Abstract
Mycalamide B (MycB) is a marine sponge-derived natural product with potent antitumor activity. Although it has been shown to inhibit protein synthesis, the molecular mechanism of action by MycB remains incompletely understood. We verified the inhibition of translation elongation by in vitro HCV IRES dual luciferase assays, ribosome assembly, and in vivo [35S]methinione labeling experiments. Similar to cycloheximide (CHX), MycB inhibits translation elongation through blockade of eEF2-mediated translocation without affecting the eEF1A-mediated loading of tRNA onto the ribosome, AUG recognition, or dipeptide synthesis. Using chemical footprinting, we identified the MycB binding site proximal to the C3993 28S rRNA residue on the large ribosomal subunit. However, there are also subtle, but significant differences in the detailed mechanisms of action of MycB and CHX. First, MycB arrests the ribosome on the mRNA one codon ahead of CHX. Second, MycB specifically blocked tRNA binding to the E-site of the large ribosomal subunit. Moreover, they display different polysome profiles in vivo. Together, these observations shed new light on the mechanism of inhibition of translation elongation by MycB.
Keywords: Mycalamide B, eukaryotic ribosome, translation elongation, tRNA
INTRODUCTION
Small molecules have played a big role in the elucidation of the structure and function of the ribosome in prokaryotes (Yonath 2005). More recently, high-throughput screening in conjunction with efforts to elucidate the mechanisms of action of antiproliferative natural products have led to the identification of several interesting inhibitors of eukaryotic translation (Novac et al. 2004; Low et al. 2005; Moore 2010; Schneider-Poetsch et al. 2010; Cencic et al. 2011). As translation plays an essential role in cell proliferation and survival, and fast-proliferating cancer cells are particularly dependent on protein synthesis, inhibitors of translation have potential in becoming useful leads in anticancer drug development.
A number of inhibitors of prokaryotic translation have been used as antibiotics in the clinic over the past few decades (Poehlsgaard and Douthwaite 2005; Yonath 2005). More recently, several inhibitors of eukaryotic protein synthesis have entered the cancer drug development pipeline with a few advancing to Phase I and Phase II clinical trials, establishing translation as a promising target for chemotherapy (Pelletier and Peltz 2007). However, most of the translation inhibitors did not succeed in clinical trials, often due to dose-limiting toxicity. It has been proposed that translation inhibitors may be more effective and less toxic when administered in conjunction with other therapeutic agents (Pelletier and Peltz 2007). Aside from their clinical potential, the discovery of specific inhibitors of eukaryotic translation has enhanced our understanding of the similarities and differences between the translational apparatus of eukaryotes and bacteria. The discovery and characterization of new inhibitors of translation will likely improve our knowledge and offer leads to develop therapeutic agents.
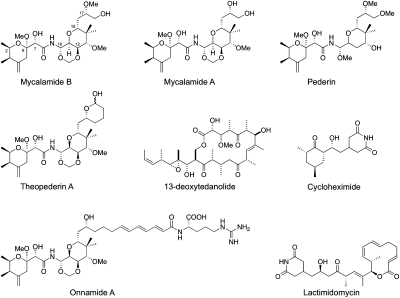
Mycalamides A and B (MycA and MycB) belong to a family of structurally related natural products of distinct origins including onnamide, pederin, and theopederins (Fig. 1). Mycalamides were originally isolated from the marine sponge of the Mycale genus off the coast of New Zealand (Burres and Clement 1989). MycB possesses potent antitumor and immunosuppressive activities, inhibiting the growth of several tumor cell lines with IC50 values in the low nanomolar range and blocking T-cell activation in vitro (Burres and Clement 1989; Galvin et al. 1993). It can also reverse the morphological changes associated with Ras-transformed NRK-cells to a normal state (Ogawara et al. 1991). Its congener MycA has a similar effect and has been shown to inhibit tumor growth in several murine allograft and human solid-tumor xenograft models (Burres and Clement 1989). Mycalamides and structurally related natural products have previously been reported to inhibit protein synthesis. For example, Pederin has been reported to inhibit translation at the translocation step (Brega et al. 1968; Barbacid et al. 1975). Recently, the structure of MycA bound to an archaeal (Haloarcula marismortui) ribosome was solved and it revealed that MycA binds to the E-site of the large ribosomal subunit (Gurel et al. 2009). Despite the structural information, however, how binding of MycA or MycB to the E-site of the large ribosomal subunit affects the function of the ribosome remains largely unknown. Furthermore, the structural study was based on an archaeal ribosome, which differs significantly from its eukaryotic counterpart.
FIGURE 1.
The structures of MycB, its congeners, and other translation inhibitors.
To elucidate the mechanism of translation inhibition by MycB in eukaryotes, a biochemical approach was taken to dissect the translation step interfered by MycB. The results confirmed that MycB primarily targets the translation elongation step in vivo and in vitro. Chemical footprinting of the large ribosomal subunit rRNA revealed that it binds to the same position in the E-site as the CCA tail of deacylated tRNA. Upon binding to the E-site, MycB prevents movement of the tRNA from the P-site to the E-site. Furthermore, MycB inhibits stress granule (SG) formation in vivo, as do other translation elongation inhibitors.
RESULTS
Inhibition of translation underlies the antiproliferative effect of MycB
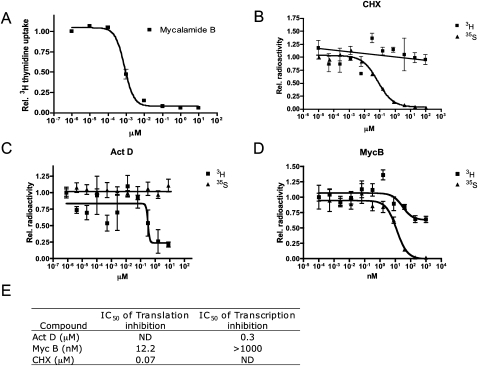
Although MycB has been reported to inhibit translation, it remains unclear whether this inhibition is responsible for its antiproliferative effect in cancer cells. To address this question, we determined the effect of MycB on both protein and RNA synthesis. We used a synthetic sample of MycB that has been previously fully characterized and structurally verified (Jewett and Rawal 2010). We found the IC50 of the synthetic MycB against HeLa cell proliferation to be ∼1 nM, corroborating the earlier reports (Fig. 2A; Burres and Clement 1989). Cells were metabolically labeled with [35S]methionine and cysteine or [3H]uridine for 2 h in the absence or presence of varying concentrations of MycB. MycB was compared with the established translation and transcription inhibitors cycloheximide (CHX) and actinomycin D (Act D), respectively (Fig. 2B–D). MycB blocked protein synthesis in vivo at ∼12 nM with little effect on transcription. Even at 1 μM, MycB suppressed RNA synthesis by <50%. We note that there is a significant difference in the observed IC50 values between cell proliferation and translation assays (Fig. 2E), which may be attributed to the different incubation times and intrinsic sensitivity of the different readouts.
FIGURE 2.
MycB selectively inhibits translation. (A) MycB inhibits HeLa cell proliferation at an IC50 of ∼1 nM. (B–D) HeLa cells were treated with different concentration of CHX (B), Act D (C), and MycB (D), and labeled with [3H]uridine and [35S]methionine and cystine for 2 h. Cells were harvested and transferred onto glass fiber filters. Remaining radioactivity was counted and plotted. (E) The IC50 values of translation and transcription inhibition of CHX, ActD and MycB.
MycB inhibits translation elongation
While MycB's effect on translation in general has been observed previously (Burres and Clement 1989), whether it interferes with the initiation or elongation phase of protein synthesis has not been determined. To distinguish between those possibilities, we took advantage of internal ribosome entry sequences (IRES) from several viruses that can direct translation of target genes by bypassing the requirement for several canonical initiation factors. While the IRES of the hepatitis C virus (HCV) enables translation initiation without a functional eIF4F complex that contains the cap-binding proteins eIF4E and initiation factors eIF4A and eIF4G (Pestova et al. 1998), the IRES of the cricket paralysis virus (CrPV) circumvents the entire initiation apparatus (Jan and Sarnow 2002).
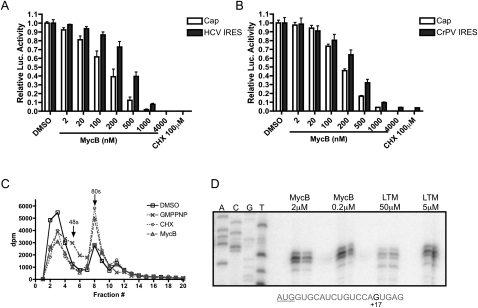
Transcribed HCV IRES and CrPV IRES dual luciferase reporters were used to perform in vitro translation assays with rabbit reticulocyte lysates. MycB dose-dependently inhibited cap-dependent translation and IRES-dependent translation of the mRNAs from both dual luciferase reporters (Fig. 3A,B). These results suggest that MycB inhibits translation at the elongation phase. Next, we carried out an in vitro translation sucrose gradient profiling in RRL to assess the distribution of ribosomal populations using the radioactivity of [32P]UTP incorporated into rabbit β-globin mRNA as readout. As shown in Figure 3C, MycB clearly blocked the ribosome after completing 80S complex formation and had a similar effect on mRNA distribution as CHX. But, it differed from the mRNA distribution pattern obtained with the nonhydrolyzable GTP analog, GMPPNP, which prevents the GTP-dependent coupling of 60S and 40S subunits, consequently resulting in an increased 48S population. The 80S peak fractions were immediately used for toeprinting with a primer hybridized 60 nt downstream from the AUG start codon of rabbit β-globin mRNA. MycB caused the 80S ribosome to stall at the start codon (or +17) position of β-globin mRNA, which is similar to the effect of lactimidomycin (LTM), another translation elongation inhibitor recently characterized by our group (Fig. 3D; Schneider-Poetsch et al. 2010). For CHX, as reported, the 80S arrests after one round of translocation at position +20 of the mRNA template (Supplemental Fig. 1). Together, these results indicated that MycB inhibits translation at the elongation phase similar to LTM and does not allow the ribosome to progress through a full elongation cycle.
FIGURE 3.
MycB inhibits translation elongation. HCV (A) and CrPV (B) IRES dual reporters were used in in vitro RRL translation assays in the presence of different concentrations of MycB and 100 μM CHX. (C) 32P-labeled β-globin mRNA was incubated with RRL in the presence of GMPPNP, CHX, and MycB, and subjected to ultracentrifugation. The radioactivity of the aliquots was counted and the data were processed with GraphPad software. (D) In a separate set of experiments 80S fractions of lysates treated with the indicated compounds were collected and utilized in toeprinting assays by primer extension. The sequencing reactions were performed with full-length β-globin and separated on the far left four lanes.
MycB inhibits eEF2-mediated ribosome translocation
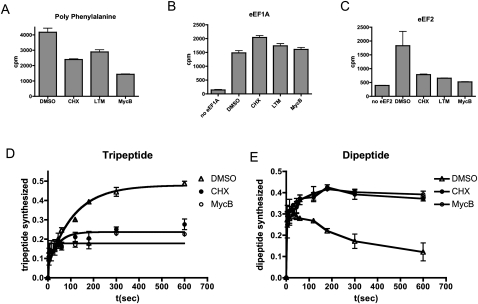
Translation elongation can be divided into three steps, beginning with the G protein eEF1A delivering aminoacyl-tRNA to the empty A-site, followed by peptidyl transfer and the eEF2-mediated peptidyl-tRNA translocation from A-site to P-site, with concomitant transfer of deacylated tRNA from P to E-site. To determine which step was affected by MycB, polyuridine-directed polyphenylalanyl synthesis was used (Fig. 4A). Using purified ribosomes, eEF1A, eEF2, GTP, and [15C]phenylalanine-charged tRNA, CHX, LTM, and MycB dramatically inhibited the synthesis of polyphenylalanine, though the percentages of inhibition were different. Furthermore, when the activity of eEF1 was measured in the presence of the above-mentioned inhibitors, none showed any inhibition (Fig. 4B), suggesting that MycB functions downstream from aminoacyl-tRNA binding to the A-site. Since eEF2 mediates peptidyl-tRNA translocation, we checked for possible inhibition of translocation by MycB, CHX, and LTM by evaluating the puromycin reactivity of A-site tRNA. MycB blocked the movement of tRNA and prevented the formation of phenylalanyl-puromycin (Fig. 4C).
FIGURE 4.
MycB inhibits translocation. (A) Polyuridine and 14C-labeled phenylalanine-charged tRNAPhe were used in in vitro reassembled translation assays in the presence of 200 μM CHX, 100 μM LTM, and 20 μM MycB. Synthesized polyphenylalanine was precipitated onto nitrocellulose filters and the radioactivity was counted. (B) eEF1A-mediated tRNA binding was measured as described above, except that a higher amount of eEF1A was applied and GTP was replaced with GMPPNP. 14C-labeled Phenylalanyl-charged tRNAPhe remaining on the ribosomes was detected after binding to a nitrocellulose membrane. (C) eEF2-mediated translocation assays were performed in the presence of the three compounds. After the reaction, the final [14C]Phe-puromycin product was extracted with ethyl acetate and counted. (D,E) Reassembled translation elongation was performed in the presence of DMSO, 2.6 μM MycB, and 174 μM CHX. The template was switched to a designated RNA encoding Met-Phe-Phe. [35S]methionine-charged tRNAMet was used to label the synthesized peptide. The final tripeptide (D) and dipeptide (E) products were separated by electrophoretic TLC, and were detected on a PhosphorImager plate and quantified using ImageQuant 5.2 software. The final values of dipeptide and tripeptide signals were normalized to the total amount of signal in the lane.
Up to this point, we had not ruled out an effect of MycB on peptide bond formation. We thus subjected MycB to a dipeptide and tripeptide formation assay using a short mRNA template coding for Met-Phe-Phe, which allows for a dynamic parsing of translation elongation. The products of the reaction were separated on an electrophoretic cellulose thin-layer chromatography plate. MycB inhibited tripeptide formation without affecting the formation of dipeptides (Fig. 4D,E). These results ruled out the possibility that MycB inhibited peptidyl transfer, corroborating our earlier observation that MycB blocked eEF2-mediated translocation.
MycB gives rise to a footprint at the E-site of the larger ribosomal subunit
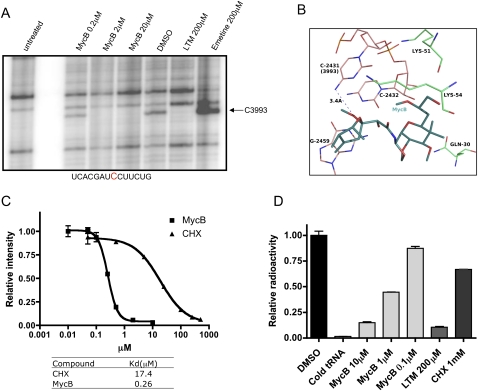
Chemical footprinting has been widely used to map the site of binding of small molecule inhibitors to the ribosome. Previously, we have utilized this method to determine specific chemical footprints of both LTM and CHX on the large eukaryotic ribosomal subunit (Schneider-Poetsch et al. 2010). In the case of MycB, the availability of an X-ray crystal structure of its complex with the ribosome already provided a detailed view of how it interacts with the ribosome, albeit with the ribosome of an archaeal organism, in the E-site. This also offered a unique opportunity to determine how well results of chemical footprinting correlate with those from an X-ray structure. Thus, ribosomes were incubated with each of the inhibitors, followed by treatment with dimethyl sulfate (DMS). Footprints were obtained with extracted ribosome RNA by primer extension using avian myoblastoma (AMV) reverse transcriptase. Thus, treatment of ribosomes with DMS caused methylation of C3993 on the 28S rRNA of the large ribosomal subunit among other bases in comparison with untreated control, giving rise to the corresponding footprint (Fig. 5A). Pretreatment of ribosomes with either MycB or LTM protected C3993 from DMS methylation, eliminating the C3993 footprint. It is worth noting that the C3993 footprint protection was also seen with CHX (Supplemental Fig. 2) but not emetine, another translation elongation inhibitor that does not bind the E site of the ribosome (Fig. 5A). We also titrated MycB in the footprinting assays to estimate its dissociation constant (Kd) in comparison with that of CHX. Ribosomes were used at 50 nM. The Kd is ∼260 nM for MycB and 17 μM for CHX (Fig. 5C; Supplemental Fig. 2), which is in agreement with the value reported previously (Schneider-Poetsch et al. 2010).
FIGURE 5.
MycB is bound to E site of the larger ribosomal subunit. (A) Ribosomes were incubated with different compounds as indicated and then methylated with DMS. Extracted rRNAs were subjected to reverse transcription. rRNA not treated with DMS was labeled as “untreated” and served as a negative control. The 32P-labeled DNA was resolved on a denaturing polyacrylamide gel and was detected by PhosphorImager. (B) Interaction of MycB with the H. marismortui ribosome (PDB ID: 3I55): A methyl group has been added onto the O-17 of MycA (original structure) to turn the structure into MycB. The numberings correspond to H. marismortui ribosome. Note the potential H-bonding interaction between the 6-OMe group of MycB and N4-C2431. This is the conserved cytidine corresponding to C3993 in the eukaryotic ribosome for which we have shown chemical footprinting evidence. (C) To measure the dissociation constants of MycB and CHX, ribosomes were incubated with serially diluted compounds and treated with DMS. Extracted rRNAs were subjected to reverse transcription. The 32P-labeled DNA was resolved on a denaturing polyacrylamide gel and was detected by PhosphorImager. The intensity of the band was quantified and plotted using GraphPad software from two independent experiments. One gel is shown in Supplemental Figure 2. (D) The E-site tRNA-binding assay was performed with purified ribosomes with the A and P site occupied by Phe-charged tRNAPhe and the acetylated Phe-charged tRNAPhe, respectively. 32P-labeled deacylated tRNAPhe was added in the presence of the compounds as indicated. Ribosomes were bound to nitrocellulose membranes and the [32P]tRNA was measured by scintillation counting.
The binding of MycB to the E-site suggested that MycB should compete for binding with deacylated tRNA. To test this prediction, MycB was first incubated with purified 80S ribosomes before 32P-labeled deacylated tRNA was added. The amount of ribosome-bound tRNA was determined by scintillation counting. MycB inhibited deacylated tRNA binding to the ribosome in a dose-dependent manner, albeit with significant background signal (data not shown). We suspected that deacylated tRNA bound nonspecifically to both A and P sites. To limit this nonspecific binding, we filled the A and P sites with Phe-charged tRNA (tRNAphe) and acetylated Phe-tRNAphe, each of which was purified by HPLC. For the same binding assay (Fig. 5D), the cold tRNA almost completely blocked binding of 32P-labeled deacylated tRNA, demonstrating that under these conditions all deacylated tRNA bound exclusively to the E-site. MycB did dose dependently prevent the deacylated tRNA from binding to the E site, as did LTM. As reported previously, CHX did not significantly compete with tRNA binding, even at 1 mM concentration (Schneider-Poetsch et al. 2010).
MycB inhibits stress granule formation through the blockade of translation elongation
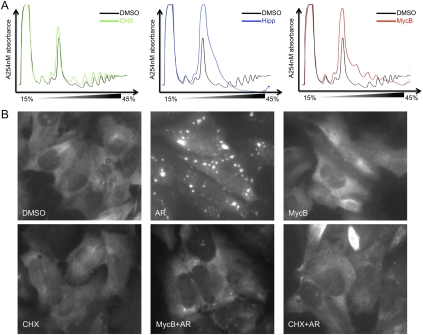
The buildup of 80S ribosomes in vitro in the presence of MycB suggested that MycB would also change the distribution of ribosomal populations in vivo. We thus analyzed the polysome profile of HEK293T cells in the presence of MycB, CHX, and hippuristanol (Bordeleau et al. 2006), a translation initiation inhibitor. As shown in Figure 6A, MycB treatment increased the amount of 80S ribosomes, and oligoribosome, and decreased polysome levels in comparison to DMSO control. As expected, hippuristanol abolished polysomes and dramatically increased an apparent 80S peak, due to its effect on translation initiation. However, in the case of translation initiation inhibitors such as hippuristanol or pateamine A, this apparent 80S fraction results from a cellular stress response (Low et al. 2005; Dang et al. 2006; Mazroui et al. 2006) and does not reflect an actual buildup of fully assembled ribosomes, as both compounds arrest translation long before ribosomal subunit joining occurs. Moreover, in the presence of MycB, the increase of 80S and oligoribosomes and the decrease of polysomes are dynamic and time dependent. After a 30-min treatment, the polysome fractions appear to be completely wiped out by MycB, but seem slightly increased under CHX treatment (Supplemental Fig. 3). The observed profile for MycB is in agreement with our expectations and underlines the mechanistic difference between MycB and CHX. As previously reported, CHX stabilizes polysomes, as it can bind to the E site in the presence of a deacylated tRNA (Schneider-Poetsch et al. 2010), thereby also halting ribsomes that have traveled further downstream from the start codon. MycB, which competes with the deacylated tRNA for binding, consequently inhibits protein synthesis primarily during the first round of elongation before any tRNA reaches the E site, but appears not to halt actively elongating ribosomes, consequently permitting ribosome runoff. We note that while all experimental data are consistent with the aforementioned ribosome runoff model, we cannot rule out an alternative possibility that the polysome depletion in the presence of MycB resulted from the dissociation of the 80S ribosome from both mRNA template and the peptidyl-tRNA upon exchange of MycB with deacylate tRNA in the E site. However, considering that CHX and MycB bind to the same position on the ribosome, it would be difficult to imagine why only one compound should lead to ribosome dissociation and not the other. Hence, the ribosomal runoff model appears to provide the more likely explanation.
FIGURE 6.
MycB affects polysome distribution and stress granule formation. (A) Polyribosome profiles determined from HEK293T cells after 1 h of treatment with 100 μg/mL CHX, 1 μM hippuristanol, and 100 nM MycB. (B) SGs induction performed in U2OS cells stably expressing GFP-G3BP in the presence of different compounds as indicated. The images were taken with an Axioskop microscope.
The formation of stress granules (SGs) reflects the status of translation in vivo and is caused by disturbing the translation initiation process, either through induction of eIF2a phosphorylation or interference with the function of eIF4A. Since MycB inhibits translation at a relatively early stage and increases the 80S peak, somewhat similar to hippuristanol or pateamine A, we further determined whether MycB induced stress granule formation or whether it interfered with their production. The induction of SGs was performed in U2OS cells stably expressing GFP-tagged G3BP, a stress-granule marker (Kedersha et al. 2005). Upon MycB treatment, no SG was observed, in contrast to the treatment with arsenite, a known inducer of SGs (Fig. 6B). This result is similar to that obtained with CHX. In addition, pretreatment with CHX or MycB blocked the induction of SGs by arsenite, which is consistent with the previous reports that blocking translation elongation can prevent the SG formation (Kedersha et al. 2000). This result was further confirmed by the ablation of processing bodies (PBs) by MycB treatment in a manner similar to CHX (Supplemental Fig. 4), as PB-formation can also be blocked by translation elongation inhibitors. The results further suggest that stress granule formation only occurs if translation initiation, but not elongation, is perturbed, and inhibition of elongation prevents SG formation.
DISCUSSION
The attainment of high-resolution structures of the ribosome has enabled rapid advances in the field of protein synthesis. In the case of antibacterial agents, it became possible to validate their mechanisms in unprecedented detail and to resolve minute details that evaded classic biochemical dissection (Yonath 2005; Moore 2010). Nevertheless, crystal structures only provide a single-frame snapshot of a dynamic process and biochemical verification is essential to fully understanding the mechanisms. Furthermore, crystallography has only recently advanced to resolving true eukaryotic ribosomes. Therefore, biochemical dissection adds credence to known structures obtained from prokaryotic organisms.
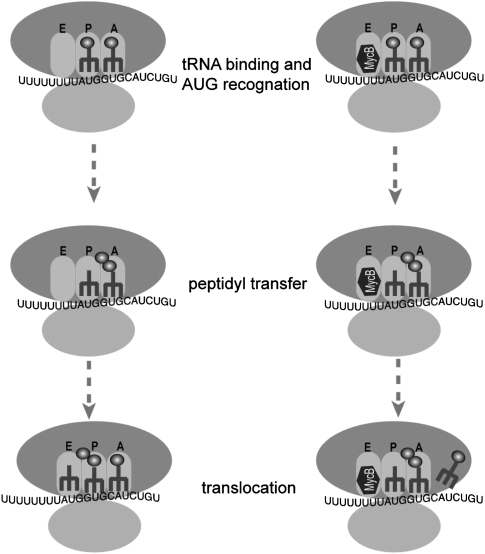
In this study we demonstrated that MycB inhibited eukaryotic translation elongation by occupying the large subunit's E site, thereby preventing translocation of deacylated tRNA from the P sie to the E site (Fig. 7). The result is consistent with the crystallographic data obtained on the closely related molecule MycA, which was shown to bind to the E-site of the large archaeal ribosomal subunit. The present findings support the notion that the E-site portion of the ribosome shares significant similarity between Achaea and eukaryotes, but not bacteria (Gurel et al. 2009).
FIGURE 7.
A model of the mechanism of action of MycB. (Left) Normal translation elongation; (right) MycB blocks deacylated tRNA progression elongation through binding to the E-site.
Three independent pieces of evidence support that MycB inhibits translation elongation. First, we were able to confirm inhibition of translation elongation, since MycB inhibited cap-dependent and initiation factor-independent IRES reporter translation. Furthermore, MycB inhibited in vitro polyphenylalanine synthesis in the absence of any initiation factor. Second, with 32P-labeled β-globin mRNA, MycB halted the translation progress after assembly of 80S ribosome in RRL, in a manner similar to CHX. As CHX is significantly smaller than MycB or LTM, it can leave enough space for tRNA shuttling from the P site to the E site. Last, MycB has a toeprint similar to LTM (Schneider-Poetsch et al. 2010), another elongation inhibitor. The two GTPases, eEF1A and eEF2, as well as the ribosome, catalyze translation elongation. MycB dose-dependently inhibited eEF2-mediated translocation, but not eEF1A's function. MycB also did not interfere with peptide bond formation itself, as evidenced by the uninhibited formation of dipeptides in the presence of MycB. Neither CHX nor MycB affected the formation of dipeptides, but clearly prevented the third amino acid from getting incorporated. At first the inhibition of tripeptide formation by CHX appears a bit surprising as CHX inhibits translation together with a deacylated tRNA. However, the reaction mixture contained a vast excess of free deacylated tRNA, which likely bound the ribosomal E site together with the small molecule, thereby allowing CHX to interrupt translation before the third peptide bond was formed.
Regarding the difference between the in vivo polysome profiles of MycB and CHX, binding of the smaller CHX to the E site can occur simultaneously during the translation process and seems not limited to early elongation alone. However, MycB appears to mainly block translation when it occupies the empty E site before or during the first cycle of elongation, but allows ongoing translation elongation to run off. This explains the observed accumulation of 80S ribosomes and oligoribosomes with a concomitant depletion of the polysome population. Since the polysome profile still shows some remaining polysomes in the presence of MycB, it is possible that the small molecule occasionally binds between release of deacylated tRNA from the E site and the next round of translocation. The induction of SGs only occurs during translation initiation and can be suppressed by translation elongation inhibitors such as emetine and CHX (Kedersha et al. 2000; Dang et al. 2006; Anderson and Kedersha 2009). In the presence of MycB, the induction of SGs by arsenite or pateamine A (data not shown) is attenuated in a manner similar to CHX, indicating that the increase in 80S ribosomes in vivo by MycB is unrelated to the induction of SGs, and that MycB is capable of suppressing the formation of SG at least as efficiently as CHX. Based on the Kd of the two inhibitors, MycB exhibited a higher affinity for the ribosome with a dissociation constant of 260 nM, while CHX binds more loosely with a Kd of ∼17 μM. We cannot rule out the possibility that this lower affinity also contributes to the different properties of the two inhibitors.
The MycB binding site identified by chemical footprinting agrees well with our mechanistic findings as well as with the reported archaeal crystal structure. The protected residue C3993 normally interacts with the 3′OH group of the deacylated tRNA (Schneider-Poetsch et al. 2010). As per the structural evidence, the 6-methoxy group of mycalamide lies within a hydrogen-bonding distance from N-4 of C3993 (C2394 in Achaea) (Fig. 5B). Although MycB's 11-O and 7-OH may also form hydrogen bonds with other atoms of the 28S/23S rRNA, we did not observe any footprint other than C3993. The yeast ribosome crystal structure has recently been resolved at 4.15Å resolution (Ben-Shem et al. 2010). A relevant snippet from the structural superimposition of MycA-bound archael ribosome and the yeast ribosome is shown in Supplemental Figure 5. Despite significant differences between the archaeal and yeast ribosomes, the E-site region where we have observed the footprint appears to superimpose remarkably well with the consensus nucleotide residues: G2459–G2793, C2431–C2764, and C2432–C2765 from 23S of Archaea and 25S of yeast large ribosomal subunits, respectively. Cytidine-2431 (archaea), and C2764 (yeast) correspond to C3993 in the 28S rRNA of the mammalian ribosome. On the other hand, ribosomal proteins L28 in bacteria, L44E in archaea, and L42 in the case of yeast occupy this region. In the MycA-bound archaeal ribosome structure, Gln30, Lys54, and Lys51 (all from L44E) appear to make contacts with MycA. In contrast, in the overlay with yeast ribosome, Ala33, Pro56–Val57, and Thr54 (all from L42) are found to fill those spots. Given the divergence in the ribosomal proteins, the additional interactions of MycB with the higher eukaryotic ribosomal protein(s) may only be mapped when further biochemical or structural evidences become available.
The ability of MycB to compete with deacylated tRNA for E-site binding in a dose-dependent manner underscores the physiological relevance of the identified binding pocket. Binding of MycB to this position did come as a surprise considering that two other translation inhibitors, CHX and LTM, also protect C3993. While the two aforementioned molecules share significant structural similarity, MycB bears no resemblance to these two glutarimide-containing inhibitors. It is interesting to note that the binding of three different classes of inhibitors—CHX, LTM, and MycB, as well as 13-deoxytedanolide—converge on the eukaryotic E site, whereas none of the known antibacterial translation inhibitors work by a comparable mechanism. That the E site of eukaryotic ribosome is susceptible to inhibition by structurally distinct inhibitors with drastically different origins—bacteria for CHX and LTM and sea sponges for MycB and 13-deoxytedanolide—suggest that this site has significantly different properties from its bacterial counterpart, which has been taken advantage of during the evolution of new bioactive natural products. It is intriguing as to why no antibiotics have been found to target the E site of the prokaryotic ribosome, given the abundance of antibiotics that target other essential processes in prokaryotes. It is possible that the very flexibility of the E site, as seen in yeast among the CHX- and LTM-resistant mutants, renders it easy to evolve resistance toward antibiotics targeting this site of the ribosome.
13-Deoxytedanolide is structurally related to the class of macrolide antibiotics that normally inhibit bacterial translation by blocking the nascent peptide tunnel, similar to erythromycin (Schroeder et al. 2007). It inhibits eukaryotic translation translocation through directly binding to ribosome large subunit. It can compete with pederin and its congeners, suggesting that it may target the same location on the 60S subunit. Interestingly, it cannot compete with CHX, which shares the same binding site with MycB and LTM (Nishimura et al. 2005). Unlike its relatives, however, 13-deoxytedanolide does not inhibit bacterial protein synthesis. Thus far, no small molecule has been reported to target the E site of eubacterial ribosome (Yonath 2005; Moore 2010). This difference in specificity may be the result of distinct differences in protein composition at the E site of bacteria on one hand and eukaryotes and Achaea on the other. Binding of MycB appears to depend on ribosomal protein L44e (L36a in eukaryotes) (Gurel et al. 2009). In its place bacterial ribosomes contain the unrelated protein L28. Pederin, theopederins, mycalamides, onnamides, and icadamides are the major pederin polyketides that share, by-and-large, a common structural subunit called pederic acid that spans from O-1 to C-10, and their structures diverge only after C-10. A pharmacophore model that was advanced to account for the cellular effect of these polyketides concluded that the pederic acid subunit is the most critical and invariable moiety that is prone to loosing activity with slight changes of substitution or stereochemistry (Soldati et al. 1966; Burres and Clement 1989; Galvin et al. 1993; Richter et al. 1997; Narquizian and Kocienski 2000; West et al. 2000; Hood et al. 2001). It is highly likely that it is this very pederic acid portion of the translation inhibitors MycA, MycB, onnamide A, pederin, and theopederin A that is responsible for conferring specificity for the ribosomal E-site (Fig. 1).
Considering the utility of antibiotics targeting protein biosynthesis and a pressing need for new clinical agents, development of a bacterial equivalent of the eukaryotic translation inhibitor LTM or MycB may prove extremely valuable. Understanding the mechanism of these inhibitors will facilitate their potential clinical application in cancer therapy.
MATERIALS AND METHODS
Reagents and cell lines
MycB and hippuristanol were synthesized as reported (Li et al. 2009; Jewett and Rawal 2010) and dissolved in DMSO. CHX, dimethyl sulfate (DMS), and actinomycin D were purchased from Sigma. Yeast tRNAphe was purchased from Chemical Block. eEF1 and eEF2 were generously supplied by Dr. Merrick at Case Western Reserve University. All short RNAs were purchased from Invitrogen. HEK293T and HeLa cells were cultured in DMEM medium supplemented with 10% FBS and were maintained in a 5% CO2 atmosphere. The vector pβ-Hb carrying β-globin cDNA was generously provided by Dr. Karen Browning at the University of Texas. The HCV IRES dual luciferase reporter vector was provided by Dr. Jerry Pelletier at McGill University and CrPV IRES dual luciferase reporter vector came from Dr. Peter Sarnow at Stanford University. U2OS cells expressing GFP-G3BP and RFP-Dcp1a were provided by Drs. Paul Anderson and Nancy Kedersha at Harvard University.
Metabolic labeling
HeLa cells were used for cellular metabolic labeling experiments. For cell proliferation assays, cells were plated onto 96 well plates at 5000 cells per well, allowed to adhere overnight, and treated with different concentrations of MycB as indicated in Figure 2 for 18 h. The cells were then labeled with [3H]thymidine (1 μCi per well) (PerkinElmer) for another 6 h. Cells were printed onto GF/C glass fiber filters using a Tomtec harvester and washed before the activity of the remainder of [3H]thymidine on the filters was counted.
For in vivo transcription and translation assays, 10,000 cells per well in 96-well plates were labeled by [3H]uridine (1 μCi per well) (PerkinElmer) or the mixture of [35S]methionine and cystine (PerkinElmer) (0.5 μCi per well) for 2 h in the presence of different compounds at increasing concentrations as indicated in Figure 2. For in vivo transcription assays, cells were harvested and processed as stated above for the cell proliferation assay. For the in vivo translation measurements, after labeling, cells were washed with PBS and lysed with RIPA buffer. The total proteins were precipitated with 5% trichloroacetic acid (TCA) and passed through a filter. After extensively washing with 5% TCA and drying, the filters were scintillation counted.
Dual luciferase reporter assay
HCV and CrPV IRES dual luciferase reporter vectors were linearized with BamHI (NEB) and transcribed using SP6 or T7 polymerase (Promega) (Novac et al. 2004). Briefly, each reaction was performed using 10 μL of the Flexi rabbit reticulocyte lysate (RRL, from Promega), 200 ng of RNA, 0.2 μL each of Met and Leu amino acid mixtures, 70 mM KCl, 2 mM DTT, and 10 U of RNaseout (Invitrogen) in 20 μL with the indicated concentration of compounds. The mixtures were incubated at 30°C for 1 h and the reaction was quenched with 20 μL of passive lysis buffer (Promega), and a 10-μL aliquot was assayed for luciferase activity according to the instructions of the manufacturer (Dual-Luciferase reporter assay system; Promega).
Polyphenylalanine synthesis, eEF1, and eEF2 assays
Both methods were described in our previous report (Schneider-Poetsch et al. 2010). Briefly, yeast tRNAphe was charged with [14C]phenylalanine and used for both assays, with a specific activity of ∼1300 cpm per picomol. 80s ribosomes were purifed from RRL. For polyphenylalanine synthesis, 8 μg of polyuridine RNA, 0.4 OD260 of ribosomes, 2 μg of eEF1A, 0.5 μg of eEF2, 10 pmol of [14C]Phe tRNAPhe in 30 mM Tris-HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 1 mM GTP, 2.1 mM phosphoenol pyruvate, and 0.3 U pyruvate kinase were incubated for 5 min at room temperature with the compounds as indicated in Figure 4. Reactions were quenched with 1 mL of cold 10% TCA and boiled for 15 min. The samples were filtered through nitrocellulose filters and washed thee times with 5% TCA before scintillation counting. For the eEF1 assays, 89 pmol of ribosomes, 200 ng of polyuridylic acid, 10 pmol of [14C]Phe tRNAPhe, 2.2 μg of eEF1 and 150 μM of GDPPNP were incubated in the buffer (20 mM Hepes-KOH at pH 7.4, 100 mM KCl, 10 mM MgCl2, and 1 mM DTT) in the presence of the compounds as indicated in Figure 4, with eEF1A and tRNA being added last. The mixtures were reacted for 10 min at 37°C and diluted with a milliliter of the same buffer and immediately filtered through nitrocellulose and counted afterward. For eEF2 assay, the reaction was set up the same way as eEF1 assay, except GTP was used instead of GDPPNP. After incubation with the compounds as shown in Figure 4, 4.5 μL of 10x buffer, 0.5 μg of eEF2, 10 μL of 10 mg/mL puromycin, and 6 μL of 15 mM GTP were added and reacted for 10 min at 37°C. The reaction was quenched with 1.4 mL of cold ethyl acetate and thoroughly vortexed. The organic phase was separated by centrifugation and 1-mL aliquots were counted after mixing with scintillation cocktail.
Chemical footprints
The ribosomal RNA footprints were performed as described before (Merryman and Noller 1998; Schneider-Poetsch et al. 2010). Briefly, the purified 80S ribosomes were incubated with the indicated compounds at increasing concentrations (CHX [500, 250, 100, 25, 5, 0.5, and 0.05 μM] and MycB [10, 2, 0.5, 0.25, 0.1, 0.05, and 0.01 μM]) and treated with 90 mM DMS at 37°C for 5 min. The rRNAs were isolated with the RNAqueous kit from Ambion. Then, isolated rRNAs were used for primer extension (primer sequence: 5′-CTGCGTTACCGTTTGAC). The final DNA products were extracted and resolved on a DNA sequencing gel. The gel was dried and exposed to a PhosphorImager.
tRNA binding assay
Deacylated tRNAPhe was labeled with 32P as described previously (Ledoux and Uhlenbeck 2008; Walker and Fredrick 2008). Briefly, A 50-μL reaction containing 1 μM tRNAPhe, 50 μM of sodium pyrophosphate, 0.2 μM tRNA nucleotidyl transferase (CCA adding enzyme), 0.3 μM [α-32P]ATP in the buffer (20 mM MgCl2 and 50 mM Tris-HCl at pH 7.5), were incubated at 37°C for 5 min. CTP (1 μM final concentration) was added with 10 U/mL of yeast inorganic pyrophosphatase and incubated for two additional min. tRNA was extracted with phenol and chloroform and purifed through G50 columns (GE Healthcare).
To make acetylated Phe-charged tRNAPhe, after the charging reaction the reaction mixture was first desalted using a PD10 desalting column (GE Healthcare) and then purified by HPLC. Purified tRNA was diluted to 1.6 μM in 250 μL of cold 200 mM NaOAc (pH 5.2). Acetic anhydride (4 μL) was added to each tube and incubated on ice for 1 h, with the process being repeated one more time. The product was precipitated by ethanol and kept at −80°C until use. A 50-μL reaction contained 2 μg of polyuridine RNA, 2 pmol ribosomes, 6 pmol each of Phe-tRNAPhe and acylated Phe-tRNAPhe in the presence of the test compounds or 4 μM cold tRNA in 30 mM Hepes-KOH (pH 7.4), 25 mM MgCl2, 100 mM KOAc, 0.25 M sucrose for 10 min. Deacylated tRNAPhe labeled with 32P (6 pmol) was added and incubated at 37°C for 5 min. The reaction was stopped by the addition of the same buffer (1 mL, 1x) before being filtered through a nitrocellulose membrane. The dried filter was assayed using a scintillation counter.
Dipeptide and tripeptide sythesis assay
The 80S initiation complexes were assembled essentially as described previously (Acker et al. 2007; Saini et al. 2009) with slight modifications. A different reaction buffer (20 mM Tris-Cl at pH 7.5, 100 mM potassium acetate at pH 7.6, 2.5 mM magnesium acetate, 0.25 mM spermidine, 2 mM DTT) was used and tRNAMet was labeled exclusively with [35S]Met. In addition, the mRNA contained the ORF sequence AUGUUCUUCUAA. After assembly, initiation complexes were layered onto reaction buffer containing 1.1 M sucrose and centrifuged for 1 h at 424,000g in a TLA-100.3 rotor. Complexes were resuspended in reaction buffer, flash-frozen, and stored at −80°C. For each elongation reaction, Phe-tRNAPhe ternary complex was prepared. Each batch of ternary complex contained the following reagents: 60 pmol eEF1A, 50 fmol Met-tRNAMet, 25.6 pmol Phe-tRNAPhe, 4 pmol eEF2, 4 pmol eEF3, 1.3 mM GTP, 0.4 mM ATP, drug at 10 times the Kd or DMSO, and 1x reaction buffer, and was incubated at 26°C for 15 min. During the incubation, initiation complex was thawed and incubated with drug at ten times the Kd or vehicle for 3 min. At the end of this incubation, the ternary complex was mixed with the initiation complex and incubated at 26°C. Aliquots were removed at the indicated times and quenched in 50 mM KOH. Reaction products were separated by electrophoresis on cellulose TLC plates (pyridine-acetate buffer at pH 2.8; 1200 V, ∼35 min). The dipeptide signal is the fraction of all peptides that are Met-Phe. In the case of tripeptide, it is the amount of tripeptide Met-Phe-Phe. [35S]Met-containing reaction products were detected by PhosphorImaging and quantified using ImageQuant 5.2 software (GE Healthcare Life Sciences). The values in Figure 4, D and E, for dipeptide and tripeptide, are normalized to the total amount of available reactive material.
Cellular polysome fractionation
HEK293T cells were treated with different compounds for 1 h, then washed once with ice-cold PBS and lysed by pipetting in TMK100 buffer (20 mM Tris-Hcl at pH 7.4, 5 mM MgCl2 100 mM KCl, 2 mM DTT, 1% Triton X-100), 100 U/mL RNasin (Promega) and protease inhibitors (Roche Diagnostics). After spinning at 10,000g (10 min) at 4°C, the supernatants were loaded onto 10 mL, 10%–40% linear sucrose gradients containing 20 mM HEPES (pH 7.4), 100 mM KCl, 5 mM MgCl2, and 2 mM DTT. Ultracentrifugation at 40,000 rpm for 2 h at 4°C was performed in a SW41Ti rotor (Beckman). Gradient profiles were monitored at 254 nm from top to bottom.
Processing bodies and induction of stress granules
U2OS cells stably expressing GFP-G3BP and RFP-Dcp1α were treated with the indicated compounds for 1 h and then fixed with 4% polyformaldehyde. The cells were examined with an Axioskop microscope (Carl Zeiss, Inc.). Images were captured with a Sensys CCD camera (Photometrics Ltd.) using IP Lab software v3.1 (Scanalytics).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are grateful to Drs. Paul Englund, Gerald Hart, and Daniel Lane for their generous provision of advice and access to special equipment. We thank Drs. Nancy Kedersha, Paul Anderson, Peter Sarnow, Jerry Pelletier, Karen Browning, and William Merrick for reagents and plasmids. We thank Shan He and Wei Shi for assistance with the performance of experiments and preparation of the manuscript. This work was supported by a discretionary fund of the Liu Lab and the Keck Foundation (J.O.L.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2624511.
REFERENCES
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR 2007. Reconstitution of yeast translation initiation. Methods Enzymol 430: 111–145 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2009. Stress granules. Curr Biol 19: R397–R398 [DOI] [PubMed] [Google Scholar]
- Barbacid M, Fresno M, Vazquez D 1975. Inhibitors of polypeptide elongation on yeast polysomes. J Antibiot (Tokyo) 28: 453–462 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M 2010. Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2: 213–220 [DOI] [PubMed] [Google Scholar]
- Brega A, Falaschi A, De Carli L, Pavan M 1968. Studies on the mechanism of action of pederine. J Cell Biol 36: 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burres NS, Clement JJ 1989. Antitumor activity and mechanism of action of the novel marine natural products mycalamide-A and -B and onnamide. Cancer Res 49: 2935–2940 [PubMed] [Google Scholar]
- Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, Qui M, Lewis I, Kurtkaya S, Dingledine R, et al. 2011. Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci 108: 1046–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 281: 32870–32878 [DOI] [PubMed] [Google Scholar]
- Galvin F, Freeman GJ, Razi-Wolf Z, Benacerraf B, Nadler L, Reiser H 1993. Effects of cyclosporin A, FK 506, and mycalamide A on the activation of murine CD4+ T cells by the murine B7 antigen. Eur J Immunol 23: 283–286 [DOI] [PubMed] [Google Scholar]
- Gurel G, Blaha G, Steitz TA, Moore PB 2009. Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui. Antimicrob Agents Chemother 53: 5010–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood KA, West LM, Northcote PT, Berridge MV, Miller JH 2001. Induction of apoptosis by the marine sponge (Mycale) metabolites, mycalamide A and pateamine. Apoptosis 6: 207–219 [DOI] [PubMed] [Google Scholar]
- Jan E, Sarnow P 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902 [DOI] [PubMed] [Google Scholar]
- Jewett JC, Rawal VH 2010. Temporary restraints to overcome steric obstacles: an efficient strategy for the synthesis of mycalamide B. Angew Chem Int Ed Engl 49: 8682–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Uhlenbeck OC 2008. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Dang Y, Liu JO, Yu B 2009. Expeditious synthesis of hippuristanol and congeners with potent antiproliferative activities. Chemistry 15: 10356–10359 [DOI] [PubMed] [Google Scholar]
- Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO 2005. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol Cell 20: 709–722 [DOI] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol Biol Cell 17: 4212–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryman C, Noller HF 1998. Foot-printing and modification-interference analysis of binding sites on RNA. In RNA-protein interactions: a practical approach (ed. Smith CWJ), p. 237–253 Oxford University Press, Oxford, New York [Google Scholar]
- Moore P 2010. Inhibitors of the large ribosomal subunit from Haloarcula marismortui. Isr J Chem 50: 36–44 [Google Scholar]
- Narquizian R, Kocienski PJ 2000. The pederin family of antitumor agents: structures, synthesis and biological activity. Ernst Schering Res Found Workshop 32: 25–56 [DOI] [PubMed] [Google Scholar]
- Nishimura S, Matsunaga S, Yoshida M, Hirota H, Yokoyama S, Fusetani N 2005. 13-Deoxytedanolide, a marine sponge-derived antitumor macrolide, binds to the 60S large ribosomal subunit. Bioorg Med Chem 13: 449–454 [DOI] [PubMed] [Google Scholar]
- Novac O, Guenier AS, Pelletier J 2004. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res 32: 902–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawara H, Higashi K, Uchino K, Perry NB 1991. Change of ras-transformed NRK-cells back to normal morphology by mycalamides A and B, antitumor agents from a marine sponge. Chem Pharm Bull (Tokyo) 39: 2152–2154 [DOI] [PubMed] [Google Scholar]
- Pelletier J, Peltz SW 2007. Therapeutic opportunities in translation. In Translational control in biology and medicine (ed. Mathews MB, Sonenberg N, Hershey JWB), pp. 855–895 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard J, Douthwaite S 2005. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol 3: 870–881 [DOI] [PubMed] [Google Scholar]
- Richter A, Kocienski P, Raubo P, Davies DE 1997. The in vitro biological activities of synthetic 18-O-methyl mycalamide B, 10-epi-18-O-methyl mycalamide B and pederin. Anticancer Drug Des 12: 217–227 [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SJ, Blaha G, Tirado-Rives J, Steitz TA, Moore PB 2007. The structures of antibiotics bound to the E site region of the 50 S ribosomal subunit of Haloarcula marismortui: 13-deoxytedanolide and girodazole. J Mol Biol 367: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati M, Fioretti A, Ghione M 1966. Cytotoxicity of pederin and some of its derivatives on cultured mammalian cells. Experientia 22: 176–178 [DOI] [PubMed] [Google Scholar]
- Walker SE, Fredrick K 2008. Preparation and evaluation of acylated tRNAs. Methods 44: 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West LM, Northcote PT, Hood KA, Miller JH, Page MJ 2000. Mycalamide D, a new cytotoxic amide from the New Zealand marine sponge Mycale species. J Nat Prod 63: 707–709 [DOI] [PubMed] [Google Scholar]
- Yonath A 2005. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem 74: 649–679 [DOI] [PubMed] [Google Scholar]