Abstract
Functional and kinetic constraints must be efficiently balanced during the folding process of all biopolymers. To understand how homologous RNA molecules with different global architectures fold into a common core structure we determined, under identical conditions, the folding mechanisms of three phylogenetically divergent group I intron ribozymes. These ribozymes share a conserved functional core defined by topologically equivalent tertiary motifs but differ in their primary sequence, size, and structural complexity. Time-resolved hydroxyl radical probing of the backbone solvent accessible surface and catalytic activity measurements integrated with structural-kinetic modeling reveal that each ribozyme adopts a unique strategy to attain the conserved functional fold. The folding rates are not dictated by the size or the overall structural complexity, but rather by the strength of the constituent tertiary motifs which, in turn, govern the structure, stability, and lifetime of the folding intermediates. A fundamental general principle of RNA folding emerges from this study: The dominant folding flux always proceeds through an optimally structured kinetic intermediate that has sufficient stability to act as a nucleating scaffold while retaining enough conformational freedom to avoid kinetic trapping. Our results also suggest a potential role of naturally selected peripheral A-minor interactions in balancing RNA structural stability with folding efficiency.
Keywords: structural homology, RNA folding, kinetic intermediates, group I introns, ribozymes
INTRODUCTION
It is well documented in the scientific literature that biological polymers like nucleic acids and proteins often fold into conserved, energetically favorable structural scaffolds to execute similar biochemical reactions (Lockless and Ranganathan 1999; Socolich et al. 2005). Consequently, homologous RNA molecules displaying striking differences in nucleotide composition, chain lengths, global structural complexities, and stabilities of the tertiary motifs, more often than not, adopt highly conserved functional cores defined by topologically equivalent tertiary interactions (Westhof and Massire 2004; Vicens and Cech 2006). While formation of specific structural features of RNA is expected to generate stable low-energy intermediates during folding, satisfying the topological constraints conferred by the conserved tertiary interactions requires sufficient conformational freedom to overcome steric barriers. It is poorly understood how RNA molecules that are designed to fold into an evolutionarily conserved core conformation balance these competing energetic constraints during folding.
A number of studies in the protein folding field, mostly using comparative Φ-value analysis (Matouschek et al. 1989; Maxwell et al. 2005), have revealed that structurally homologous proteins fold through similar folding transition state ensembles (TSE) (Maxwell et al. 2005). Do RNA molecules sharing a phylogenetically conserved structural core fold through similar pathways and intermediates too? The answer to this question remains elusive because of the absence of quantitative comparative analyses of structurally homologous RNA molecules folding under identical conditions. Meta-analysis of the folding rates of topologically diverse RNAs revealed that for slow folding molecules, folding rates are inversely proportional to structural complexity. In contrast, folding rates are independent of topological complexity for fast folding molecules (Sosnick and Pan 2004). From the limited information comparing folding of a few RNA classes, it is generally believed that a correlation between folding rate and topology depends upon whether the rate-limiting step involves disruption of non-native interactions or small local conformational changes that act as nucleation scaffolds (Treiber and Williamson 2001a; Sosnick 2008).
The routinely used Φ-value analysis in protein folding makes a fundamental assumption that a structural perturbation does not change the folding pathways, which in turn renders it unsuitable for characterizing the folding TSEs of RNA. A well-established tenet in RNA folding is that even subtle perturbations in the stability of the native state alter the dominant folding pathways (Shcherbakova and Brenowitz 2005; Laederach et al. 2006). Therefore, in order to address the relationship between topologically conserved tertiary interactions and folding pathways in RNA, we adopted a complementary quantitative approach that critically compares structural-kinetic folding models resolved from time-resolved hydroxyl radical (•OH) footprinting experiments on structurally homologous RNA molecules. Structural-kinetic folding models, by providing insight into the dominant folding pathways, structural features of the folding intermediates, and their solution lifetimes (Laederach et al. 2006; Shcherbakova and Brenowitz 2008), have helped elucidate the contributions of both physical and intrinsic factors that influence RNA folding mechanisms (Laederach et al. 2007).
We describe, validate, and critically compare the folding mechanisms of three group I intron ribozymes that catalyze their own excision from precursor RNA molecules in vivo, cleave an oligonucleotide substrate with multiple turnovers in vitro, have solved crystal structures, and are derived from three phylogenetically divergent organisms: Tetrahymena thermophila—a ciliate protist (Guo et al. 2004), Azoarcus sp. BH72—a purple bacterium (Adams et al. 2004), and Twort sp.—a bacteriophage (Golden et al. 2005). The Tetrahymena structure, spanning ∼2/3 of the full length, contains the conserved core, but lacks the peripheral domains P2–2.1 and P9.1–9.1a–9.2.; a well-validated tertiary structure model derived from phylogenetic covariation analysis (Michel and Westhof 1990; Lehnert et al. 1996) complements the missing information about the global geometries of the peripheral domains.
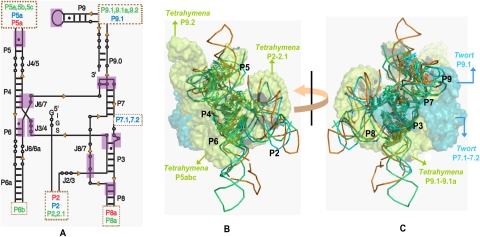
The structures of the three ribozymes reveal superimposable catalytic active sites formed by the juxtaposition of two sets of coaxially stacked helices and stabilized by conserved tertiary contacts that are typical of the group I intron architecture (Fig. 1B,C; Woodson 2005; Hougland 2006; Vicens and Cech 2006). Azoarcus, the smallest of the three ribozymes, has just the core architecture, while Tetrahymena, the largest of the three, has extensive peripheral domains buttressing the core. The Twort ribozyme is of intermediate size and complexity. Despite significant differences in their structural complexities and the total number of tertiary contacts, 10 homologous tertiary interactions are sufficient to define the conserved core architecture of each molecule (Fig. 1A). If the conserved interactions defining the native state topology dictate the choice of folding pathways, we expect to see a common kinetic folding mechanism for the three ribozymes.
FIGURE 1.
The conserved core structure of Group I introns. (A) Conserved secondary structure depicted using the conventional nomenclature (Burke et al. 1987). The additional intron-specific peripheral structural elements and their sites of insertion are indicated within the dotted boxes and are coded by cyan, orange, and lime colors representing Twort, Azoarcus, and Tetrahymena, respectively. The purple boxes indicate the positions where •OH footprints corresponding to the 10 topologically equivalent tertiary contacts are observed. (B,C) Superposition of the crystal structures of the three ribozymes (Twort-1Y0Q.pdb, Azoarcus-1ZZN.pdb, Tetrahymena-1X8W.pdb). The superimposed structures are either viewed from an angle such that the P4–P6–P5abc domains appear in the front (B) or from the opposite side, such that the P9.1–P9.2 and P7.1–P7.2 domains appear in the front. (C) The introns are color coded as in A. The backbones of the conserved core regions are shown as ribbons. The nucleotides within the 10 homologous tertiary contacts are shown as sticks. The peripheral domains of Twort (P7.1–7.2 and P9.1) and Tetrahymena (P5abc, P2–2.1, and P9.1–9.1a–9.2) are shown as semitransparent space-filled surfaces in cyan and lime, respectively. Since the peripheral structures are missing in the Tetrahymena crystal structure (1X8W), these domains were obtained from the phylogenetically derived 3D model (TtLSU.pdb) and the structure of the P4–P6 domain (1GID.pdb), and spatially oriented by superposing these structures on 1X8W.pdb. B and C were generated in PyMOL (DeLano 2009).
RESULTS
Choice of a common folding condition
Since RNA folding pathways are extremely sensitive to solution conditions (Kwok et al. 2006; Laederach et al. 2007), we conducted our folding comparisons under a condition compatible with all three ribozymes (10 mM potassium cacodylate, 0.1 mM EDTA, 100 mM KCl, at 37°C) with the reaction initiated by a saturating concentration of MgCl2, 10 mM. This ionic strength and temperature has been used for several RNA folding studies (Zarrinkar and Williamson 1994; Fang et al. 1999; Rook et al. 1999; Russell and Herschlag 2001; Kwok et al. 2006), and thus facilitates comparison of the present work with these other analyses. The moderate-to-high net G+C content of the ribozymes (37% for Twort, 44% for Tetrahymena, and 60% for Azoarcus) results in stable secondary and tertiary structures under our solution conditions (Banerjee et al. 1993; Tanner and Cech 1996; Rangan et al. 2003; Vicens et al. 2008). These ribozymes are fully catalytically active without the need of additional protein cofactors for structural stability (Herschlag and Cech 1990a,b; Tanner and Cech 1996; Golden et al. 2005), observations confirmed under our experimental conditions by trans oligonucleotide substrate cleavage analysis (Fig. 5A–C, below; Supplemental Figs. S5–S7).
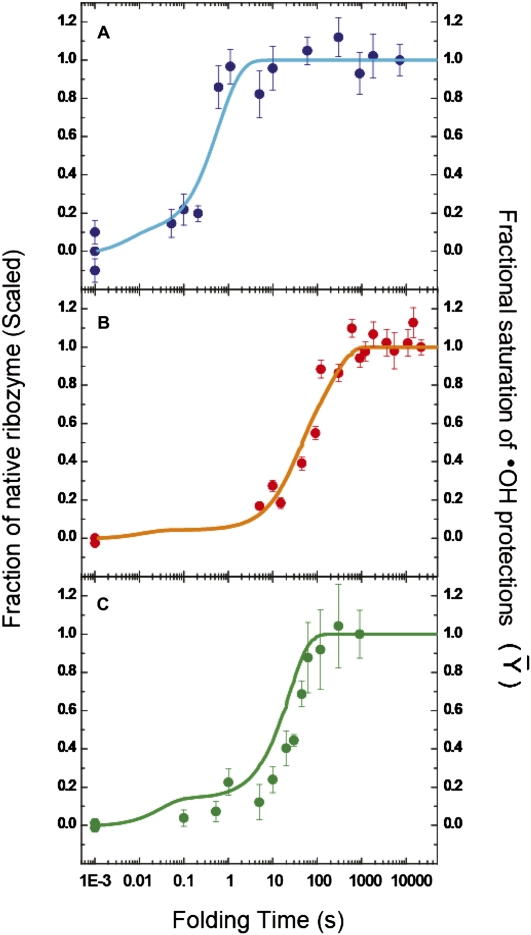
FIGURE 5.
Correspondence between •OH footprinting and activity assays. The time-dependent evolutions of the native ribozyme species, as measured by enzymatic activity assays (filled circles with error bars), and the time-dependent appearance of the folded ribozyme species, as derived form KinFold analysis of time-resolved •OH footprinting data (solid lines), for the Twort (A: blue), Azoarcus (B: orange), and Tetrahymena (C: green) ribozymes, are depicted. The activity assays are described in Supplemental Figures S5, S6, and S7. The left ordinate, in all three panels, represents the fraction of substrate cleaved during the initial product burst phase. The fraction native at each time point is scaled either with respect to a prefolded control (incubated at 50°C—for the Twort and Azoarcus ribozymes) or with respect to the 15-min time point (end of the first phase—for the Tetrahymena ribozyme). The right ordinate represents the extent to which a RNA backbone region has become solvent inaccessible, as compared with the solvent accessibility of the same region in the fully folded control (Fractional Saturation). The abscissa represents the time for which the ribozyme is allowed to fold, after addition of Mg2+ ions, prior to the addition of the oligonucleotide substrate and guanosine (in the enzymatic activity assays) or the addition of •OH generating reagents (in time resolved •OH footprinting experiments).
The presence of 100 mM K+ engenders a relaxed initial state ensemble in which the repulsive columbic forces between the negatively charged backbones of the RNA helices are screened, allowing for greater conformational flexibility (Russell et al. 2000; Rangan and Woodson 2003; Takamoto et al. 2004). This condition also recapitulates the physiological ionic milieu in which almost all RNA molecules have evolved to fold and function (Cayley et al. 1991), and helps avoid the idiosyncratic Mg2+-mediated folding behavior observed for both the Tetrahymena (Sclavi et al. 1998) and the Azoarcus (Chauhan and Woodson 2008) ribozymes under conditions of very low ionic strength.6
Since the folding of the Twort ribozyme has not been quantitatively studied, we performed equilibrium •OH footprinting and analytical ultracentrifugation analyses to verify, under these experimental conditions, that the native structure is fully formed in 10 mM Mg2+ (Supplemental Fig. S1A,B), and that its initial Mg2+ free state is a relaxed ensemble (Supplemental Fig. S1C).
The temporal order of formation of homologous tertiary contacts is not conserved
Following initiation of the folding reaction by addition of 10 mM MgCl2, we probed the backbone solvent accessibility change for each nucleotide of an RNA by time resolved •OH footprinting. A total of 24 positions in the Twort, 19 in the Azoarcus, and 33 in the Tetrahymena ribozymes were analyzed. For each of these positions, we confirmed that the observed decrease in •OH radical reactivity upon folding correlates with the formation of a native tertiary contact by comparing reactivity profiles of the equilibrium folded states with the solvent accessibilities of the C4′ atom within the crystal structures (Supplemental Figs. S1A, S2A,B).
The time-progress curves determined for each of these positions report the formation rate of the corresponding tertiary contact. Three statistically significant temporal clusters were obtained for each ribozyme (Fig. 2A–C) using the KinFold software (Laederach et al. 2006, 2007; Martin et al. 2009). These clusters identify groups of positions with common time evolutions. The cluster centroids, representing the average time dependence of formation of the tertiary contacts pertaining to a cluster, are noticeably different for the three ribozymes (Figure 2A–C). For the smallest (Azoarcus) and largest (Tetrahymena) of the ribozymes studied, the fastest tertiary contacts reach their native extent of protection within 50 to 100 msec, while the slowest contacts require ∼200 sec (Fig. 2B,C). The long time spans contrast sharply with those of the medium-sized Twort ribozyme, whose fastest and slowest tertiary contacts reach the native state in ∼500 msec and ∼5 sec, respectively, leading to a compressed folding time span (Fig. 2A). A correlation between size and folding rate is not observed for these homologous ribozymes.
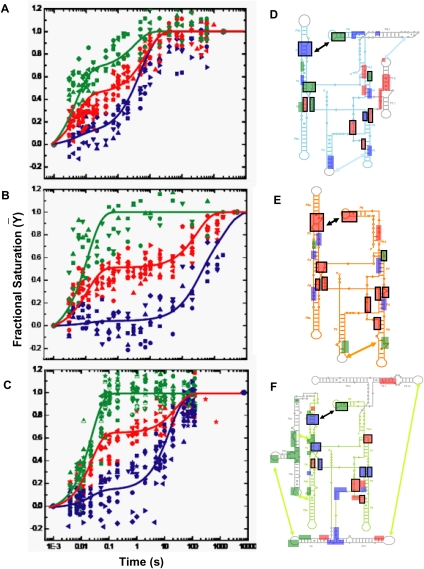
FIGURE 2.
Clustering the kinetic progress curves derived from time resolved •OH footprinting during Mg2+ mediated folding of (A) Twort, (B) Azoarcus, and (C) Tetrahymena ribozymes. The 24 progress curves of Twort, 19 of Azoarcus, and 33 of Tetrahymena, each representing the time-dependent formation of a tertiary contact, statistically clustered into three groups—fast (green), medium (red), and slow (blue). The cluster centroids, plotted as solid lines though the clusters, represent the average kinetic behavior of a group and are used in the KinFold analysis. For cluster affiliations of the tertiary contacts please refer to Supplemental Table S1. Hydroxyl radical footprints are shown as boxes, colored according to their cluster affiliations, on the secondary structures of the three ribozymes—(D) Twort, (E) Azoarcus, and (F) Tetrahymena. The core helices (P4–P5–P6 and P3–P7–P8) and the substrate docking site (P1) are shown in cyan, orange, and lime for D, E, and F, respectively; while intron-specific peripheral domains, as described in Figure 1A, are shown in gray. Boxes representing the homologous tertiary interactions, depicted in Figure 1A, are indicated by black outlines.
Plotting the clustered residues on the secondary structure diagrams of the three ribozymes reveals a striking observation; the 10 homologous tertiary contacts defining the conserved native core structure of group I intron ribozymes form at widely different rates and belong to different temporal clusters in each ribozyme (Fig. 2D–F; Supplemental Table S1). This observation is counter to our expectation that homologous tertiary contacts would display a conserved temporal order of formation. This insight is “model independent” in that it does not depend on the kinetic modeling of the time progress curves that is described below.
Similarities among the kinetic folding models reveal conserved dominant folding pathways
The clustered kinetic progress curves of each ribozyme were analyzed with the KinFold software, yielding the kinetic models that best describe folding (Fig. 3A,C,E). In simulations conducted without bound reverse rates, the reverse rates are close to zero and poorly determined due to the progression of all our reactions to full completion (Supplemental Table S2A–C). We chose to bind the reverse rates along each transition pathway to zero in the models we report, as this constraint reduces the dimensionality of the optimization problem and yields overall lower errors on the forward rates. Numerical simulation of the possible folding pathways leading to the native state reveals the relative flux of folding molecules flowing through each pathway and identifies the pathways that are most highly populated (Fig. 3B,D,F; Laederach et al. 2006, 2007).
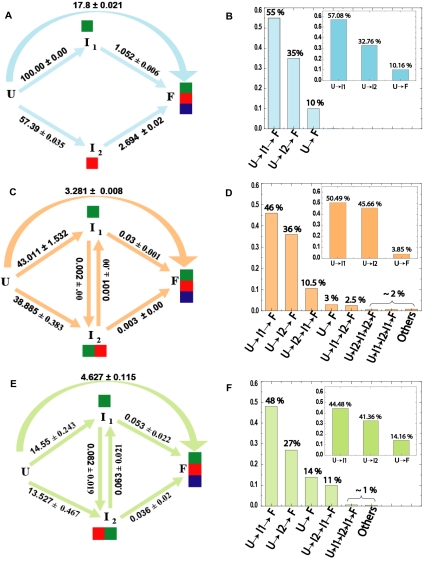
FIGURE 3.
Kinetic folding models and flux analysis. Folding model topologies (left) and flux of folding molecules through the dominant pathways (right) for the Twort (A,B), Azoarcus (C,D), and Tetrahymena (E,F) ribozymes, respectively, as derived from KinFold analysis. U, I, and F refer to the unfolded, intermediate, and folded species, respectively. The resolved microscopic rate constants are indicated for each step of a folding pathway. In the three left panels, the green, red and blue colored boxes, associated with the intermediates (I1, I2) and the folded species (F), represent the cluster affiliation of tertiary contacts formed in the corresponding species. In the three right panels, each histogram represents the percentage of folding molecules traversing the pathway indicated below it. The insets of B, D, and F show the initial flux partitioning for the corresponding ribozymes.
The kinetic models of the three ribozymes share a common theme; the initial ensemble of folding molecules immediately partitions into three parallel pathways. Two pathways lead to intermediates, U→I1 and U→I2, while the third leads directly to the native state (U→F). The initial folding flux partitions predominantly through U→I1 (Twort: 57.1%, Azoarcus: 50.5%, and Tetrahymena: 44.5%; insets of Fig. 3, B,D,F). The U→I2 pathway is the second most highly populated (Twort: 32.8%, Azoarcus: 45.7%, and Tetrahymena: 41.4%). The small remainder of the folding flux is destined to the direct pathway (Twort: 9.7%, Azoarcus: 3.9%, and Tetrahymena: 14.2%). We refer to the intermediates as ITwort, IAzoarcus, and ITetrahymena for the respective folding pathways.
As we follow Twort folding from the intermediates to the final state, we see that almost all of the initial U→I1Twort and U→I2Twort fluxes are channeled through to F (Fig. 3B vs. insets); there is very little exchange between I1Twort and I2Twort. The dominant pathway to the native state (U→I1Twort→F, 55%) is through the intermediate comprised only of the fastest forming tertiary contacts (green cluster). U→I2Twort→F is the second most populated pathway (35%); its intermediate is exclusively comprised of the tertiary contacts affiliated to the medium temporal cluster (red). The remaining 10% of the folding flux follows the direct pathway. The close resemblance between the initial flux partitioning and the total flux through the dominant pathways suggests that the Twort RNA molecules are directed from the onset of folding into distinct channels mutually separated by high-energy barriers throughout the folding reaction.
In both the Azoarcus (Fig. 3C) and the Tetrahymena (Fig. 3E) ribozymes, the most dominant pathways are again U→I1→F and U→I2→F. As observed for I1Twort, I1Azoarcus and I1Tetrahymena are comprised of the tertiary contacts pertaining only to the fastest cluster (green). In contrast, the nature of I2 differs; the tertiary contacts defining I2Twort are exclusively the medium (red) cluster, while those defining I2Azoarcus and I2Tetrahymena include both the fastest (green) and the medium (red) clusters.
Another difference is that pathways, in addition to the ones observed in the Twort ribozyme are significantly populated. These pathways are those that involve interconversion of the intermediates I1 and I2. We note that for both Azoarcus and Tetrahymena a small but significant fraction (∼10% ± 1%) of U→I2 flux is diverted to I1 (i.e., U→I2→I1→F as opposed to U→I2→F) (Fig. 3D,F vs. insets). Moreover, while the rates of I1Tetrahymena→F and I2Tetrahymena→F conversions are comparable to the rate of I1Tetrahymena↔I2Tetrahymena interconversion (Fig. 3E), the I1Azoarcus→F conversion is an order of magnitude faster than both I2Azoarcus→F and I1Azoarcus↔I2Azoarcus (Fig. 3C). Finally, while U→F is the third dominant pathway in Tetrahymena (14%) (Fig. 3F), Azoarcus’ third most populated pathway is U→I2Azoarcus→I1Azoarcus→F (10.5%) (Fig. 3D). Together, these observations suggest that while I2 is very stable in both Tetrahymena and Azoarcus, it is a deeper folding trap in Azoarcus.
The folding behavior of the three ribozymes was also analyzed at the lower temperature of 25°C. Identical kinetic model topologies were resolved. The only observed difference was an overall slowing of the folding process (data not shown). Detailed studies of the temperature dependence of these folding reactions are being conducted and will be presented elsewhere.
Structural character of the predicted folding intermediates
Since the time–progress curves that comprise each cluster are associated with specific nucleotides, structural characteristics of the folding intermediates can be inferred by mapping their locations on the native structures of the ribozymes (Laederach et al. 2006). We emphasize that the time-progress curves only report the rate at which a site becomes inaccessible to solvent; hydroxyl radical protections do not directly identify pairwise contacts. We interpret the data to indicate formation of a native contact only when pairs of time-progress curves, for protections observed in the native structure to form a tertiary contact, are coincident. Alternatively, where coincidence of •OH progress curves for native pairwise contacts is not observed, we interpret these cases to reflect transient interactions.
In the structures shown in Figure 4, we color nucleotides that are predicted by the kinetic model to be protected in an intermediate, by the color of the affiliated cluster. Our present analysis addresses only protections that are present in the intermediates; topological constraints and constraints on “unfolded” stretches of polynucleotide are explored in a detailed modeling analysis of the novel Twort and Azoarcus intermediate structures that will be published separately (C Chen, S Mitra, M Jonikas, M Brenowitz, and A Laederach, in prep.). The intermediate structures inferred for the Tetrahymena ribozyme are identical to those previously published. The time evolution of the unfolded, intermediates, and the native species, calculated from the resolved structural-kinetic models, are shown alongside the representative predicted structured regions of each folding intermediate.
FIGURE 4.
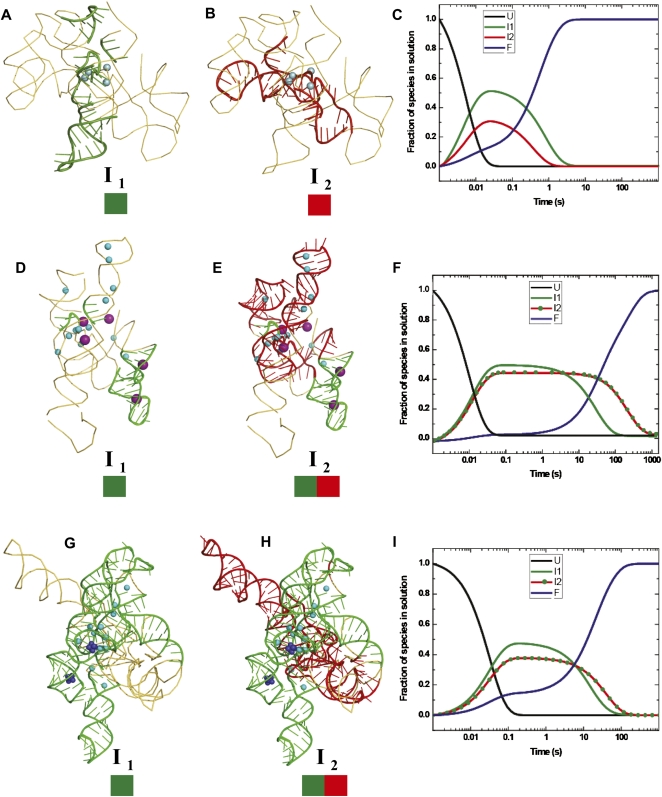
Mapping the temporal clusters of tertiary interactions onto the crystal structures of the Twort (A,B) and Azoarcus (D,E) ribozymes, and the biochemically validated phylogenetic model of the Tetrahymena (G,H) ribozyme. The coordinates of the structures were obtained from the following PDB entries: 1Y0Q (Twort), 1ZZN (Azoarcus), and TtLSU (Tetrahymena). The positions of divalent and monovalent ions (when known in the crystal structures) are indicated by blue and purple spheres, respectively. The green and red colored boxes drawn below the intermediates (I1 and I2) represent the clusters of tertiary contacts formed in each species. The schematic models of the intermediates were generated in PyMOL (DeLano 2009). The three right panels show the lifetimes in solution of the unfolded (U: black line), intermediate (I1: green line, I2: red line or red line with green dots), and the folded (F: blue line) species, as derived from KinFold analysis, for the Twort (C), Azoarcus (F), and Tetrahymena (I) ribozymes. The two deep blue clusters of spheres in the P4–P6 domain of the Tetrahymena ribozyme represent the two cobalt hexamine(III) molecules observed in the crystal structure (1GID.pdb).
I1Twort is characterized by •OH protection of nucleotides corresponding to the conserved triple helical junctions (J3/4 and J6/7), the L9 GNRA tetraloop, and the G binding site (Fig. 4A). These represent only one-quarter of the total number of protections observed in the native state (six out of 24). It is important to note that the protection corresponding to the P5 receptor half of the L9–P5 tetraloop–receptor interaction is not observed until much later in the folding reaction. This observation suggests that the L9 tetraloop transiently forms non-native contacts early in folding that are reorganized during the later steps.
I2Twort is characterized by protections corresponding to the P7.1–P7.2 peripheral domain, long-range contacts between the P6 and P3 domains, the central J8/7 stretch of unpaired residues, and the 3′ side of the P7 and the 5′ side of the P9.0 domains (Fig. 4B). These protections together represent only one-third of all the native state protections (eight out of 24). What is striking about the Twort intermediates is that the protections observed in one intermediate are not present in the other. Most of the protections that define I1Twort and I2Twort form around the specifically bound Mg2+ visible in the Twort crystal structure.
The slowest forming 10 protections in Twort folding correspond mostly to the peripheral interactions that include the P5 tetraloop receptor half of the conserved L9–P5 tetraloop receptor interaction, the long range L2–P8 A-minor motif interaction, the P9–P9.1 peripheral domain, and the conserved catalytic core involving residues on the 5′ sides of the P3 and P7 helices (Supplemental Figs. S3A, S4A). Therefore, the critical local core interactions assemble in the Twort ribozyme before the more global long-range peripheral interactions are established.
Both Twort intermediates are short-lived, with only a ∼20-msec lifespan at their peak concentrations. The native tertiary structure appears at a rate of 1.71 ± 0.04 s−1 (Fig. 4C). These characteristics suggest that both the Twort intermediates are transient nucleation scaffolds rather than energetically stable modules.
The Azoarcus intermediates differ from those of Twort in both structure content and time evolution. I1Azoarcus displays four of the total 19 protections observed in the native Azoarcus ribozyme. These protections correspond to nucleotides in the P2 GAAA tetraloop, the 11-nucleotide (11 nt) P8 canonical receptor, the J5/4 junction, the helix P1, and the guanosine binding site (Fig. 4D). I2Azoarcus is characterized by protections corresponding to the conserved triple helical junctions J3/4 and J6/7, the L9 GAAA tetraloop, the P5 11-nt canonical receptor, the upper part of the P3 helix, and the unpaired J8/7 segment, in addition to all of the I1Azoarcus protections (Fig. 4E). Thus, the well-structured I2Azoarcus possess almost two-thirds of the protections observed in the native state (14 of 19 contacts). As discussed in more detail below, the fastest forming protections in I1Azoarcus are located around specific K+ binding sites. In contrast, the protections belonging to the medium temporal cluster in I2Azoarcus are formed around sites of specifically bound Mg2+ (Fig. 4D,E).
The slowest forming Azoarcus protections correspond to nucleotides in the J6/6a segment, the minor groove of the P3 helix, the J4/5 junction, the P1 helix, and the core protections involving the unpaired J8/7 and the P4 and P7 helices (Figs. 3B, 4B). Together, these protections comprise only 26% of the native state protections (five out of 19). In the Azoarcus ribozyme, the majority of the peripheral interactions are established before the critical core contacts are assembled.
The native tertiary structure of the Azoarcus ribozyme appears much more slowly than Twort, with a rate constant of 0.013 (± 0.001) s−1 (Fig. 4F). Both I1Azoarcus and I2Azoarcus reach their peak concentrations in ∼40 msec (Fig. 4F). Unlike the Twort intermediates, their lifetimes are long; up to 760 msec for I1Azoarcus and ∼20 sec for I2Azoarcus. Therefore, I1Azoarcus and especially I2Azoarcus are significantly stabilized.
I1Tetrahymena exhibits all of the protections pertaining to the P5abc subdomain (Fig. 4G), which include the two Type I/II A-minor interactions, the metal ion core (P5a)—receptor (P4) interaction, and the GAAA tetraloop (L5b)-11 nt canonical receptor (J6a/6b) interaction. This intermediate also shows protections of nucleotides in the conserved L9 tetraloop and those consistent with formation of the long range L2–L5c loop–loop interaction (P14) (Fig. 4G). Almost half of the protections observed in the native Tetrahymena ribozyme are detected in I1Tetrahymena (15 of 33).
I2Tetrahymena shows the protections observed in I1Tetrahymena and those that correspond to the long range L9.1a–L2.1 loop–loop interaction (P13), the G binding site, P6 and P3 helices, and the J8/7 and J1/2 junctions (Fig. 4H). The 23 protections formed in I2Tetrahymena constitute ∼70% of the 33 contacts that define the ribozyme's native conformation. Mapping these contacts onto the available crystal structures (Cate et al. 1996; Guo et al. 2004) reveals that most of the protections observed in both I1Tetrahymena and I2Tetrahymena form around K+ and Mg2+ binding sites.
The slowest forming protections in the Tetrahymena ribozyme involve nucleotides corresponding to the core interactions represented by the conserved triple helix (J3/4), interaction of the J6/6a with the minor groove of P3, contact between the P2–2.1 domain and J2/3 segment, and finally, the receptor half of the L9–P5 tetraloop receptor interaction. These protections comprise the remaining ∼30% of the native state protections (Supplemental Figs. S3C, S4C). As the Azoarcus ribozyme, the majority of the Tetrahymena peripheral interactions are established before the critical core contacts are assembled. The Tetrahymena native tertiary structure appears at a rate of 0.048 (± 0.002) s−1 (Fig. 4I). I1Tetrahymena and I2Tetrahymena remain at their peak concentrations between ∼100 msec and 1 sec (Fig. 4I), which reflects their moderate energetic stabilization, slightly more for I2Tetrahymena than for I1Tetrahymena.
A common behavior of the all three ribozymes is that the dominant path to the native state involves fast nucleation of a few key tertiary interactions unencumbered by additional structure formation. In the Azoarcus and Tetrahymena ribozymes, both of which contain a peripheral GAAA tetraloop and its canonical 11-nt receptor, protections corresponding to this interaction are among the fastest to appear during tertiary folding. These data reflect early organization of this interaction around K+ ions that are site-specifically bound to this motif. In the Twort ribozyme lacking the canonical receptors of the peripheral GNRA tetraloops, nucleation seems to proceed around site-specifically bound Mg2+ ions near the two triple helical junctions (J3/4 and J6/7) and near the guanosine binding site. Most importantly, the protections consistent with formation of stable peripheral contacts appear before protections corresponding to the catalytic core are evident in the Azoarcus and the Tetrahymena ribozymes. This folding hierarchy is in clear contrast to the behavior of the Twort ribozyme, in which formation of these groups of protections occurs in concert.
Ribozyme activity assays confirm time-dependent evolution of the native states
We performed oligonucleotide substrate cleavage assays to independently follow the formation of the catalytically active native state (Supplemental Figs. S5–S7). The time-dependent appearances of catalytic activity of the Twort and the Azoarcus ribozymes are in an excellent agreement with the rates of formation of their native tertiary structures predicted by the structural-kinetic models (Fig. 5A,B). In contrast, the time evolution of the Tetrahymena ribozyme's full catalytic activity lags the formation of native state predicted by the structural kinetic model (Supplemental Fig. S7A,B). At the end of the time scale for activity measurements, commensurate with the end points of the footprinting experiment, the ribozyme activity reaches only 30% as compared with a prefolded control at 50°C. This result was expected, since under folding conditions comparable to those used herein, the Tetrahymena ribozyme acquires 25%–50% of its full activity at the end of the measured time scale (Russell and Herschlag 1999; Treiber and Williamson 2001b; Russell et al. 2002; Uchida et al. 2003). The remaining fraction is trapped into a long-lived misfolded state denoted “M” (Russell and Herschlag 2001). “M” is a nearly identical topological isomer of the native structure “N” that is indistinguishable by •OH footprinting (Russell et al. 2000, 2006).
Extending our catalytic activity measurement of the Tetrahymena ribozyme to several hours yielded results consistent with the above-referenced studies; an initial phase with an amplitude of ∼30% is in close agreement with the rate of formation of the folded RNA predicted by the structural-kinetic model (Fig. 5C; Supplemental Fig. S7C). A second phase with an amplitude of ∼70%, appears very slowly, unaccompanied by further •OH cleavage reactivity (Supplemental Fig. S7A). This result is consistent with conversion of “M” to the “N” state. Since “M” and “N” possess nearly identical tertiary structures, we conclude that the rate of folding of the Tetrahymena ribozyme, calculated from the structural-kinetic model, is directly comparable to the rate of the first phase of native ribozyme formation. Therefore, for all the three ribozymes, the structural-kinetic models accurately describe the major conformational events associated with native tertiary structure formation.
DISCUSSION
The three homologous RNA molecules compared in this study fold into a common core conformation to execute the same biological function despite significant disparities in terms of their global structures, highly divergent phylogenetic backgrounds, and distinctly different locations in the genomic context (rRNA, tRNA, and mRNA) (Cech 1990; Reinhold-Hurek and Shub 1992; Landthaler and Shub 1999). Our combined approach of chemical probing, ribozyme activity measurements, and kinetic modeling enable quantitative comparison of their folding mechanisms and reveal basic principles of RNA folding.
Phylogenetic covariation analyses have revealed how a few tertiary contacts, despite sequence dissimilarities, are geometrically conserved in evolutionary divergent homologous RNA molecules to generate similar core architectures, regardless of the presence or absence of additional nonconserved peripheral domains (Gutell et al. 1986; Michel and Westhof 1990; Lehnert et al. 1996). It is generally believed that peripheral domains (i.e., the P5abc subdomain in the Tetrahymena ribozyme) of large RNA molecules specifically stabilize the native core structures (Engelhardt et al. 2000; Johnson et al. 2005); in the absence of such domains, strong tertiary interactions such as the two canonical GAAA tetraloops and their cognate 11-nt canonical receptors present in the Azoarcus ribozyme, impart unusually high stability to the folded structure (Tanner and Cech 1996). The stability conferred by peripheral domains, strong canonical tertiary contacts, or both, is expected to generate stable folding intermediates. However, the compromised conformational dynamics of stable intermediates could generate steric obstacles early during folding, and thus impede the overall folding rate.
Our comparative analysis reveals that large RNA molecules fold efficiently through energetically balanced pathways involving nucleation of minimally stabilized transient structural scaffolds that are poised for capture of native tertiary structure. We outline a set of general principles that dictate the choice of the dominant pathways on all three folding landscapes.
Less-structured folding intermediates offer more efficient folding routes
The majority of molecules in the three ribozymes preferentially fold not through the direct pathway, but through multiple structurally less-constrained pathways. This observation suggests that the energy barriers along the direct pathway are high enough so that nucleation of a minimal set of interactions is necessary for guiding the molecules into pathways with lower energy barriers. These pathways are presumably those that balance enthalpy gain with the entropy loss associated with progressive structure formation. All three structural-kinetic models identify the most populated pathway as U→I1→F. The intermediate I1 is structured either minimally (∼25% of the native contacts are formed in Twort and Azoarcus) or, at the most, moderately (∼45% in Tetrahymena). The majority of the remaining formation of native structure occurs during the transition from the I1 intermediate to the native state.
We conclude from this observation that formation of an early structural scaffold narrows down conformational heterogeneity without imposing too many steric constraints. This balance between “order and chaos” allows the ensemble to retain sufficient freedom to fully explore the available conformational space. A similar theme of a late bias toward the native state has been recently revealed in protein folding. For structurally homologous proteins that fold through an intermediate, and hence, through two TSEs, the late TSE of each protein resemble the other's much more than their early TSE (Calosci et al. 2008). The observation implies that while the native state uniquely defines the reaction coordinate during the later stages of folding, the earlier stages retain much more conformational freedom on the free energy landscape (Calosci et al. 2008).
Interestingly, the structurally most complex Tetrahymena ribozyme, which populates long-lived misfolded intermediates under some experimental conditions, displays significantly higher I1 structuring compared with Twort and Azoarcus. Sixty seven percent of this structuring (10 out of 15) is attributable to contacts that specifically stabilize the independently folding P5abc subdomain. This observation suggests that early folding of the P5abc subdomain constrains the conformational ensemble that in turn restricts alternate pathways that could favor either correct folding or lead to faster resolution of misfolded species. Consistent with this hypothesis, the earliest steps of folding of the EΔP5abc version of the Tetrahymena ribozyme that lacks P5abc, are greater than fivefold faster than those observed in the folding of the wild-type ribozyme (Russell et al. 2007). EΔP5abc folding involves fewer and different tertiary interactions as compared with the wild-type ribozyme. Even though EΔP5abc misfolds to the catalytically inactive “M” state, refolding from the “M” to the “N” state is two orders of magnitude faster as compared with the wild type (Russell et al. 2007).
Overall folding speed is governed by folding intermediate stability
A “mutual tertiary structure capture” model, where folding proceeds through parallel pathways involving moderately stable structural scaffolds, was proposed based on the effect of destabilization of the tetraloop–tetraloop receptor interaction in the P4–P6 domain of the Tetrahymena ribozyme (Treiber and Williamson 2001b). Twort ribozyme folding reflects a natural occurrence of this mechanism, in which long-lived kinetic intermediates that are often a kinetic trap are avoided. Twort is therefore the fastest folding ribozyme of the three analyzed under our quasiphysiological experimental conditions. The Twort intermediates are characterized by formation of several key interactions whose moderate stability is evidenced by their short lifetimes.
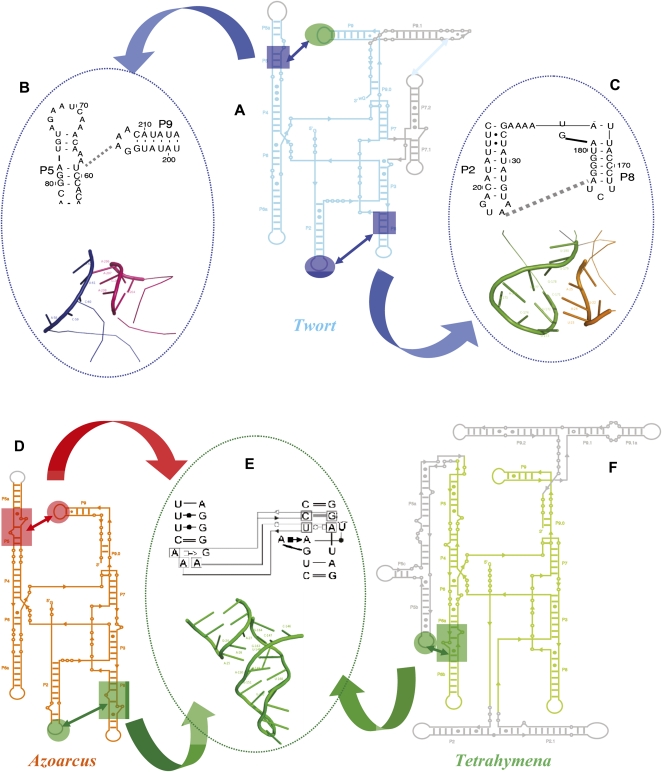
The short-range interactions surrounding the central metal ion-binding sites are efficiently organized in this ribozyme before the periphery completely closes upon the core through the Type I/II A-minor interactions between the L9 and L2 tetraloops and their respective receptors in P5 and P8 (Fig. 6A–C; Supplemental Figs. S3A, S4A). Thus, the late imposition of spatial constraints through long-range tertiary interactions is a folding strategy that appears to be favored only in the absence of strong peripheral tertiary interactions.
FIGURE 6.
Noncanonical and canonical receptors in group I introns. The two tetraloops in the Twort ribozyme (A), the GUAA (L2) and the GAAA (L9) dock into an A-minor motif in P8 and a half receptor in P5, respectively (shown in their secondary and tertiary forms in B and C, respectively). These two noncanonical tetraloop receptor interactions are colored blue to indicate their affiliations to the slow cluster. The Azoarcus (D) and the Tetrahymena (F) ribozymes, containing the canonical GAAA tetraloops and their cognate 11-nt receptors (represented in its secondary and tertiary structure forms in E), are shown. The L9–P5 and L2–P8 tetraloop–receptor interactions in the Azoarcus are colored red and green, respectively, to indicate their cluster affiliations. Similarly, the L5b–J6a/6b tetraloop–receptor interaction in the Tetrahymena ribozyme is shown in green to indicate its cluster affiliation.
In contrast, what happens when high-affinity interactions are formed in early folding intermediates (Fig. 6E)? The answer to this question comes from the Azoarcus ribozyme, in which two GAAA tetraloop–canonical receptor interactions stabilize its global fold (Fig. 6D) and from the Tetrahymena ribozyme that uses a GAAA tetraloop–canonical receptor interaction to stabilize its early forming and independently folding P4–P6 domain (Fig. 6F). I1Azoarcus is stabilized by formation of the L2-GAAA tetraloop-P8 canonical receptor contact (Fig. 6F), an exceptionally high-affinity tertiary interaction (Costa and Michel 1995). I2Azoarcus is stabilized by a second, identical, high-affinity interaction between the L9-GAAA tetraloop and its receptor in P5. These two interactions, along with other tertiary contacts, firmly lock both ends of the molecule, and thus strongly stabilize I2Azoarcus. I1Azoarcus with only one such interaction still retains some conformational freedom.
Our initial state contains a physiologically relevant monovalent cation type and concentration, 100 mM K+ (Cayley et al. 1991). K+ binds specifically to the adenosine platforms (AA) in the tetraloop receptor of the Tetrahymena ribozyme (Basu et al. 1998). Of the five specifically bound K+ ions observed in the Azoarcus crystal structure (Stahley et al. 2007), two are within the L2–P8 tetraloop receptor (one to the L2 loop and the other to the AA platform in the P8 receptor) and one is coordinated by residues in the G binding site and the J5/4 segment. The fact that these are the fastest forming I1 interactions in both Azoarcus and Tetrahymena suggests that under physiological conditions, stabilization by cations of secondary structure modules participating in tertiary interactions strongly influences RNA folding kinetics.
We propose that very stable intermediates such as those observed for the Azoarcus and the Tetrahymena ribozymes are ensembles of low-energy structures in which most of the peripheral native/native-like tertiary contacts are present. Therefore, rearrangements necessary to generate the native conformation of the catalytic core are impeded by steric constraints arising from the severely compromised conformational dynamics of the intermediates. This is especially true for intermediates like I2Azoarcus, which is locked at both ends. The prolonged lifetimes of I1Azoarcus and I2Azoarcus are likely to arise from slow conformational search and/or slow interconversion that therefore reduce the overall folding speed of the Azoarcus ribozyme.
The conclusion that long-lived intermediates, populated and stabilized by the synergistic effects of monovalent and divalent cations on strong tetraloop receptor interactions, retard folding of the Azoarcus ribozyme, is consistent with previous studies conducted in a lower ionic strength buffer (∼8 mM Na+); all of the native tertiary contacts are observed to form in <50 msec under this very low-ionic strength condition (Rangan et al. 2003; Chauhan and Woodson 2008). The importance of the strong tertiary interactions like canonical tetraloop–tetraloop receptors on kinetic folding pathways, especially in the absence of peripheral domains, is underscored by the observation that destabilization of these interactions through mutations or backbone nicks slows the overall folding kinetics of the Azoarcus ribozyme under the low-salt initial condition (Chauhan and Woodson 2008; Chauhan et al. 2009). Such perturbations most likely introduce alternate folding pathways.
I1Tetrahymena and I2Tetrahymena are also stabilized by a network of strong tertiary interactions that increase the probability of forming trapped states. It has been shown that the commitment to fold to the native state is made late along the folding pathway of the Tetrahymena ribozyme (Russell and Herschlag 2001), and misfolded states presumably arise from well-structured intermediates such as I2Tetrahymena, whose conformational freedom is compromised. Among the fastest-forming interactions that cooperatively stabilize the P5abc subdomain (Sattin et al. 2008) present in the Tetrahymena intermediates are Type I/II A minor interactions and a high-affinity GAAA tetraloop–canonical receptor interaction.
Although these peripheral interactions pose kinetic impediments, their importance is underscored by the observation that the independently folding P5abc subdomain stabilizes the native structure of the ribozyme over a misfolded state by ∼6 Kcal/mol (Johnson et al. 2005). Therefore, as opposed to the Azoarcus ribozyme, where early tertiary structure capture by the GAAA tetraloop–tetraloop receptor interactions most likely generates kinetic traps, the same interaction in the Tetrahymena ribozyme stabilizes an early folding subdomain, which, in turn, thermodynamically discriminates between native and non-native structures.
A proposed role of naturally selected peripheral A-minor interactions in RNA folding
Our results suggest that the folding mechanism of homologous RNA molecules cannot be accurately predicted simply based on the topology of the conserved native core structure. The nature of the unpaired adenosines that mediate long-range minor groove contacts in structured RNA through Type I/II A- minor interactions (Gutell et al. 2000; Doherty et al. 2001; Lee et al. 2006) play a key role in determining pathway preference and folding speed. The binding efficiency of GAAA tetraloop–canonical 11-nt receptors present in the Azoarcus and the P5abc subdomain of the Tetrahymena ribozymes, is 50-fold higher than that of the homologous A-minor interactions (GUAA tetraloops–CCU:AGG receptor) found in the Twort ribozyme (Costa and Michel 1997).
The relative efficiency of these interaction classes correlates directly with the predicted lifetime of the folding intermediates and inversely with the overall folding speed; the Twort ribozyme folds more than an order of magnitude faster than the Azoarcus and the Tetrahymena ribozymes. This observation suggests that a tradeoff to highly stable native folds is a reduction of the speed with which the final structure forms, and a greater temporal spread between the earliest and the latest steps in folding (Fig. 4, cf. F, I, and C). The existence of this tradeoff predicts a preferential occurrence of high-efficiency A-minor interactions under selective pressures that demand structural integrity over speed; the removal of such pressures should allow for a more generous selection of A-minor interactions with a wider range of affinities and specificities. Two questions are posed by this hypothesis: (1) Is such a biased distribution observed in nature? (2) What selective pressure influences the natural occurrence of A-minor interactions, at least for the group I introns?
We propose that for group I introns the natural selection of peripheral A-minor tertiary motifs with different relative efficiencies could be a direct consequence of the extent of structuring in the adjoining regions of their corresponding precursor RNA transcripts. Evidence for biased distribution of A-minor interactions comes from several independent observations. The Tetrahymena and Azoarcus group I introns belong to the IC1 and IC3 subgroups that occur almost exclusively in highly structured noncoding transcripts such as preribosomal and pretransfer RNAs (Cannone et al. 2002). High-affinity interactions within these introns help them specify a unique thermodynamically stable structure by avoiding competition from the surrounding tertiary interactions. It has been shown that the independently folding P5abc subdomain, an integral part of which is the GAAA tetraloop-canonical 11-nt receptor, stabilizes the native structure of the ribozyme over a misfolded state by ∼6 Kcal/mol (Johnson et al. 2005).
Consistent with this hypothesis, it has been shown that in the IC3 group I introns, 88.7% of L2 and 98.6% of the L9 peripheral tetraloops belong to the GNRA family; all of these GNRA loops are, in fact, GAAA (Prathiba and Malathi 2008). The binding affinity of a GAAA tetraloop for its 11-nt canonical receptor is up to a 1000-fold higher than the affinity of other GNRA tetraloops for the same or closely related receptor motifs (Ikawa et al. 2001; Geary et al. 2008).
A comprehensive comparative analysis of natural and in vitro-selected GNRA tetraloop receptors suggests that the preferred occurrence of the receptors with the highest specificities and affinities could be an evolutionary adaptation to avoid alternate tertiary folds (Geary et al. 2008). That the natural splicing efficiency of IC1 and IC3 introns has evolved as a subtle balance between the stabilization of a unique conformation over competing interactions and the kinetic impediments imposed by high-affinity interactions is perhaps best exemplified by the fact that the splicing of the Tetrahymena intron is more efficient in its natural pre-rRNA context as compared with its splicing in an artificially engineered pre-mRNA context (Hagen and Cech 1999).
In contrast, the Twort ribozyme belongs to the IA2 group I intron subgroup that, as known to date, occur exclusively in significantly less-structured protein-coding bacteriophage mRNA (Cannone et al. 2002). Almost all of the IA2 introns have either GUAA or GUGA as their peripheral L2 and L9 tetraloops (Prathiba and Malathi 2008), which dock into noncanonical receptors. The need to fold and splice out quickly from a cotranscriptionally translated mRNA could have disfavored the selection of tertiary interactions that potentially generate long-lived intermediates in IA2 introns.
In fact, it has been demonstrated that in the IA2 intron containing mRNAs, continuous cotranscriptional translation is essential for efficient splicing of the introns, presumably by avoiding misfolding (Sandegren and Sjoberg 2007). Also consistent with our hypothesis, mutational analysis of a bacteriophage T4 IA2 group I intron showed that perturbation of the naturally selected noncanonical L9–P5 or L2–P8 interactions severely impaired the intron's ability to self-splice in vivo (Brion et al. 1999).
Our studies support the following general conclusions: (1) Selection pressures phylogenetically divergent homologous RNA sequences to fold into identical catalytically active core conformations structured by conserved patterns of tertiary contacts. The energetic stabilities of conserved and nonconserved tertiary motifs vary between homologous RNA molecules depending upon the intron's location within the natural genomic context. (2) The greater thermodynamic stability of a native conformation over competing nonnative structures, achieved through selection of strong tertiary interactions, comes at the expense of slower folding to the catalytically active conformation due to formation of long-lived intermediates. (3) Unique kinetic intermediates associated with a given overall native state topology offer energetically efficient routes to fold to the correct functional conformation. Efficient folding is achieved by balancing the gain in structural stability due to tertiary contact formation with the probability of misfolding due to loss of conformational freedom.
MATERIALS AND METHODS
RNA preparation and labeling
The three ribozymes were in vitro transcribed using the MEGAscript T7 Kit (Ambion) from three linear DNA templates encoding nucleotides 22–409 of the Tetrahymena ribozyme (Zaug et al. 1988), 4–204 of the Azoarcus ribozyme (L-3; R Russell, pers. comm.), and 9–251 of the Twort ribozyme (Golden et al. 2005), each of which contained a T7 promoter sequence immediately upstream of the ribozyme sequence. The transcribed RNA molecules were purified either on a 5% denaturing PAGE or by the MEGAclear kit (Ambion). Each ribozyme was radioactively labeled with 32P at either the 5′ end (Zaug et al. 1988) or the 3′ end (Huang and Szostak 1996). Labeled RNA molecules were purified by 5% denaturing PAGE, gel eluted, precipitated, and dissolved in CE buffer (10 mM Potassium Cacodylate, 0.1 mM EDTA, at pH 7.3). The following oligo substrates, for activity assays, were ordered from Dharmacon: 5′-CAUACGGCC-3′ (Azoarcus), 5′-GCUUAAAAA-3′ (Twort), and 5′-CCCUCUAAAAA-3′ (Tetrahymena); substrates were labeled at the 5′ end as described (Zaug et al. 1988).
Time-resolved and equilibrium hydroxyl radical (•OH) footprinting
Time-resolved •OH footprinting was performed as described (Shcherbakova et al. 2006; Shcherbakova and Brenowitz 2008). Briefly, the 32P-labeled RNA samples were denatured at 95°C for 2 min in “CEK Buffer” (10 mM Potassium Cacodylate, 0.1 mM EDTA at pH 7.3, and 100 mM KCl) and cooled to 37°C. Folding was initiated by rapidly mixing the RNA with a folding buffer containing 20 mM MgCl2 (final Mg2+ concentration after mixing is 10 mM) and 80 mM H2O2, using the two sample syringes in a Kintek quench flow apparatus, and allowed to age for the desired time period in the mixing chamber. •OH cleavage was initiated by adding a Fe-EDTA solution [1 mM Fe(NH4)2(SO4)2 and 1.1 mM EDTA] into the mixing chamber through the quench syringe, allowed to proceed for 5 msec, and the reaction mix was expelled into a tube containing 100% ethanol. The temperature of the mixing chamber and the reaction loops, contained within a box, was maintained by circulating water through the box from a temperature-regulated bath (Neslab RTE-111). Reaction products were precipitated and separated by 8% denaturing PAGE. The dried gels were visualized by Storm PhosphorImaging (GE), and band intensities were quantitated by the SAFA (Das et al. 2005) at single nucleotide resolution. The band intensities were normalized and plotted to identify nucleotides that are at least 20%–30% protected from •OH cleavage between the unfolded (without Mg2+) and the folded (pre-equilibrated sample in 10 mM Mg2+ at 50°C) controls. The normalized bands intensities of protected residues were scaled to fractional saturation, 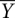 , by
, by
where p denotes the integrated density of the bands and plower and pupper represent the lower and upper limits to the transition, respectively. plower denotes the average of the zero-time point values (initial state without Mg2+, 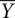 = 0), whereas pupper denotes the average value of the pre-equilibrated controls (folded state in 10 mM Mg2+,
= 0), whereas pupper denotes the average value of the pre-equilibrated controls (folded state in 10 mM Mg2+, 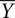 = 1). Fractional values of
= 1). Fractional values of 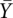 , for each tertiary contact, indicate the extent of progress along the folding reaction coordinate as a function of time.
, for each tertiary contact, indicate the extent of progress along the folding reaction coordinate as a function of time.
Equilibrium •OH footprinting was performed as described in Takamoto et al. (2004) and in the Supplemental legend to Figure S1C.
Analytical ultracentrifugation (AUC)
AUC studies were performed in a Beckman XL-I analytical ultracentrifuge with a An-60 Ti 4 hole rotor. The scans were analyzed with the software DCDT+ (Philo 2006). SEDNTERP was used to estimate the density and viscosity of the buffer. The partial specific volume 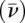 and the hydration of the RNA molecule were assumed to be 0.53cm3/gm and 0.59, respectively.
and the hydration of the RNA molecule were assumed to be 0.53cm3/gm and 0.59, respectively.
Catalytic activity assays
Details of the assays are described in the Supplemental figures (S5–S7). Briefly, 0.5 μM of unlabeled ribozyme was denatured at 95°C for 2 min in CEK buffer and cooled to 37°C. Folding was initiated by adding MgCl2 (final concentration 10 mM) and allowed to proceed for the desired time period. For slower time points (>5 sec) RNA and MgCl2 were hand mixed, whereas for faster time points (<5 sec) mixing was performed in the Kintek quench flow apparatus. At the end of the folding time point, the folding reaction mix was added to the cleavage reaction mix (CEK at pH7.0, 10mM Mg2+, trace amounts 32P-labeled substrate, 1 μM unlabeled substrate, 0.5 μM Guanosine). Aliquots were removed at intervals of 10–15 sec and quenched by gel loading buffer II Ambion (95% Formamide, 18 mM EDTA, and 0.025% SDS, Xylene Cyanol, and Bromophenol Blue). Reaction products were separated by 20% denaturing PAGE, dried gels visualized by Storm PhosphorImaging (GE), and quantitated by Imagequant (GE).
Structural-kinetic modeling
The time progress curves were analyzed by KinFold as described (Laederach et al. 2006, 2007; Martin et al. 2009). Briefly, the sets of time-progress curves of each ribozyme are statistically clustered using Gap statistics built in the KinFold algorithm (http://simtk.org/home/KinFold). Next, all possible kinetic models are critically tested to determine the best-fit or “winning” kinetic topology (with lowest RMS) and the corresponding rate constants. The reaction flux through each intermediate is calculated from the best-fit rate constants, providing a reasonable estimate of the partitioning among pathways from the initial to the final molecular ensemble. The structural models were generated in PyMOL (DeLano 2009).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article. In addition, see the “Intron type vs. exon” table at http://www.rna.ccbb.utexas.edu/SAE/2C/Distributions/g12tte.php (section 2: structure and evolution, subsection 2C: introns—Gr I/II intron distributions).
ACKNOWLEDGMENTS
We thank Rick Russell for his advice on the activity assays and Jesse Stombaugh and the RNA Ontology Consortium for discussions about RNA tertiary interaction maps. These studies were supported by grants from the Institute of General Medical Sciences (RO1-GM085130, PO1-GM066275, R00-GM079953, and U54GM072970) and the Institute of Mental Health (R21-087336) of the National Institutes of Health.
Footnotes
Folding of the Tetrahymena ribozyme from low salt conditions (20 mm Na+) leads to the formation of ∼75% of misfolded species (Russell and Herschlag 1999) and also produces mutually heterogeneous behavior between several tertiary contact partners. Thus, such low ionic strength conditions are unsuitable for our comparative studies. Under conditions comparable to our initial state, the Azoarcus and Tetrahymena ribozymes exist as a compact ensemble in which the native secondary structure is stabilized, while only a few tertiary contacts are at the most partially formed (Rangan and Woodson 2003; Takamoto et al. 2004).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2694811.
REFERENCES
- Adams P, Stahley M, Kosek A, Wang J, Strobel S 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50 [DOI] [PubMed] [Google Scholar]
- Banerjee AR, Jaeger JA, Turner DH 1993. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry 32: 153–163 [DOI] [PubMed] [Google Scholar]
- Basu S, Rambo RP, Strauss-Soukup J, Cate JH, Ferre-D'Amare AR, Strobel SA, Doudna JA 1998. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat Struct Biol 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Brion P, Schroeder R, Michel F, Westhof E 1999. Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: a comparative study. RNA 5: 947–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Belfort M, Cech TR, Davies RW, Schweyen RJ, Shub DA, Szostak JW, Tabak HF 1987. Structural conventions for group I introns. Nucleic Acids Res 15: 7217–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calosci N, Chi CN, Richter B, Camilloni C, Engstrom A, Eklund L, Travaglini-Allocatelli C, Gianni S, Vendruscolo M, Jemth P 2008. Comparison of successive transition states for folding reveals alternative early folding pathways of two homologous proteins. Proc Natl Acad Sci 105: 19241–19246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2 doi: 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J, Gooding A, Podell E, Zhou K, Golden B, Kundrot C, Cech T, Doudna J 1996. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273: 1678–1685 [DOI] [PubMed] [Google Scholar]
- Cayley S, Lewis BA, Guttman HJ, Record MT Jr 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol 222: 281–300 [DOI] [PubMed] [Google Scholar]
- Cech TR 1990. Self-splicing of group I introns. Annu Rev Biochem 59: 543–568 [DOI] [PubMed] [Google Scholar]
- Chauhan S, Woodson SA 2008. Tertiary interactions determine the accuracy of RNA folding. J Am Chem Soc 130: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Behrouzi R, Rangan P, Woodson SA 2009. Structural rearrangements linked to global folding pathways of the Azoarcus group I ribozyme. J Mol Biol 386: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F 1995. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J 14: 1276–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J 16: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB 2005. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL 2009. The PyMOL Molecular Graphics System, http://www.pymol.org. DeLano Scientific LLC, Palo Alto, CA
- Doherty EA, Batey RT, Masquida B, Doudna JA 2001. A universal mode of helix packing in RNA. Nat Struct Biol 8: 339–343 [DOI] [PubMed] [Google Scholar]
- Engelhardt MA, Doherty EA, Knitt DS, Doudna JA, Herschlag D 2000. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry 39: 2639–2651 [DOI] [PubMed] [Google Scholar]
- Fang XW, Pan T, Sosnick TR 1999. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat Struct Biol 6: 1091–1095 [DOI] [PubMed] [Google Scholar]
- Geary C, Baudrey S, Jaeger L 2008. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res 36: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden BL, Kim H, Chase E 2005. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol 12: 82–89 [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR 2004. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell 16: 351–362 [DOI] [PubMed] [Google Scholar]
- Gutell RR, Noller HF, Woese CR 1986. Higher order structure in ribosomal RNA. EMBO J 5: 1111–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Cannone JJ, Shang Z, Du Y, Serra MJ 2000. A story: unpaired adenosine bases in ribosomal RNAs. J Mol Biol 304: 335–354 [DOI] [PubMed] [Google Scholar]
- Hagen M, Cech TR 1999. Self-splicing of the Tetrahymena intron from mRNA in mammalian cells. EMBO J 18: 6491–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D, Cech TR 1990a. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 29: 10159–10171 [DOI] [PubMed] [Google Scholar]
- Herschlag D, Cech TR 1990b. Catalysis of RNA cleavage by the Tetrahymena thermaphila ribozyme. 2. Kinetic description of the reaction of an RNA substrate that forms a mismatch at the active site. Biochemistry 29: 10172–10180 [DOI] [PubMed] [Google Scholar]
- Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D 2006. How the Group I intron works: A case study of RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Huang Z, Szostak JW 1996. A simple method for 3′-labeling of RNA. Nucleic Acids Res 24: 4360–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa Y, Nohmi K, Atsumi S, Shiraishi H, Inoue T 2001. A comparative study on two GNRA-tetraloop receptors: 11-nt and IC3 motifs. J Biochem 130: 251–255 [DOI] [PubMed] [Google Scholar]
- Johnson TH, Tijerina P, Chadee AB, Herschlag D, Russell R 2005. Structural specificity conferred by a group I RNA peripheral element. Proc Natl Acad Sci 102: 10176–10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok L, Shcherbakova I, Lamb J, Park H, Andresen K, Smith H, Brenowitz M, Pollack L 2006. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J Mol Biol 355: 282–293 [DOI] [PubMed] [Google Scholar]
- Laederach A, Shcherbakova I, Liang MP, Brenowitz M, Altman RB 2006. Local kinetic measures of macromolecular structure reveal partitioning among multiple parallel pathways from the earliest steps in the folding of a large RNA molecule. J Mol Biol 358: 1179–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M 2007. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc Natl Acad Sci 104: 7045–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Shub DA 1999. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc Natl Acad Sci 96: 7005–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Gutell RR, Russell R 2006. The UAA/GAN internal loop motif: a new RNA structural element that forms a cross-strand AAA stack and long-range tertiary interactions. J Mol Biol 360: 978–988 [DOI] [PubMed] [Google Scholar]
- Lehnert V, Jaeger L, Michel F, Westhof E 1996. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol 3: 993–1009 [DOI] [PubMed] [Google Scholar]
- Lockless SW, Ranganathan R 1999. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286: 295–299 [DOI] [PubMed] [Google Scholar]
- Martin JS, Simmons K, Laederach A 2009. Exhaustive enumeration of kinetic model topologies for the analysis of time-resolved RNA folding. Algorithms Mol Biol 2: 200–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A, Kellis JJ, Serrano L, Fersht A 1989. Mapping the transition state and pathway of protein folding by protein engineering. Nature 340: 122–126 [DOI] [PubMed] [Google Scholar]
- Maxwell KL, Wildes D, Zarrine-Afsar A, De Los Rios MA, Brown AG, Friel CT, Hedberg L, Horng JC, Bona D, Miller EJ, et al. 2005. Protein folding: defining a “standard” set of experimental conditions and a preliminary kinetic data set of two-state proteins. Protein Sci 14: 602–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Westhof E 1990. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol 216: 585–610 [DOI] [PubMed] [Google Scholar]
- Philo JS 2006. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem 354: 238–246 [DOI] [PubMed] [Google Scholar]
- Prathiba J, Malathi R 2008. Group I introns and GNRA tetraloops: remnants of ‘The RNA world’? Mol Biol Rep 35: 239–249 [DOI] [PubMed] [Google Scholar]
- Rangan P, Woodson SA 2003. Structural requirement for Mg2+ binding in the group I intron core. J Mol Biol 329: 229–238 [DOI] [PubMed] [Google Scholar]
- Rangan P, Masquida B, Westhof E, Woodson SA 2003. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci 100: 1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Shub DA 1992. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357: 173–176 [DOI] [PubMed] [Google Scholar]
- Rook MS, Treiber DK, Williamson JR 1999. An optimal Mg(2+) concentration for kinetic folding of the tetrahymena ribozyme. Proc Natl Acad Sci 96: 12471–12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Herschlag D 1999. New pathways in folding of the Tetrahymena group I RNA enzyme. J Mol Biol 291: 1155–1167 [DOI] [PubMed] [Google Scholar]
- Russell R, Herschlag D 2001. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol 308: 839–851 [DOI] [PubMed] [Google Scholar]
- Russell R, Millett IS, Doniach S, Herschlag D 2000. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat Struct Biol 7: 367–370 [DOI] [PubMed] [Google Scholar]
- Russell R, Zhuang X, Babcock H, Millett I, Doniach S, Chu S, Herschlag D 2002. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci 99: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D 2006. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol 363: 531–544 [DOI] [PubMed] [Google Scholar]
- Russell R, Tijerina P, Chadee AB, Bhaskaran H 2007. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry 46: 4951–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L, Sjoberg BM 2007. Self-splicing of the bacteriophage T4 group I introns requires efficient translation of the pre-mRNA in vivo and correlates with the growth state of the infected bacterium. J Bacteriol 189: 980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattin BD, Zhao W, Travers K, Chu S, Herschlag D 2008. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc 130: 6085–6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA 1998. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279: 1940–1943 [DOI] [PubMed] [Google Scholar]
- Shcherbakova I, Brenowitz M 2005. Perturbation of the hierarchical folding of a large RNA by the destabilization of its Scaffold's tertiary structure. J Mol Biol 354: 483–496 [DOI] [PubMed] [Google Scholar]
- Shcherbakova I, Brenowitz M 2008. Monitoring structural changes in nucleic acids with single residue spatial and millisecond time resolution by quantitative hydroxyl radical footprinting. Nat Protoc 3: 288–302 [DOI] [PubMed] [Google Scholar]
- Shcherbakova I, Mitra S, Beer R, Brenowitz M 2006. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res 34: e48 doi: 10.1093/nar/gkl055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R 2005. Evolutionary information for specifying a protein fold. Nature 437: 512–518 [DOI] [PubMed] [Google Scholar]
- Sosnick TR 2008. Kinetic barriers and the role of topology in protein and RNA folding. Protein Sci 17: 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnick TR, Pan T 2004. Reduced contact order and RNA folding rates. J Mol Biol 342: 1359–1365 [DOI] [PubMed] [Google Scholar]
- Stahley MR, Adams PL, Wang J, Strobel SA 2007. Structural metals in the group I intron: a ribozyme with a multiple metal ion core. J Mol Biol 372: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamoto K, Das R, He Q, Doniach S, Brenowitz M, Herschlag D, Chance M 2004. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J Mol Biol 343: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Tanner M, Cech T 1996. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA 2: 74–83 [PMC free article] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR 2001a. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol 11: 309–314 [DOI] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR 2001b. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J Mol Biol 305: 11–21 [DOI] [PubMed] [Google Scholar]
- Uchida T, Takamoto K, He Q, Chance M, Brenowitz M 2003. Multiple monovalent ion-dependent pathways for the folding of the L-21 Tetrahymena thermophila ribozyme. J Mol Biol 328: 463–478 [DOI] [PubMed] [Google Scholar]
- Vicens Q, Cech TR 2006. Atomic level architecture of group I introns revealed. Trends Biochem Sci 31: 41–51 [DOI] [PubMed] [Google Scholar]
- Vicens Q, Paukstelis PJ, Westhof E, Lambowitz AM, Cech TR 2008. Toward predicting self-splicing and protein-facilitated splicing of group I introns. RNA 14: 2013–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Massire C 2004. Structural biology. Evolution of RNA architecture. Science 306: 62–63 [DOI] [PubMed] [Google Scholar]
- Woodson SA 2005. Structure and assembly of group I introns. Curr Opin Struct Biol 15: 324–330 [DOI] [PubMed] [Google Scholar]
- Zarrinkar P, Williamson J 1994. Kinetic intermediates in RNA folding. Science 265: 918–924 [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Grosshans CA, Cech TR 1988. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry 27: 8924–8931 [DOI] [PubMed] [Google Scholar]