Abstract
We previously showed that the insertion of a hammerhead ribozyme (Rz) in a critical intronic position between the EDA exon and a downstream regulatory element affects alternative splicing. Here we evaluate the effect of other intronic cotranscriptional cleavage events on alternative pre-mRNA processing using different ribozymes (Rz) and Microprocessor target sequences (MTSs). In the context of the fibronectin EDA minigene, intronic MTSs were cleaved very inefficiently and did not affect alternative splicing or the level of mature transcripts. On the contrary, all hammerhead Rz derivatives and hepatitis δ Rz were completely cleaved before a splicing decision and able to affect alternative splicing. Despite the very efficient Rz-mediated cleavage, the levels of mature mRNA were only reduced to ∼40%. We show that this effect on mature transcripts occurs regardless of the type and intronic position of Rzs, or changes in alternative splicing and exon definition. Thus, we suggest that intron integrity is not strictly required for splicing but is necessary for efficient pre-mRNA biosynthesis.
Keywords: ribozymes, alternative splicing, cotranscriptional processing, Microprocessor target sequence
INTRODUCTION
A typical mammalian protein-coding gene transcribed by RNA polymerase II (Pol II) contains relatively short exonic sequences separated by long introns, which average 3 kb but are frequently longer than 10 kb. Introns are removed from pre-mRNAs by the spliceosome and, according to EST data, >90% of human genes undergo alternative splicing (Pan et al. 2008; Wang et al. 2008). The efficiency and accuracy of pre-mRNA splicing depend on complex interactions on the nascent pre-mRNAs. Small nuclear ribonuclear proteins assemble on splice sites along with splicing factors such as SR proteins and hnRNPs that interact with exonic and/or intronic regulatory sequences (Maniatis and Reed 2002). Several lines of evidence indicate that pre-mRNA splicing and transcription are coupled events. Nuclear imaging studies in Drosophila and polytene chromosomes indicated that intron looping can occur while the transcript is tethered to the chromatin template (Beyer et al. 1981; Beyer and Osheim 1988). During transcription, splicing factors are recruited to actively transcribed genes (Gornemann et al. 2005; Lacadie et al. 2006; Listerman et al. 2006), and the accumulation of spliceosomal components requires Pol II with an intact C-terminal domain (CTD) (Misteli and Spector 1999). Pol II transcripts are more efficiently processed than T7 transcripts both in in vivo and in vitro assays in which transcription and splicing are linked (Das et al. 2006; Hicks et al. 2006; Natalizio et al. 2009). Published data support a key role for Pol II and specifically for its CTD in the coupling of transcription and pre-mRNA processing (McCracken et al. 1997), including a direct contact with splicing factors (Morris and Greenleaf 2000; Rosonina et al. 2005). Recently, SR proteins and components of U1 snRNP have been suggested to be involved in coupling transcription to pre-mRNA processing via the CTD (Das et al. 2007). Coupling of pre-RNA processing and transcription also has an influence on alternative splicing. Promoter type, rate of elongation, transcriptional activators, and chromatin-remodeling factors have been shown to affect alternative splicing (Kornblihtt 2005, 2007; Luco et al. 2010).
Coupling of pre-RNA processing and transcription is thought to facilitate processing of intronic sequences through exon tethering by Pol II (Dye et al. 2006). The existence of a molecular tether between the nascent pre-mRNA and RNA Pol II transcripts comes from the original observation that RNAP II transcripts derived from β-globin are efficiently spliced in vivo when the intron is cotranscriptionally cleaved by a hammerhead ribozyme (Dye et al. 2006). In addition, evidence that intronic cotranscriptional cleavage does not affect splicing has been reported in yeast (Lacadie et al. 2006). Exon tethering is also consistent with the fact that several introns contain pri-miRNAs hairpins, which have been reported to be cotranscriptionally cleaved by the Microprocessor without affecting the splicing of adjacent constitutive exons (Kim and Kim 2007; Morlando et al. 2008). The excision of pri-miRNAs from introns involves a specific complex called the Microprocessor, containing Drosha, an RNase III–like enzyme, and its cofactor DGCR8 (Denli et al. 2004). However, it is not known if the Microprocessor complex crops pri-miRNAs cotranscriptionally with a different efficiency and if this affects the regulation of alternative splicing.
In contrast to the less efficient hammerhead Rz, δ hepatitis Rz was recently reported to inhibit splicing in the β-globin system (Fong et al. 2009), suggesting that exon tethering of Pol II to rescue splicing of a severed transcript depends on the relative activity of the Rz in comparison to the rate of splicing (Fong et al. 2009). A rapid cotranscriptional cleavage of the β-globin intron, as provided by the δ hepatitis Rz, but not by the hammerhead Rz, aborts pre-mRNA processing with a nearly complete inhibition of gene expression.
We previously showed that, in contrast to the constitutive β-globin, two alternatively spliced exons (the fibronectin EDA and α-TM) are more sensitive to cotranscriptional cleavage mediated by an Rz and more prompt to changes in their pattern of splicing. In particular, placing the N117 derivative of a Schistosoma mansoni (Sm) hammerhead Rz between the EDA exon and its negative intronic downstream regulatory element (DRE) affects EDA alternative splicing (Gromak et al. 2008). The N117 Rz was selected for its extremely high cleavage activity in vivo (Yen et al. 2004), and, when placed within an intron of an expression vector, it did not apparently inhibit the expression of a reporter gene (Yen et al. 2004). When the N117 Rz was positioned in the middle of the relatively long introns flanking the EDA exon and outside the DRE, it did not affect the regulation of alternative splicing (Gromak et al. 2008). Thus, in the case of alternative splicing, the continuity of the nascent transcript between the EDA exon and DRE is essential for the alternative splicing of the fibronectin EDA minigene. In addition, the DRE has been recently shown to affect the relative order of intron removal (de la Mata et al. 2010). As cotranscriptional cleavage between the EDA and its regulatory element affects alternative splicing, this system provides a useful model to evaluate exon tethering and investigate the effect of different Rzs and MTSs on pre-mRNA processing.
In this study, using the EDA alternative splicing minigene system, we evaluated intronic cleavage events mediated by different Rzs and Microprocessor target sequences and their effect on alternative splicing regulation. We show that all Rzs, in contrast to MTSs and consistent with their rapid cotranscriptional cleavage, affect EDA alternative splicing. MTS are processed less efficiently but, interestingly, showed a variable residual cleavage activity. On the other hand, we observed that all Rzs reduce the mRNA levels to ∼40% regardless of the distance from splice sites, alternative splicing pattern and exon definition, and relative cleavage efficiency. We suggest that in our system the disruption of the intron integrity is not completely rescued by Pol II–mediated exon tethering and that defective transcripts are to some extent subjected to abortive processing.
RESULTS
The hammerhead ribozyme interrupts the continuity of the nascent transcript and facilitates splicing of downstream introns in a DRE-dependent manner
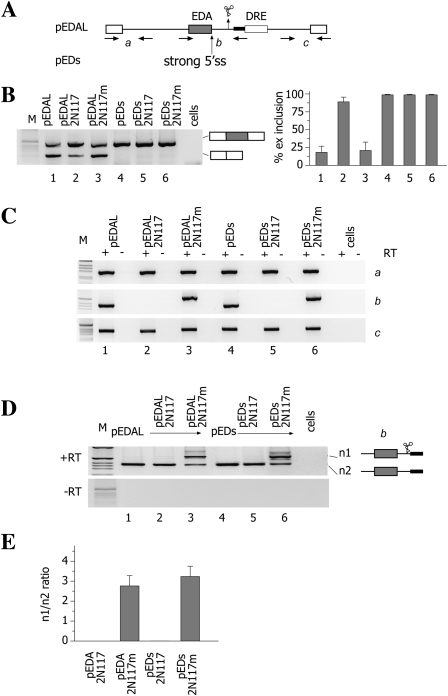
We previously showed that the insertion of the N117 hammerhead Rz between the alternatively spliced EDA exon and its downstream regulatory element (DRE) induces exon inclusion (Gromak et al. 2008). To study in more detail the cleavage efficiency of the hammerhead Rz, we have analyzed the nascent transcripts by means of RT-PCR. To amplify nascent pre-mRNAs across the Rz cleavage site and distinguish them from endogenous fibronectin transcripts, we inserted a short 25-nt-long spacer within the intron (Fig. 1A, pEDAL and pEDAs minigenes derivatives). As shown in Figure 1B, the spacer insertion did not modify the previously reported splicing pattern, whereas the wild-type Rz pEDAL2N117 induced exon inclusion, and the inactive Rz had no effect on spliced mRNA (Fig. 1B, lanes 1–3). Analysis of the pre-mRNAs showed that the active Rz had no significant effect on the abundance of upstream and downstream nascent transcripts (Fig. 1C, lanes 1–3, a and c products, respectively), while the nascent unspliced pre-mRNA across the Rz was completely absent (Fig. 1C, lanes 1–3, b product). To confirm the absence of nascent pre-mRNA across the Rz, we transfected pEDAL alone or together with pEDAL2N117 or pEDAL2N117m constructs, followed by amplification of the b pre-mRNA. Cotransfection of pEDAL with the inactive mutant Rz minigene pEDAL117m, followed by amplification of the b fragment, showed the two nascent pre-mRNAs originating from the two minigenes, as expected. The upper band incorporates the mutant Rz, whereas the lower band derives from the empty pEDAL minigene (Fig. 1D, lane 3). On the contrary, cotransfection of pEDAL along with pEDALN117 did not show any upper band derived from the active Rz that is consistent with its high cleavage efficiency (Fig. 1D, lane 2). We also evaluated the abundance of the nascent pre-mRNA in the pEDs minigenes (Fig. 1A) in which the 5′ss of the exon was strengthened, rendering the exon constitutively included (Fig. 1B, lanes 4–6). Again, amplification across the active Rz did not generate any product (Fig. 1C,D, lane 5), whereas the amplification of the upstream and downstream sequences (a and c products) was not affected (Fig. 1C, lanes 4–6). We also tested in vitro the activity of N117Rz in the context of the pEDA intron, and we it found it to be extremely effective, cleaving the RNA with an almost 100% efficiency (Supplemental Fig. 1). The absence of any nascent transcript deriving from the minigenes containing the wild-type hammerhead N117 Rz indicates that this element is very efficiently cleaved before splicing and that it interrupts the continuity of the nascent transcript.
FIGURE 1.
Cleavage efficiency mediated by the N117 hammerhead ribozyme affects EDA alternative splicing. (A) Diagram of the pEDAL and pEDs minigenes. The position of the N117 ribozyme is indicated. To distinguish amplification products in cotransfection experiments, two active or mutant N117 hammerhead ribozymes were inserted in tandem between the EDA exon and the DRE. The DRE is indicated, and the thick line represents the 25-nt-long spacer (see text). The primers used in RT-PCR analysis to amplify nascent transcripts are indicated. (B) Analysis of mature spliced transcripts. RT-PCR analysis of total RNA from Hep3B cells expressing different minigenes was performed, and the resulting amplification products were resolved on 2% agarose gel. The two splicing products are schematically indicated. (M) The 1-kb molecular weight marker. (Histogram on the right) The percentage of EDA exon inclusion (±SD) based on at least three independent duplicate experiments. (C) Analysis of nascent transcripts. Amplification of nascent mRNA with (+) and without (−) RT resolved on 2% agarose gel. The identity of the amplified fragments (a, b, and c) is reported on the right of each panel. (D) Analysis of nascent b transcript across the Rzs. The indicated minigenes (0.45 μg) were cotransfected with pEDAL (0.15 μg) (lanes 1–3) or with pEDs (0.15 μg) (lanes 4–6) in Hep3B cells and the b fragment amplified from total RNA. The identity of b bands originating from pEDAL or pEDS (n1) and other minigenes (n2) is reported. (Lower panel) The PCR control, without reverse transcriptase (−RT). (E) Ratio of the intensity of the n1/n2 bands in D expressed as the mean ±SD of three independent experiments done in duplicate. Quantification of pre-mRNA was done using a fluorescent primer.
To understand why the Rz-mediated cleavage of the nascent transcript induces exon inclusion, we initially considered the possibility that 5′–3′ exonucleases can cotranscriptionally degrade the DRE. However, silencing of the Xrn2 5′–3′ exonuclease did not affect the splicing pattern in minigenes with either wild-type or mutant Rzs, indicating that a cotranscriptional degradation of the DRE is not implicated in the changes of EDA alternative splicing (Supplemental Fig. 2).
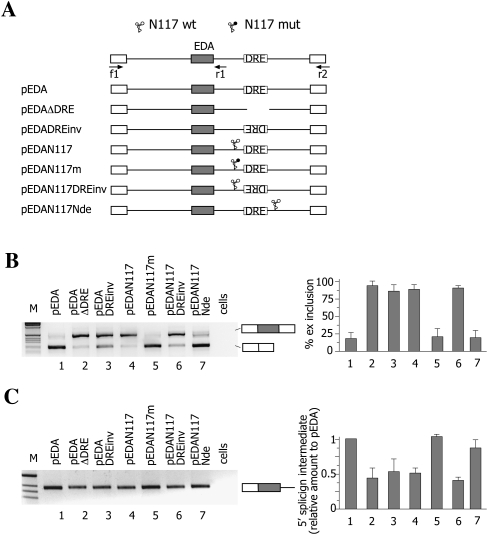
An alternative explanation for the effect of the Rz on mature transcript could be a change in the splicing efficiency of the intron that contains the Rz. Previous studies both in the natural chromosomal context and in minigene systems have shown that there is a preferential removal of the intron downstream from the EDA exon before the upstream intron has been removed (Pandya-Jones and Black 2009; de la Mata et al. 2010). Interestingly, deletion of the DRE was shown to affect the relative rate of the downstream intron excision, decreasing the amount of the 5′-splicing intermediate (de la Mata et al. 2010). To evaluate the possible effect of the Rz-mediated interruption of the nascent transcript on the intron splicing efficiency, we analyzed the 5′-splicing intermediate by semiquantitative RT-PCR amplification. Consistent with the previous observation, deletion or inversion of the DRE in our minigene system reduces the amount of 5′-splicing intermediate (Fig. 2C, lanes 2,3). The diminished level of the 5′-splicing intermediate reflects an increase in the splicing efficiency of the downstream intron. A similar effect was observed when the active Rz, but not the inactive one, was inserted between the exon and the DRE (Fig. 2C, lanes 4 and 5, respectively). On the contrary, insertion of the Rz after the DRE did not affect the amount of 5′ intermediate (Fig. 2C, lane 7), indicating that the Rz reduces the 5′-splicing intermediate only when it cleaves the nascent transcript between the EDA exon and the DRE. Thus, the interruption of the continuity of the nascent transcript mediated by the Rz facilitates splicing of the downstream intron in a DRE-dependent manner.
FIGURE 2.
The N117 ribozyme facilitates splicing of the downstream intron in a DRE-dependent manner. (A) Diagram of the pEDA minigenes. The position of the ribozyme (wild-type [WT] or mutant) and of the DRE sequence is indicated. (Arrows) The locations of primers used in RT-PCR analysis. Exonic sequences are boxed. (B) Analysis of mature spliced transcripts. Minigenes (0.5 μg) were transfected into Hep3B cells and the pattern of splicing evaluated using f1 and r2 primers. (M) The 1-kb molecular weight marker. The identity of the splicing products is schematically reported on the right of the panel. The histogram shows the percentage of EDA exon inclusion. Data represent the mean ± SD of three independent experiments performed in duplicates. (C) Analysis of 5′ splicing intermediates. Minigenes (0.5 μg) were transfected into Hep3B cells along with 0.1 μg of pCF1 plasmid, which was used as control for normalization of transfection and reverse transcriptase efficiencies. RT-PCR was carried out with a1 and r1 primers and amplification products resolved on 2% agarose gel. The identity of the band is indicated on the right of the panel. The histogram shows the relative amount of EDA 5′-splicing intermediates expressed as normalized ratio to pEDA. Calculations show mean ± SD at two independent experiments done in duplicate. Real-time PCR was carried out on pCF1 transcript to assess transfection efficiency (see Materials and Methods).
The intronic hammerhead N117 ribozyme modulates alternative splicing in different minigene contexts
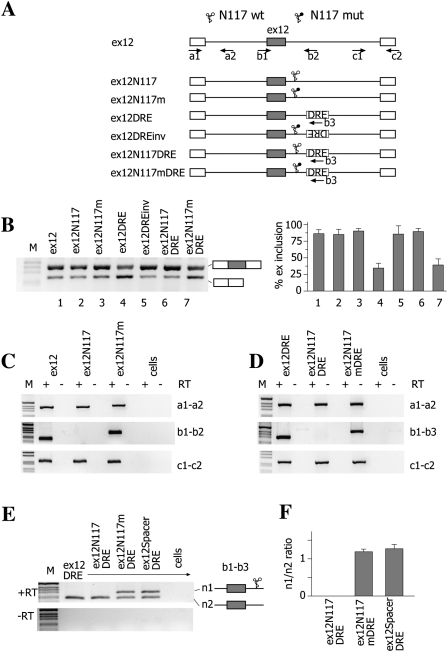
To further explore the effect of the Rz on alternative splicing, we tested both the N117 Rz and the DRE regulatory element in the context of the CFTR exon 12 (Fig. 3A). Transient transfection of the normal CFTR minigene produced a splicing pattern in which ∼85% of the exon is included in the final mRNA (Fig. 3B, lane 1; Pagani et al. 2003). Cloning of the DRE downstream from the CFTR exon 12 reduced the percentage of exon inclusion to ∼40% (Fig. 3B, lane 4), whereas the DRE in inverted orientation did not change the splicing pattern (Fig. 3B, lane 5). The insertion of an active Rz, but not of an inactive one, between the CFTR exon 12 and the DRE restored the original splicing pattern, suggesting that the continuity of the nascent pre-mRNA molecule is required for the DRE-dependent splicing inhibition also in the case of CFTR (Fig. 3B, lanes 6 and 7, respectively). On the other hand, in the absence of an intronic DRE, wild-type and mutant Rzs alone did not affect the splicing pattern (Fig. 3B, lanes 2 and 3, respectively), consistent with the absence of a natural downstream intronic splicing regulatory element in CFTR exon 12.
FIGURE 3.
The N117 hammerhead ribozyme affects CFTR exon 12 alternative splicing in a DRE-dependent manner. (A) Schematic representation of the CFTR exon 12 minigenes. The positions of wild-type (WT) and mutant N117 ribozymes and of DRE are indicated. (Arrows) The locations of primers used in RT-PCR analysis. Exonic sequences are boxed. (B) Analysis of mature spliced transcripts. Minigenes were transfected into Hep3B cells, and RT-PCR products amplified with a1 and c2 primers were resolved on a 2% agarose gel. Exon 12 inclusion or exclusion forms are indicated. (M) The 1-kb molecular weight marker. The histogram shows the percentage of CFTR exon 12 inclusion (±SD) based on at least three independent duplicate experiments. (C,D) Analysis of nascent transcripts. Ex12 minigenes were transfected in Hep3B cells and analyzed with the indicated pairs of primers reported on the right of each panel. RT(−) is the control in absence of reverse transcriptase. (M) The 1-kb molecular weight marker. (E) Analysis of the b1–b3 pre-mRNA transcript. The indicated minigenes (0.45 μg) were cotransfected with pTB ex 12 DRE (0.15 μg). Total RNA was amplified with b1 and b3 primers, and the identity of the two resulting products (n1 and n2) is indicated. (Lower panel) The amplification without reverse transcriptase (−RT). (F) Quantification of pre-mRNA n1 and n2 ratio expressed as mean ± SD of two independent experiments done in duplicate.
To study the cleavage efficiency of the hammerhead Rz in the CFTRex12 context, we have analyzed the pre-mRNAs: The nascent transcripts showed that the active Rz had no significant effect on pre-mRNA abundance of upstream and downstream nascent transcripts (Fig. 3C,D, lanes 1–3, a1–a2 and c1–c2 products), whereas the nascent pre-mRNA across the Rz was completely absent (Fig. 3C, lanes 2, b1–b2; Fig. 3D, b1–b3). We confirmed the absence of nascent pre-mRNA across the active Rz through cotransfection experiments in which no amplification of the b1–b3 product was observed (Fig. 3E,F). Furthermore, the mutant Rz and a control minigene (ex12 spacer DRE) showed a band of similar intensity, indicating that no residual cleavage activity is present in the mutant Rz, as also evaluated by quantitative analysis (Fig. 3F). On the other hand, the small difference in the isoform ratio between the ex12DRE and ex122N117mDRE is not due to the residual activity of the mutant Rz but possibly as a consequence of a minor effect of the transcribed mutant Rz sequences on the DRE.
We also evaluated the effect of the N117 Rz in the context of the EDA combined with a strong downstream intronic silencer consisting of a GAA repeat expansion (Baralle et al. 2008). The wild-type (WT) Rz but not the mutant was able to rescue EDA exon skipping, and analysis of nascent intermediates did not show any PCR product derived from amplification of the WT Rz (Supplemental Fig. 3; Supplemental Data). These results, obtained with different minigene systems, are entirely consistent with the hypothesis that the Rz-mediated interruption of the continuity of nascent RNA between the exon and the intronic splicing regulatory element affects alternative splicing regulation.
Both hammerhead and hepatitis δ ribozymes efficiently cleave nascent mRNA and affect EDA alternative splicing regulation
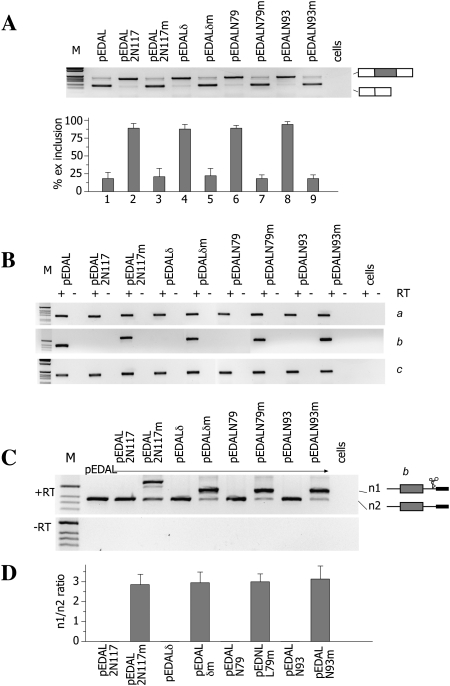
To clarify the role of the catalytic activity of different Rzs on alternative splicing regulation, we evaluated the hepatitis δ Rz, which has been suggested to be more active in vivo when compared to the hammerhead Rz (Fong et al. 2009), as well as derivatives of the hammerhead itself (Yen et al. 2004). The hammerhead derivatives were specifically developed in vivo and showed different cleavage efficiencies in comparison to the original hammerhead, with 1401 ± 228–, 225 ± 70–, and 53 ± 13–fold increase for N117, N79, and N93, respectively (Yen et al. 2004). In addition, transcripts containing the N79 ribozyme were recently shown to be completely cleaved in vivo in transfected cells (Chen et al. 2009). The Rzs or their corresponding inactive variants were inserted between the EDA exon and the DRE, and the resulting minigenes were analyzed for the EDA splicing pattern and nascent pre-mRNA. In all cases, only active Rzs induced EDA exon inclusion (Fig. 4A) and showed no nascent pre-mRNA (Fig. 4B, b product). Quantitative analysis in cotransfection experiments confirmed the absence of nascent products (Fig. 4C,D). Furthermore, the pre-mRNA abundance across the upstream or downstream exon junctions was not significantly affected (Fig. 4B, lanes 1–3, a and c products). Thus, we did not detect in our system significant differences in the cleavage activity between hammerhead and hepatitis δ Rzs. More precisely, both types of Rzs possess a sufficient cleavage activity in vivo that leads to equally efficient interruption of the nascent transcript and comparable changes in the EDA alternative splicing pattern.
FIGURE 4.
Effects of different ribozymes on EDA alternative splicing and nascent transcripts. (A) Analysis of mature spliced transcripts. The indicated minigenes derived from pEDAL (Fig. 1A) contain different Rzs or their corresponding inactive variant inserted between the EDA and the DRE. Minigenes were transfected into Hep3B cells, total RNA was extracted, and its splicing pattern was analyzed by RT-PCR. (M) The 1-kb molecular weight marker. The identity of the corresponding amplified bands is shown. The histogram shows the percentage of EDA exon inclusion, and data are the mean ± SD of three independent experiments performed in duplicate. (B) Analysis of nascent transcripts. Nascent transcripts derived from transfections of the indicated minigenes in cells were analyzed by RT-PCR using the specific pairs of primers indicated in Figure 1A. The identity of the bands (a, b, c) is shown. (C) Analysis of nascent b pre-mRNA transcript across the ribozyme. Minigenes (0.45 μg) were cotransfected with pEDAL (0.15 μg). Total RNA was amplified with the specific pair of primers indicated in Figure 1A. (M) The 1-kb molecular weight marker. The identity of the bands (n1 and n2) is shown. (Lower panel) A control PCR, without reverse transcriptase (−RT). (D) Quantification of n1 and n2 ratio of C expressed as the mean ± SD of two independent experiments done in duplicate.
Microprocesssor target sequences are cleaved less efficiently and do not affect EDA alternative splicing
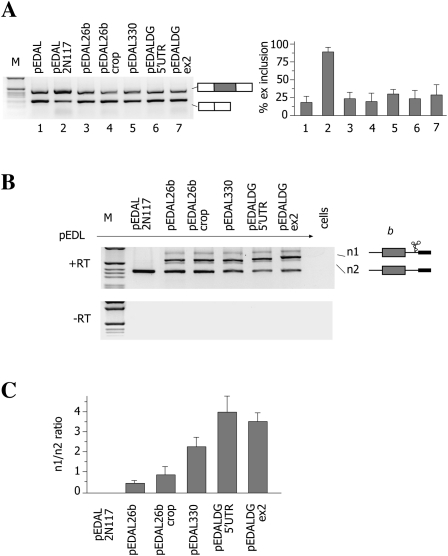
As primary miRNA transcripts are processed cotranscriptionally (Kim and Kim 2007; Morlando et al. 2008; Pawlicki and Steitz 2008), we evaluated the effect of different Microprocessor target sequences (MTSs) on the regulation of EDA alternative splicing. We hypothesized that if the cropping by the Microprocessor is as fast as by Rzs, it will affect EDA alternative splicing. We tested miR-26b and miR-330 pri-miRNA hairpins, a cropping-deficient miR-26b mutant (miR-26bcrop) (Kim and Kim 2007), and two MTS hairpins of DGCR8 mRNA. The last two sequences are located in the 5′ UTR region and in exon 2 of DGCR8, respectively, and are the target for the Microprocessor complex and act in a DGCR8 autoregulatory feedback to degrade their own mRNA (Han et al. 2009; Shenoy and Blelloch 2009; Triboulet et al. 2009). Different MTS hairpin sequences, along with ∼60 bp of corresponding natural flanking regions, were inserted between the EDA exon and its intronic regulatory element. The resulting minigenes were transfected in Hep3b cells for the analysis of the splicing pattern (Fig. 5A) and evaluation of miRNA production. Evaluation of miRNA production revealed that transfection of the miR-26b and miR-330 minigenes, but not of the defective miR-26bcrop, produced the corresponding mature miRNAs (Supplemental Fig. 4): This indicated that in pEDA minigene context, the pri-miRNA hairpins are correctly processed by the Microprocessor complex. Analysis of the splicing pattern showed that, in contrast to N117 Rz, which increases the percentage of exon inclusion, none of the MTSs affected the EDA splicing pattern significantly, suggesting an inefficient cotranscriptional cleavage of nascent pre-mRNA (Fig. 5A). As MTSs did not affect EDA alternative splicing when inserted between the exon and the intronic regulatory element, it is possible that their relative cotranscriptional cleavage activity is reduced in comparison with the activity of Rzs. To study the efficiency of cleavage mediated by different MTSs, we evaluated the abundance of nascent pre-mRNA across the MTSs in cotransfection experiments. Each minigene, containing MTSs or Rzs, was cotransfected with the control pEDAL construct, and the resulting RNAs were amplified to obtain the b product. Two major bands were identified, the lower one resulting from amplification of pEDA-derived transcripts and the higher one from amplification of MTS- or Rz-derived splicing intermediates. While no upper band was evident in the case of pEDAL2N117 due to its very efficient Rz cleavage, in all the other cases 5′-splicing intermediate and nascent pre-mRNA products were detected (Fig. 5B). Interestingly, analysis of nascent pre-mRNA revealed different amounts of transcripts derived from MTSs (Fig. 5B), suggesting a different relative cleavage efficiency of the MTSs. To evaluate the ratio between the intensity of the two bands, we performed RT-PCR using a fluorescent oligonucleotide. The highest ratio between the two bands was observed for miR-330 and DGCR8 minigenes, whereas pEDA26b showed the lowest ratio (Fig. 5C). In comparison to miR-26b, miR-26bcrop showed an increase in the ratio, compatible with a reduced but not completely abolished activity in processing of the nascent mRNA by the Microprocessor complex. No transcripts were observed from minigenes with wild-type N117 Rz. Altogether these results indicate that the Rzs are rapidly and more efficiently cleaved than the MTSs; this could explain the lack of effect of MTSs on the splicing pattern of the alternative EDA exon (Fig. 5A). However, among the MTSs, miR-26b is cotranscriptionally processed faster than the DGCR8 and miR-330 hairpins.
FIGURE 5.
Effects of Microprocessor target sequences (MTSs) on EDA alternative splicing and pre-mRNA processing. (A) Analysis of mature spliced transcripts. The indicated minigenes derived from pEDAL (Fig. 1A) contain different Microprocessor target sequences inserted between the EDA and the DRE. Minigenes were transfected in Hep3B cells and total RNA analyzed by RT-PCR to detect mature transcripts. (M) The 1-kb molecular weight marker. The identity of the corresponding amplified bands is indicated. The histogram shows the percentage of EDA exon inclusion. Data represent the mean ± SD of three independent experiments performed in duplicate. (B) Analysis of pre-mRNA transcripts. The indicated minigenes (0.45 μg) were cotransfected with pEDAL (0.15 μg) in Hep3B cells, and the nascent b fragment was amplified with the specific pair of primers indicated in Figure 1A. The identity of the b bands originating from pEDAL (n1) or other minigenes (n2) is indicated. (Lower panel) A control PCR, without reverse transcriptase (−RT). (C) Quantification of n1 and n2 ratio expressed as the mean ± SD of two independent experiments done in duplicate.
Different Rzs but not MTSs reduce mRNA abundance regardless of exon definition, alternative splicing pattern, and intronic position
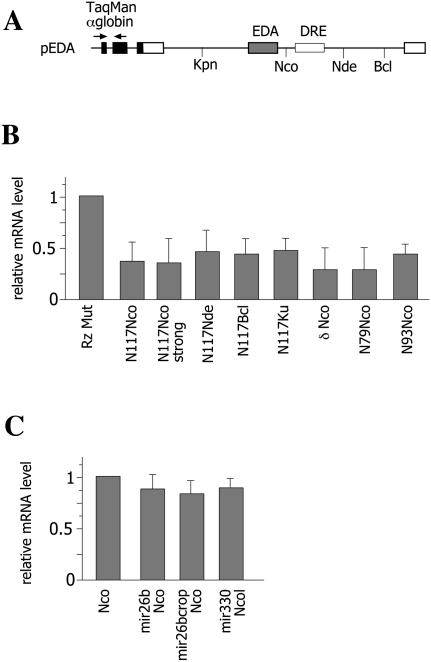
As the relative cotranscriptional cleavage efficiency has been recently shown to severely affect processing of α-globin transcripts (Fong et al. 2009), we have evaluated how insertion of the different Rzs or the MTSs in the fibronectin intron can affect the mRNA levels in vivo. Considering the competition between the Rz cleavage and splicing efficiency, as previously proposed (Fong et al. 2009), one can speculate that the differences in the amount of mature transcripts produced can depend on the cleavage efficiency, on the intronic position of the Rz, or on alternative splicing pattern. To quantify the total amount of spliced mRNA derived from the minigenes, we used an RT-PCR TaqMan assay, which amplifies the spliced α-globin portion common to all the minigenes. After normalization for transfection and RT efficiency, we compared the total amount of spliced mRNA produced for each wild type and its corresponding inactive ribozyme. Even when the N117 hammerhead ribozyme in the Nco position was completely cleaved (Fig. 2, pEDALN117 construct), it did not entirely reduce the amount of mature mRNA (Fig. 6B). In fact, relative to the inactive Rz, ∼40% of expression is retained. Interestingly, the hammerhead N79 and N93 derivatives and the hepatitis δ Rz showed a similar level of expression when inserted at the same position (Fig. 6). We also evaluated the role of exon definition, studying the N117 Rz in the context of the pEDs minigene in which, due to a strong 5′ss, the EDA exon is completely included (Fig. 2, pEDs construct). Also in this case, we observed ∼40% of mRNA levels. Lastly, we studied the role of the intronic position since the N117 Rz at the Nco site is only 30 bp downstream from the EDA exon. We evaluated the Rz-inserted 433 bp and 900 bp downstream at the Nde, Bcl positions as well as at the Kpn site, which is 622 bp upstream of the alternatively spliced EDA exon (Fig. 6A). The analysis of relative mRNA levels again gave a constant reduction to 40% levels. As the active Rz induced the exon inclusion only when inserted into the NcoI site (Fig. 1), with no effect on splicing pattern when introduced into other intronic positions (Fig. 2B, pEDAN117Nde construct; Gromak et al. 2008), this means that the reduced level of mRNA is not due to the changes associated with alternative splicing mediated by the Rzs. On the other hand, miR-26b and miR-330 pri-miRNAs are very inefficiently cleaved and do not significantly affect the amount of mRNAs (Fig. 6C). Altogether these data indicate that, in the FN minigenes, a constant amount of transcript is processed regardless of exon definition, alternative splicing pattern, or distance between the Rz cleavage site and the flanking exons.
FIGURE 6.
Analysis of gene expression in ribozyme- and MTS-derived minigenes. (A) Diagram of the fibronectin EDA minigene. Fibronectin exons (white boxes); α-globin exons (black boxes); introns (lines). The unique restriction sites in the introns used for wild-type (WT) or mutant ribozyme insertion are indicated. (Arrows) The positions of the TaqMan α-globin amplification primers. Minigenes were transfected in HeLa cells, and their expression levels were monitored by real-time PCR using a specific α-globin TaqMan assay. Each minigene (0.5 μg) was cotransfected with the control pCF1 (0.1 μg), which was used for normalization of transfection and reverse transcriptase efficiencies. (B) Relative expression levels of mRNAs produced from minigenes carrying WT or mutant ribozymes. The relative WT Rzs levels were normalized to the corresponding mutant Rz levels, and data are shown as the mean of at least two transfection experiments done in duplicate. Error bars represent the SEM. (C) Relative expression levels of mRNAs produced from miRNA-derived minigenes. The relative miRNA levels were normalized to the pEDA Nco empty vector levels, and data are shown as the mean of at least two transfection experiments done in duplicate. Error bars represent the SEM.
DISCUSSION
Intronic sequences in Pol II–derived pre-mRNA transcripts are cotranscriptionally processed by the spliceosome and by the Microprocessor to generate mature mRNAs and miRNAs, respectively. We have used different ribozymes and Microprocessor target sequences to study the effect of intronic cotranscriptional cleavage events on pre-mRNA processing and regulation of alternative splicing. Consistent with the exon tethering model (Dye et al. 2006), an Rz-mediated interruption of the continuity of the nascent transcript affects alternative splicing when this occurs between the exon and its downstream splicing regulatory element, in the fibronectin EDA minigene (Fig. 1) and in similar minigene systems (Fig. 3; Supplemental Fig. 3). On the other hand, MTSs are cleaved very inefficiently and do not affect either alternative splicing (Fig. 5) or the level of mature transcripts (Fig. 6). The DRE deletion itself affects the order of intron removal as previously reported (de la Mata et al. 2010), and we show here that the Rz-mediated interruption of the continuity of the nascent transcript facilitates splicing of the downstream intron in a DRE-dependent manner increasing exon inclusion (Fig. 2). When the Rz was inserted in other intronic positions, neither alternative splicing of mature mRNA nor splicing of the downstream intron was affected (Fig. 2).
Insertion of an artificial hammerhead Rz in the short intron of the constitutively included α-globin minigene was originally reported to have no effect on the abundance of mature mRNA or on splicing, suggesting that exons are efficiently kept in place by tethering to the Pol II (Dye et al. 2006). However, this exon-tethering model has been recently challenged by the observation that a different type of ribozyme, the hepatitis δ, aborts cotranscriptional pre-mRNA processing in a similar system, resulting in severe damage of the nascent transcript (Fong et al. 2009). This effect was related to a faster catalytic activity of the hepatitis δ relative to the hammerhead, suggesting that there may be a competition between cleavage by the Rz and splicing efficiency (Fong et al. 2009). We tested different types of Rzs (hammerhead and hepatitis δ), and all of them, when inserted between the EDA and the DRE, induced exon inclusion (Fig. 4A). Through the amplification of nascent transcripts across the Rz insertions, we did not detect significant differences in their catalytic activity (Fig. 4C,D), suggesting that all Rzs are efficiently cleaved before the splicing decision. In comparison to the α-globin construct, the EDA minigene contains relatively longer introns, and the central exon, being alternatively spliced, is less defined. These aspects might explain the comparable cleavage efficiency of the Rzs in the fibronectin minigene and their similar effect on the pattern of EDA alternative splicing.
The Rz-mediated cleavage of intronic sequences is associated with a constant reduction in the level of mature mRNAs with ∼40% of mRNA produced (Fig. 6B). However, we did not observe a relationship between different amounts of mature transcripts and type of Rz, intronic position, or strength of the 5′-splice site (Fig. 6B). If intronic cleavage occurs before the engagement of Pol II to the successive exon, this will result in the release of transcripts from RNA Pol II and will therefore abort cotranscriptional pre-mRNA processing. In our alternatively spliced system, with relatively long introns, the constant effect of the Rzs on the level of mature transcripts indicates that cleavage of the nascent transcript can affect pre-mRNA processing to a certain extent. As this effect was independent of the alternative splicing pattern, EDA exon definition, Rzs type, and intronic position, a competition between the Rz cleavage and splicing efficiency is not involved. Thus, the Rz-dependent disruption of intron integrity is not completely rescued by Pol II–mediated exon tethering. A certain amount of severed transcript is probably not tethered by the CTD of Pol II and is consequently subjected to abortive pre-mRNA processing, or the spliced transcript is not properly exported from the nucleus. The efficiency of rescue of interrupted intronic transcripts might be gene-specific and dependent on Pol II processivity. Pol II elongation rates and/or its CTD phosphorylation status might regulate exon tethering in a transcript-specific manner, thus facilitating processing of long intronic sequences. Intriguingly, as splicing of long introns in Drosophila is facilitated by components of the pre-exon junction complex (EJC) (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010), this complex might be involved in modulating exon tethering by Pol II in a transcript-specific manner.
Using the alternatively spliced EDA system, we have also evaluated to what extent different MTSs are cropped before splicing and what effect they have on alternative splicing regulation. Several lines of evidence indicate that pri-miRNAs are cropped cotranscriptionally (Kim and Kim 2007; Morlando et al. 2008; Pawlicki and Steitz 2008). In our minigene system, MTSs showed a significantly lower cleavage activity in comparison to the Rzs (Fig. 5) and do not affect the splicing pattern when inserted between the EDA exon and the DRE, suggesting that Microprocessor cropping occurs after transcription of the downstream regulatory element. This supports the idea that miRNA processing by the Drosha complex is a relatively slow process, as previously suggested (Kim and Kim 2007; Morlando et al. 2008). On the other hand, we cannot exclude that some components of the Microprocessor complex might remain after cropping on the pre-mRNA, maintaining the exon and the DRE in close proximity. In this last case, MTS-mediated cleavage would not disrupt the interaction between the EDA exon and the DRE. Through the evaluation of the levels of pre-mRNA transcripts in cotransfection experiments, we observed that MTSs are processed with different efficiencies (Fig. 5). These differences could be important for pri-miRNA maturation and might be due to the relative affinity of pri-miRNAs for the Microprocessor complex, as reported to occur in vitro (Han et al. 2006) and/or to some specific regulatory mechanisms. Biosynthesis of a number of miRNAs is regulated at the level of pri-miRNA processing, with the involvement of RNA binding proteins. Several proteins interact with Drosha and/or with the pri-miRNA sequence and can either inhibit or promote the processing efficiency of individual miRNAs at the level of pri-mRNA cropping (Guil and Caceres 2007; Piskounova et al. 2008; Viswanathan et al. 2008; Trabucchi et al. 2009). As Drosha cleavage occurs cotranscriptionally (Kim and Kim 2007; Morlando et al. 2008; Pawlicki and Steitz 2008), it is conceivable that the regulation of biogenesis of some miRNAs at the level of Microprocessor processing occurs early on the nascent transcripts. The inefficient processing of two hairpins in the 5′ UTR and coding regions of the DGCR8 mRNAs might be related to the unique role of the Microprocessor in direct destabilization of a coding mRNA (Han et al. 2009; Shenoy and Blelloch 2009; Triboulet et al. 2009). In this case, it might be possible that the Microprocessor complex degrades the coding mRNA after intron splicing.
In conclusion, we show that efficient cleavage of nascent introns mediated by different Rzs partially reduces the amount of mature mRNA and, when inserted at critical positions between the exon and its regulatory element, affects alternative splicing. Intronic Microprocessor target sequences are cleaved inefficiently relative to Rzs and do not change the pattern of splicing but retain some individual residual activity that can be important for a regulated cotranscriptional cropping. The comparable effect of Rzs on mRNA levels, which is independent from the Rz type, the intronic distance from the neighboring exons, and exon definition, suggests that severed nascent transcripts are inefficiently processed to the mature mRNA. The different efficiencies in processing of severed nascent transcripts can be due to the contribution of some splicing factors like SR proteins. These proteins, involved in coupling RNA Pol II to pre-mRNA splicing (Das et al. 2007), might modulate Pol II interaction with exonic splicing enhancers and regulate exon tethering in a transcript-specific manner.
MATERIALS AND METHODS
Cell culture, transfection, and reverse transcription-PCR analysis
Hep3B and HeLa cells (5 × 106) were grown under standard conditions and were transiently transfected with 0.5 μg of plasmid DNA with the Effectene Transfection Reagent (QIAGEN) kit according to the manufacturer's instructions. Cells were harvested after 24 h, and RNA was isolated using TRIReagent (Ambion). RT-PCR was performed as previously described (Pagani et al. 2003; Baralle et al. 2008). For 5′-splicing intermediate and nascent pre-mRNAs analysis, total RNA was treated with Dnase–RNase-free and loaded on RNA cleanup columns (QIAGEN). Cotransfection experiments were performed with 0.15 μg of pEDA or pEDs, along with 0.45 μg of the indicated minigenes. Quantification of band intensities was performed using a fluorescent primer. Amplifications were performed in the linear range; the resulting products were run on a capillary electrophoresis with a fluorescent internal MW standard and analyzed with Peak Scanner software (Applied Biosystems). The intensity of bands in agarose gels has been determined with ImageJ software as previously reported (Goina et al. 2008).
siRNA transfections
siRNA transfections were performed in HeLa cells using Oligofectamine reagents (Invitrogen). The sense strand for Xrn2 RNA interference oligonucleotides (Dharmacon) used for silencing was 5′-aagaguacagaugaucauguu-3′. Silencing was performed as previously described with two rounds of siRNA transfections (Goina et al. 2008). Cells were harvested and divided in two parts for RNA and protein extractions. Protein extract was analyzed in Western Blotting for Xrn2 (a generous gift from N. Gromak) and tubulin as a control.
Quantitative RT-PCR
For expression level, each minigene (500 ng) was cotransfected with the control pCF1 (100 ng), which was used for normalization of transfection and reverse transcriptase efficiencies. Real-time PCR amplification of pCF1 was performed with Syber green supermix (Bio-Rad) with CFex1_248dir and CFex3_292rev that gave a spliced product of 233 bp. For quantitative RT-PCR on total mRNA, the minigene expression was detected with an HBA1 TaqMan gene expression assay (Applied Biosystems). For semiquantitative RT-PCR analysis of the 5′-splicing intermediates, minigenes were amplified with f1 and r1 primers loaded on agarose gel, and the band intensity was evaluated with the ImageJ software.
Quantification of hsa-mir-26b and hsa-mir-330 expression was performed with TaqMan MicroRNA Assays (Applied Biosystems).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by the AIRC grant IG10387. We thank Natalia Gromak (Sir William Dunn School of Pathology, University of Oxford, Oxford, UK) for the Xrn2 antibody and Marcos Morgan (ICGEB, Trieste, Italy) for the pCMV26b plasmid. We thank also Rodolfo Garcia and F.E. Baralle for helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2514811.
REFERENCES
- Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrancois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, et al. 2010. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143: 251–262 [DOI] [PubMed] [Google Scholar]
- Baralle M, Pastor T, Bussani E, Pagani F 2008. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am J Hum Genet 83: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Bouton AH, Miller OL Jr 1981. Correlation of hnRNP structure and nascent transcript cleavage. Cell 26: 155–165 [DOI] [PubMed] [Google Scholar]
- Chen X, Denison L, Levy M, Ellington AD 2009. Direct selection for ribozyme cleavage activity in cells. RNA 15: 2035–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR 2010. First come, first served revisited: Factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ 2006. Exon tethering in transcription by RNA polymerase II. Mol Cell 21: 849–859 [DOI] [PubMed] [Google Scholar]
- Fong N, Ohman M, Bentley DL 2009. Fast ribozyme cleavage releases transcripts from RNA polymerase II and aborts co-transcriptional pre-mRNA processing. Nat Struct Mol Biol 16: 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goina E, Skoko N, Pagani F 2008. Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol Cell Biol 28: 3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Gromak N, Talotti G, Proudfoot NJ, Pagani F 2008. Modulating alternative splicing by cotranscriptional cleavage of nascent intronic RNA. RNA 14: 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF 2007. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14: 591–596 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ 2006. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol 4: e147 doi: 10.1371/journal.pbio.0040147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN 2007. Processing of intronic microRNAs. EMBO J 26: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR 2005. Promoter usage and alternative splicing. Curr Opin Cell Biol 17: 262–268 [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR 2007. Coupling transcription and alternative splicing. Adv Exp Med Biol 623: 175–189 [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev 20: 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822 [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3: 697–705 [DOI] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ 2008. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol 15: 902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 275: 39935–39943 [DOI] [PubMed] [Google Scholar]
- Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA 2009. The carboxyl-terminal domain of RNA polymerase II is not sufficient to enhance the efficiency of pre-mRNA capping or splicing in the context of a different polymerase. J Biol Chem 284: 8692–8702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Stuani C, Tzetis M, Kanavakis E, Efthymiadou A, Doudounakis S, Casals T, Baralle FE 2003. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum Mol Genet 12: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA 2008. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol 182: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI 2008. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283: 21310–21314 [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE 2010. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143: 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ 2005. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 25: 6734–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A, Blelloch R 2009. Genomic analysis suggests that mRNA destabilization by the Microprocessor is specialized for the auto-regulation of Dgcr8. PLoS ONE 4: e6971 doi: 10.1371/journal.pone.0006971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Chang HM, Lapierre RJ, Gregory RI 2009. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 15: 1005–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D'Amato RJ, Mulligan RC 2004. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431: 471–476 [DOI] [PubMed] [Google Scholar]