, 20 mM imidazole, 0.1% IGEPAL, 1 mM PMSF, 1 mM B-mercaptoethanol (BME), and 10 ug/mL DNase. Lysate was then centrifuged using a Beckman JA-20 rotor at 44,000g. Supernatant was then subjected to Ni-NTA resins (QIAGEN) purification, and fusion protein was eluted in 20 mM Tris-HCl (pH 8.0), 350 mM NaCl, 250 mM imidazole, and 1 mM BME. For further purification of wild-type and mutant forms of hR6CAT, Smt3 fusion proteins were cleaved by Ulp1 overnight at 4°C and fractionated by size exclusion chromatography (HiLoad 26/60 Superdex 75 pg; GE Biosciences) in Sizing Buffer: 20 mM Tris-HCl (pH 8.0), 350 mM NaCl, and 1 mM BME. For further purification of yR6CAT, Smt3 is cleaved by Ulp1 overnight at 4°C, fractionated by size exclusion chromatography (HiLoad 26/60 Superdex 200 pg; GE Biosciences) in Sizing Buffer, and then additionally purified with a linear gradient via a monoQ column (GE Biosciences) with Buffer A: 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM BME and Buffer B: 20 mM Tris-HCl (pH 8.0), 1000 mM NaCl, and 1 mM BME. For further purification of Smt3-hR6FL, the Smt3-hR6FL fusion protein was also fractionated using size exclusion chromatography (HiLoad 26/60 Superdex 200 pg; GE Biosciences) in Sizing Buffer. The purification of yR6FL has been described previously (Greimann and Lima 2008). For each protein sample, peak fractions were pooled, concentrated, and flash-frozen prior to biochemical assays. Human hR6CAT protein samples used for crystallization were further desalted to 10 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 1 mM BME and concentrated to ∼20 mg/mL.
Crystallization and structure determination
Using the vapor diffusion/hanging drop method, the D313N and Y436A point mutants of hR6CAT appeared at 18°C after 2 d in crystallization buffer (6% PEG 6000, 1.5 M NaCl, and 0.4 mM YCl3) and grew to a length of ∼200 μm in 2 wk. The crystal containing D313N that diffracted X-rays to 2.5 Å resolution (denoted RRP6-D313N1 in Table 1) was soaked in a solution containing 10 mM MgCl2 and/or 10 mM GMP for 4 h and cryo-protected by the addition of 3.5 M NaCl prior to snap-freezing in liquid nitrogen. The crystal containing D313N that diffracted X-rays to 2.7 Å (denoted RRP6-D313N2 in Table 1) and the crystal containing Y436A (denoted RRP6-Y436A in Table 1) were soaked in solution containing 10 mM MgCl2 and/or 10 mM GMP for 4 h and transferred at 4°C stepwise in increments of 5% to a final concentration of 25% glycerol before snap-freezing in liquid nitrogen. Diffraction data sets were collected at the Advanced Photon Source (APS) beamline 24-IDC and 31-ID. Data were integrated, scaled, and merged using HKL2000 (Otwinowski and Minor 1997). The data obtained from RRP6-D313N1 were used to solve the structure by molecular replacement using the Saccharomyces cerevisiae Rrp6 structure (PDB 2HBJ) as a search model using MOLREP after truncating side chains to alanine that were not conserved in the human sequence. The model was manually rebuilt and inspected using O (Jones et al. 1991) and refined using CNS 1.3 (Brunger et al. 1998). The location of yttrium ions was confirmed by inspection of anomalous difference electron density maps. Magnesium ions were placed in the model based on the observation that these ions were coordinated in octahedral geometry by protein ligands and/or water atoms. All graphics depicting structure were generated using Pymol (DeLano 2002).
RNA degradation assays
RNA oligonucleotides were synthesized with a 5′ fluorescein and HPLC-purified (Invitrogen and Dharmacon). The 49-nt poly(A), generic, AU-rich type RNA oligonucleotides have the sequences
AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA,
AAUUAUUUAUUAUUUAUUUAUUAUUUAUUUAUUUAUUAUUUAUUUAUUA,
AUGCAUCUAGAGGGCCCUAUUCUAUAGUGUCACCUAAAUGCUAGAGCUC,
respectively. The six GNRA stem–loop containing with 5, 10, 20, 29, 33, and 39 3′ nucleotide extensions have the following sequences:
UUAUUAUUUAUUUAUUAUUUAUUAGGGCGGGCGCAAGCCCGCCCAUUUA,
AUUUAUUUAUUAUUUAUUAGGGCGGGCGCAAGCCCGCCCUUAUUAUUUA,
AUUUAUUUAGGGCGGGCGCAAGCCCGCCCUAUUUAUUAUUUAUUAUUUA,
GGGCGGGCGCAAGCCCGCCCAUUAUUUAUUAUUUAUUAUUUAUUAUUUA,
GGGCGGGCGCAAGCCCGCCCAUUAUUUAUUAUUUAUUAUUUAUUAUUUAUUUA, and
GGGCGGGCGCAAGCCCGCCCAUUAUUUAUUAUUUAUUAUUUAUUAUUUAUUAUUAUUUA,
respectively. GNRA stem–loop-containing RNAs were heated at 80°C and cooled to room temperature prior to determining activities. Each RNA is labeled at the 5′ end with fluorescein to enable detection. Exoribonuclease activities were performed in a 130-μL reaction mixture of 10 mM Tris-HCl (pH 8.0), 10 mM DTT, 50 mM KCl, 5 mM MgCl2, 1 U/μL RNase inhibitor (NEB), 10 nM RNA, and 5 nM protein (unless otherwise specified) at 37°C. Single-point assays of wild-type and mutant hR6CAT were conducted for 90 min. Time-course assays were conducted by removing 10-μL aliquots at each interval (0, 8, 15, 30, 60, 120 min; 0, 0.5, 1, 2, 4, 8, 15 min; or 90 min). Reactions were quenched by the addition of 10 μL of loading buffer (95% formamide, 20 mM EDTA, 1% DNA loading dye [Bio-Rad]) and heated for 30 sec at 80°C. Samples (10 μL each in volume) were loaded onto a 15% polyacrylamide–8 M urea gel (Invitrogen) for electrophoresis. Gels were imaged using a Fuji FLA-5000 scanner.
DATA DEPOSITION
Structure factors and coordinates have been deposited in the PDB with accession codes 3SAF, 3SAG, and 3SAH.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work is based in part on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the SGX Collaborative Access Team (SGX-CAT) beamline facilities at Sector 31 of the Advanced Photon Source was provided by SGX Pharmaceuticals, who constructed and operated the facility when these data were collected. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. K.J. is a fellow of the American Cancer Society and the Michigan Cancer Research Fund (PF-10-236-01-RMC). This work was supported in part by NIH grant GM079196 (to K.J., Q.L., and C.D.L.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2763111.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D 1999a. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P 1999b. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev 13: 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841 [DOI] [PubMed] [Google Scholar]
- Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH 2008. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA 14: 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese LS, Steitz TA 1991. Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J 10: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluthner M, Bautz FA 1992. Cloning and characterization of the cDNA coding for a polymyositis-scleroderma overlap syndrome-related nucleolar 100-kD protein. J Exp Med 176: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluthner M, Mahler M, Muller DB, Dunzl H, Bautz FA 2000. Identification of an alpha-helical epitope region on the PM/Scl-100 autoantigen with structural homology to a region on the heterochromatin p25β autoantigen using immobilized overlapping synthetic peptides. J Mol Med 78: 47–54 [DOI] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139: 547–559 [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem 273: 13255–13263 [DOI] [PubMed] [Google Scholar]
- Brouwer R, Vree Egberts WT, Hengstman GJ, Raijmakers R, van Engelen BG, Seelig HP, Renz M, Mierau R, Genth E, Pruijn GJ, et al. 2002. Autoantibodies directed to novel components of the PM/Scl complex, the human exosome. Arthritis Res 4: 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP 2005. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 [DOI] [PubMed] [Google Scholar]
- Callahan KP, Butler JS 2008. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res 36: 6645–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS 2010. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem 285: 3540–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Canavan R, Bond U 2007. Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res 35: 6268–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Shaw RJ, Wills MA, Belostotsky DA 2000. Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J Biol Chem 275: 33158–33166 [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JL, Stead JA, Feigenbutz M, Jones RM, Mitchell P 2011. The C-terminal region of the exosome-associated protein Rrp47 is specifically required for box C/D small nucleolar RNA 3′-maturation. J Biol Chem 286: 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M Jr 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL 2002. The PyMOL molecular graphics system. http://www.pymol.org DeLano Scientific, San Carlos, CA [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Estevez AM, Kempf T, Clayton C 2001. The exosome of Trypanosoma brucei. EMBO J 20: 3831–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G 2003. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep 4: 889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomproix N, Hernandez-Verdun D 1999. Effects of anti-PM-Scl 100 (Rrp6p exonuclease) antibodies on prenucleolar body dynamics at the end of mitosis. Exp Cell Res 251: 452–464 [DOI] [PubMed] [Google Scholar]
- Greimann JC, Lima CD 2008. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol 448: 185–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P, Kufel J 2008. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell 32: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2008. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta 1779: 239–246 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Januszyk K, Lima CD 2010. Structural components and architectures of RNA exosomes. In RNA exosome (ed. Jensen TH), Vol. 702, pp. 9–28 Landes Bioscience and Springer Science, New York [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L, Fang F, Butler JS, Sherman F 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci 101: 8581–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM 1993. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lebreton A, Seraphin B 2008. Exosome-mediated quality control: Substrate recruitment and molecular activity. Biochim Biophys Acta 1779: 558–565 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E 2005. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol 12: 575–581 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Conti E 2008a. Structural organization of the RNA-degrading exosome. Curr Opin Struct Biol 18: 709–713 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E 2008b. Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell 29: 717–728 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Brodersen DE, Jensen TH 2009. Origins and activities of the eukaryotic exosome. J Cell Sci 122: 1487–1494 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A 2011. The eukaryotic RNA exosome: Same scaffold but variable catalytic subunits. RNA Biol 8: 61–66 [DOI] [PubMed] [Google Scholar]
- Mahler M, Raijmakers R 2007. Novel aspects of autoantibodies to the PM/Scl complex: Clinical, genetic and diagnostic insights. Autoimmun Rev 6: 432–437 [DOI] [PubMed] [Google Scholar]
- Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE 2006. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci 103: 11898–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′ → 5′ degradation. Mol Cell 11: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol 23: 6982–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov V, Mushegian AR, Koonin EV, Bork P 1997. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem Sci 22: 417–418 [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876 [DOI] [PubMed] [Google Scholar]
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA 1985a. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313: 762–766 [DOI] [PubMed] [Google Scholar]
- Ollis DL, Kline C, Steitz TA 1985b. Domain of E. coli DNA polymerase I showing sequence homology to T7 DNA polymerase. Nature 313: 818–819 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Phillips S, Butler JS 2003. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9: 1098–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CC, Campbell JL 2007. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics 175: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Pruijn GJ 2007. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res 35: 2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH 2008. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33: 501–510 [DOI] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D 2007. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell 27: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D 2009. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res 37: 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ 2010. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J 29: 2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead JA, Costello JL, Livingstone MJ, Mitchell P 2007. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res 35: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF 2000. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 8: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Dziembowski A 2010. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA 16: 1692–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29: 2342–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189 doi: 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A 2007. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci 104: 16844–16849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Stevenson AL, Kearsey SE, Watt S, Bahler J 2008a. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol 28: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jia H, Jankowsky E, Anderson JT 2008b. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA 14: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Zuo Y, Wang Y, Malhotra A 2005. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure 13: 973–984 [DOI] [PubMed] [Google Scholar]