Abstract
Elucidation of natural product biosynthetic pathways provides important insights about the assembly of potent bioactive molecules, and expands access to unique enzymes able to selectively modify complex substrates. Here we show full reconstitution in vitro of an unusual multi-step oxidative cascade for post-assembly line tailoring of tirandamycin antibiotics. This pathway involves a remarkably versatile and iterative cytochrome P450 monooxygenase (TamI) and an FAD-dependent oxidase (TamL), which act co-dependently through repeated exchange of substrates. TamI hydroxylates tirandamycin C (TirC) to generate tirandamycin E (TirE), a heretofore unidentified tirandamycin intermediate. TirE is subsequently oxidized by TamL, giving rise to the ketone of tirandamycin D (TirD), after which a unique exchange back to TamI enables successive epoxidation and hydroxylation to afford, respectively, the final products tirandamycin A (TirA) and tirandamycin B (TirB). Ligand-free, substrate- and product-bound crystal structures of bicovalently flavinylated TamL oxidase reveal a likely mechanism for the C-10 oxidation of TirE.
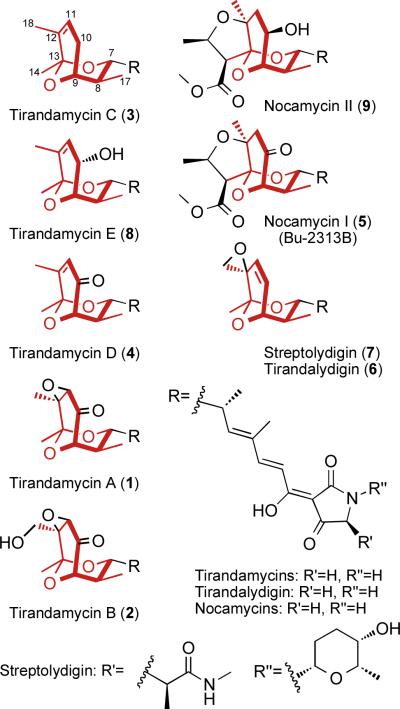
The antibiotic tirandamycin is one of several dienoyl tetramic acid natural products containing an intriguing and often heavily tailored bicyclic ketal moiety. Recently, we have expanded the family of known tirandamycins to four compounds, tirandamycin A–D (TirA–D, 1–4), that differ in the extent of oxidative tailoring of the bicyclic ketal (Fig. 1), which was found to be a key determinant of potency against vancomycin-resistant Enterococcus faecalis (VRE)1. Similar modifications are apparent in the ketone of nocamycin (5), and the epoxide of tirandalydigin (6) and streptolydigin (7), but TirB is unique in that it contains a ketone (C-10), epoxide (C-11/C-12), and an additional hydroxyl (C-18) group (Fig. 1)2. Our interest in the formation and tailoring of this pharmacophore prompted a search for the gene cluster encoding tirandamycin biosynthesis in Streptomyces sp. 307-9, resulting in identification of a hybrid polyketide synthase (PKS)/non-ribosomal peptide synthetase (NRPS) system3. Flanking the PKS-NRPS genes were coding regions for two predicted oxidative tailoring enzymes: a cytochrome P450 homolog TamI that we viewed as a likely candidate for installation of the epoxide and the C-18 hydroxyl group, and a flavoprotein homolog TamL that we hypothesized to be involved in formation of the C-10 ketone. Disruption of tamI led to exclusive accumulation of TirC3, which is devoid of oxidative modifications, suggesting that TamI is responsible for the first step in the tirandamycin tailoring pathway. This finding motivated our interest to establish the precise role of TamI and other oxidative enzymes involved in introduction of the C-10 keto, C11-C12 epoxide, and C-18 hydroxyl functional groups through in vitro analysis and reconstitution of the TirC → TirB enzymatic system. Finally, the troika of ligand-free, substrate and product-bound x-ray structures of TamL provide compelling evidence toward a mechanism for the C-10 keto group installation.
Figure 1.
Tetramic acid natural products bearing a bicyclic ketal moiety (red) with varying degrees of oxidative modification. Tirandamycin E is described within this study; all other compounds have been previously reported1,2. Nocamycin I and Bu-2313B are synonymous.
Results
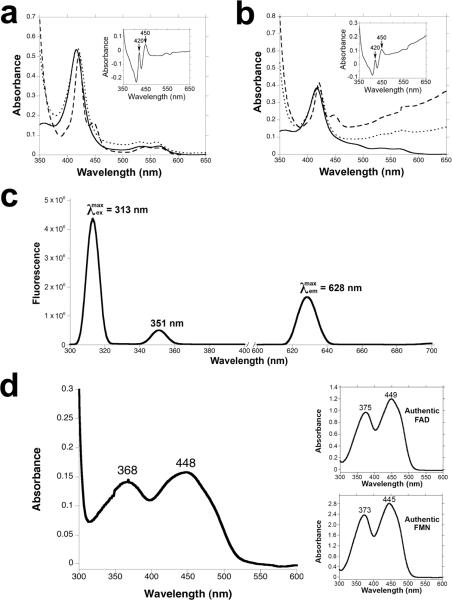
We began by examining the reactions catalyzed by TamI, a predicted cytochrome P450 monooxygenase (Fig. S1), on each of the intermediates that were previously isolated from fermentation broths of Streptomyces sp. 307-91. The N-terminal His8-tagged recombinant TamI was heterologously overexpressed and purified from Escherichia coli (Fig. S2) to yield an orange-red enzyme solution that was characterized by UV-visible spectroscopy using standard techniques4. The sodium dithionite reduced enzyme solution displayed an absorbance peak at 420 nm with a 450 nm peak arising after bubbling of the solution with CO (Fig. 2a).
Figure 2.
Spectral analysis of tirandamycin tailoring enzymes. (a) UV-vis absorption spectrum for purified TamI P450 enzyme in oxidized form (solid line), sodium dithionite reduced form (dotted line), and CO reduced form (dashed line); insets show CO-bound reduced difference spectra. (b) UV-vis absorption spectrum for purified TamI-RhFRED fusion. (c) Fluorescence excitation and emission spectra of purified TamL flavoprotein. (d) UV-vis absorption spectrum for purified TamL; inset shows UV-vis absorption spectrum of authentic FAD and FMN cofactors.
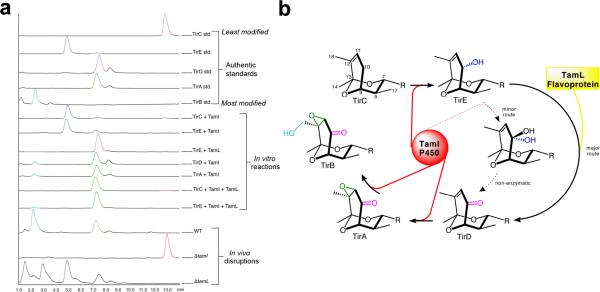
To test the hypothesis that TamI installs the C-11/C-12 epoxide and C-18 hydroxyl, the recombinant enzyme was incubated with the putative substrate TirA in reaction buffer containing spinach ferredoxin/ferredoxin-NADP+ reductase as heterologous redox partners, and NADPH. LC-MS analysis of reaction supernatants revealed a small but reproducible conversion of TirA to TirB due to hydroxylation at C-18. Under the same reaction conditions but with TirD as substrate we observed complete conversion of the substrate with predominant formation of TirA, and a minor amount of TirB (Fig. 3a). Together these experiments demonstrate that the penultimate and final steps of tirandamycin biosynthesis are a C-11/C-12 epoxidation and a C-18 hydroxylation, respectively.
Figure 3.
Elucidation of individual steps in the tirandamycin oxidative cascade. (a) in vitro reconstitution of TamI and TamL mediated oxidation steps, and metabolite profiles of Streptomyces sp. 307-9 wild type and mutant strains. Peaks were identified by comparison to authentic standards and MS detection of anticipated species. Relative zoom of the bottom three traces: wild type 4.1×, ΔtamI 1×, ΔtamL 17.5×. (b) Complete oxidative cascade scheme; the predominant TamI P450 reactions are highlighted in red with the intervening TamL oxidation reaction highlighted in yellow; the dashed arrows represent a minor activity toward the TamL-independent route. The R group is that shown in Figure 1. TamI mediates the initial hydroxylation of TirC at C-10, to form TirE, after which TamL oxidizes TirE to the ketone. Subsequent epoxidation and hydroxylation, both catalyzed by TamI, completes the cascade.
Next, we tested the possible role of TamI in formation of the C-10 ketone by incubating TirC with recombinant P450 enzyme, and observed formation of a very small amount of TirA (Fig. 3a), in addition to a new species that had not been previously identified from fermentation broths. The new compound, tirandamycin E (TirE, 8), possessed a mass and polarity consistent with a single hydroxylation of TirC. To characterize TirE by NMR, we required several milligrams of product prepared enzymatically, which was impractical using the existing reaction procedure with spinach ferredoxin/ferredoxin-NADP+ reductase. To overcome this limitation, we employed a recently developed strategy to generate a self-sufficient biosynthetic P450 reaction system by fusing TamI to the FMN/Fe2S2 containing RhFRED reductase domain from Rhodococcus sp.5, thus obviating the need for costly exogenous redox partners. Using this system we conducted a preparative scale conversion of TirC to TirE, purified the product and assigned the structure by NMR analysis (Table S1, Fig. S3–S9), which confirmed the presence of a hydroxyl group at C-10 of the bicyclic ketal moiety. The stereochemistry was assigned based on the observation of through-space deshielding between the C-10 hydroxyl and the H-7 and H-17 protons, and by comparison of NOESY data between TirC and TirE, which confirmed the disappearance of the pro-S proton at C-10.
These results demonstrate that one major activity of TamI is to catalyze hydroxylation at C-10, but the low levels of TirA formation indicated that TamI is capable of further reaction to generate the corresponding ketone. It is likely that formation of this keto group by TamI proceeds through sequential hydroxylations at C-10 to form a geminal-diol that exists (in equilibrium) predominantly in the ketone form of TirD (Fig. 3b). This route resembles that used by P450s for keto-group formation in doxorubicin biosynthesis6, or to install a carboxylate at a primary carbon atom as in the biosynthesis of artemisinic acid7. However, because this activity for TamI was low we reasoned that another oxidative enzyme in the pathway might catalyze the predominant route to TirD. Thus, we examined the ability of TamL, a predicted flavin-dependent oxidoreductase, to catalyze this reaction. Alignments revealed a sequence motif for the bicovalent attachment of FAD to invariant histidine and cysteine residues (Fig. S10)8,9. This suggested that TamL may utilize a mode of cofactor binding similar to glucooligosaccharide oxidase (GOOX)10, aclacinomycin oxidoreductase (AknOx)11, glycopeptide hexose oxidase (Dbv29)12, hexose oxidase (HOX)12,13 and berberine bridge enzyme (BBE)9,14,15, all of which belong to the p-cresol methylhydroxylase scaffold family16. These proteins show high sequence similarity in the flavin binding site, whereas few residues are conserved in their substrate binding domains due to the structural diversity of their substrates (Fig. S10).
We overexpressed and purified recombinant His8-tagged TamL in E. coli (Fig. S2) to yield a deep yellow protein solution with fluorescence excitation/emission and UV-visible absorption spectra consistent with a flavoprotein (Fig. 2c and 2d)8,17. Presence of a covalently bound co-factor was supported by boiling the enzyme solution and observing that the yellow color was retained with denatured protein, and released cofactor was absent from the supernatant as determined by LC-MS. Acid hydrolysis treatment of the protein released AMP, which confirmed FAD (and not FMN) as the cofactor (Fig. S11)18, and MS analysis of the intact protein corroborated the presence of a single FAD per enzyme molecule (Fig. S12).
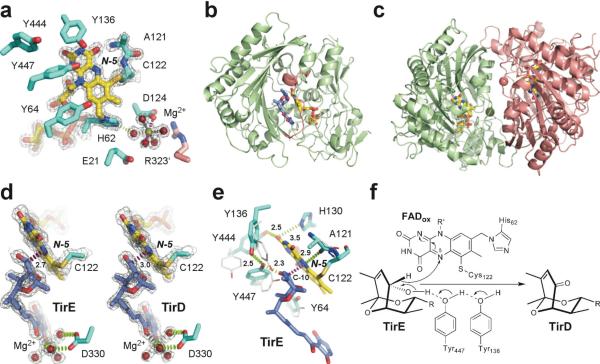
X-ray structure analysis of TamL revealed that FAD is attached covalently to the enzyme via bonds between the C-6 atom of the isoalloxazine ring and the sulfur atom of invariant cysteine C122, as well as the 8α-methyl group to the N-1 atom of H62, resulting in 6-S-cysteinyl, 8α-N1-histidyl FAD (Fig. 4a). The spacious substrate binding cleft of TamL is open to the bulk solvent and is built largely of a seven-stranded antiparallel β-sheet flanked by five α-helices (Fig. 4b). The mouth of the cleft is surrounded by the residues 323–336 (pink in Fig. 4b), which are the most variable among the closest TamL homologs from both sequence and structural perspective (Fig. S10). This region participates in substrate binding, and mediates formation of tightly associated dimers in the crystal, as observed for other members of the p-cresol methylhydroxylase superfamily16 (Fig. 4c). The 1680 Å2 dimerization interface in the substrate-free TamL is additionally stabilized by two solvated Mg2+ cations, bound to D124 of each protein monomer. Five other Mg2+ coordinates are occupied by water molecules (Fig. 4a) making multiple H-bonding interactions with the protein.
Figure 4.
Ligand-free and substrate/product-bound TamL. (a) Catalytic and the Mg2+-binding sites in the substrate-free TamL with covalently bound FAD (yellow sticks) (PDB ID 2Y08). Residues from the same monomer are in cyan, from the symmetry-related monomer in pink. (b) Tirandamycin (blue sticks) in the active site of TamL (PDB ID 2Y3R). Residues 323–336 at the mouth of the substrate binding cleft are in pink. (c) Ribbon representation of TamL dimer formed by the green and pink monomers related by the non-crystallographic symmetry (PDB ID 2Y08). The Mg2+ atoms (spheres of matching colors) stabilize dimerization interface. (d) Interactions between the C-10 site of oxidation in TirE and the N-5 locus in FAD are highlighted in magenta dash line defining an angle with the N-5/N-10 flavin atoms of 110°. Installation of the keto group in TirD flattens the ketal ring pulling C-10 away from the N-5 atom by 0.3 Å (PDB ID 2Y3R). (e) Superimposition of the amino acid residues in the tirandamycin-bound (cyan sticks) compared to substrate-free (grey sticks) TamL. (f) Mechanism of dehydrogenation at C-10 in TirE. In all panels, oxygen atoms are in red, nitrogen in blue, sulfur in dark yellow, magnesium in green. Electron density 2Fo-Fc map (gray mesh) is contoured at 1.5 σ. Distances are in Angstroms.
In the substrate-bound form (Fig. 4b), Mg2+ was found chelated by two adjacent carbonyl groups of the tirandamycin tetramic acid moiety and its 3-acyl substituent with the four other coordination sites occupied by the water molecules mediating contacts with the side chain of D330 (Fig. 4d). Location of the metal center in both structures appears to exclude its direct involvement in catalysis. This was consistent with the observation that pre-incubation of TamL with EDTA does not change its enzymatic activity. However, Mg2+ may play a stabilizing role in the monomer or dimer structure, facilitate substrate binding, or assist in covalent flavinylation through H-bonding between the Mg2+ aqueous shell and the FAD attachment site at H62 (Fig. 4a).
Biochemical analysis of recombinant TamL demonstrated complete conversion of purified TirE to TirD, thus confirming its role in oxidizing the C-10 hydroxyl of its presumed natural substrate to a ketone (Fig. 3). Furthermore, when reactions containing TamI P450 and TirC were amended with TamL, we observed nearly complete conversion to TirA (bearing the C-11/C-12 epoxide) with no residual TirE detected, indicating that oxidation of TirE by TamL affords the C-10 ketone more effectively than TamI alone. These in vitro results are supported by our previous in vivo gene disruption experiments3, and are entirely consistent with initiation of the oxidative cascade by TamI. To confirm the role of TamL in vivo, we created a ΔtamL mutant and observed accumulation of the TirE biosynthetic intermediate from this strain, demonstrating that the TamI alone is unable to catalyze conversion to TirD in vivo (Fig. 3a).
To better understand the interplay of TamI and TamL in this oxidative cascade, we obtained the kinetic parameters for each reaction (Table 1). Consistent with the above-described in vitro qualitative results (Fig. 3a), TamI was most efficient in catalyzing the initial hydroxylation of TirC and epoxidation of TirD (kcat/Km 19.3 and 3.6 μM−1 min−1, respectively), compared to the inefficient final hydroxylation of TirA (kcat/Km 5.8 × 10−4 μM−1) which resulted from both a substantially lower kcat and higher Km. Moreover, TamL-mediated oxidation of TirE to the corresponding ketone was very rapid (kcat 4.08 × 103 min−1), highlighting the significance of this enzyme in overcoming a biochemical challenge that is poorly met by TamI alone.
Table 1.
Kinetic parameters for reaction of TamI-RhFRED and TamL with tirandamycin intermediates.
| Enzyme (Substrate) | Km (μM) | kcat (min−1) | kcat/Km (μM−1min−1) |
|---|---|---|---|
| TamI-RhFRED (TirA) | 189.7 ± 25.1 | 0.11 ± 0.01 | 0.00058 |
| TamI-RhFRED (TirC) | 2.1 ± 0.3 | 40.5 ± 0.8 | 19.3 |
| TamI-RhFRED (TirD) | 23.2 ± 3.7 | 83.8 ± 3.4 | 3.6 |
| TamL (TirE) | 174.1 ± 37.0 | 4082.7 ± 375.7 | 23.4 |
To establish the mechanism of catalysis, we determined the x-ray structures of TamL alone (PDB ID code 2Y08) and of the TamL-tirandamycin complex (PDB ID codes 2Y3R, 2Y4G and 2Y3S). Given the high catalytic activity of TamL, the product, TirD, rather than the substrate, TirE, would be expected to occupy the active site in the crystals. However, the active site electron density map for the 2Y4G and 2Y3S structures unambiguously matched TirE, which was finally fitted in both monomers in the asymmetric unit in these structures. In contrast, TirD better satisfied the electron density in the monomers B and D in the 2Y3R structure and therefore, TirE was fitted in chains A and C, and TirD in chains B and D in 2Y3R (Fig. 4d). Collectively, the structures revealed that tirandamycins are bound in the active site with the bicyclic ketal moiety facing FAD and the tetramic acid moiety extending toward the mouth of the substrate binding cleft (Fig. 4b). The ketal ring positioned for oxidation runs parallel to the plane of the isoalloxazine ring with the C-10 hydrogen atom pointing directly at N-5 of FAD (distance of 2.7 Å) (Fig. 4d, e). The geometry of this disposition is consistent with the hydride ion transfer proposed for many flavin-dependent oxidases19,20. This mechanism requires concomitant abstraction of the proton from the C10-OH group, which is likely achieved through the interactions with the invariant Y447 activated by the highly conserved Y136 residue, as proposed for the TamL homolog Dbv2921 (Fig. 4e, f).
The TamL-TirD/TirE co-crystal structure provides considerable mechanistic insights into the TirD → TirE oxidation process. In addition to the repositioning of the 323–336 loop running across the opening of the substrate binding cleft (highlighted pink in Fig. 4b), binding of substrate causes conformational changes near FAD that may facilitate both hydride abstraction and subsequent re-oxidation of cofactor by molecular oxygen through stabilization of certain electronic forms of FAD. One major event clearly observed in the 2Y3R structure includes flipping of the main chain carbonyl group of A121 toward the isoalloxazine ring, which ideally positions the carbonyl oxygen H-bond acceptor (within 2.9 Å) to transiently stabilize the reduced rather than oxidized state of the co-factor (Fig. 4e). Assuming that the inversion of the A121 carbonyl group happens each time the hydride ion is transferred to the N-5 locus of FAD, it should occur with high precision to maintain the fast rate of the reaction. We propose that the covalent attachment of FAD to the nearby C122 may serve to ensure the precision of this interaction.
In addition, rotation of the Y444 side chain brings its OH group within the H-bonding distance (2.5 Å) of the O–2 locus of FAD (Fig. 4e), which may increase the oxidative power of the co-factor22. Other tyrosine residues Y64, Y136, Y323, Y447, and the main chain at G134–G137 undergo adjustments to accommodate the incoming substrate. As a result, the OH group of Y447 that apparently serves as a base to abstract a proton from C10-OH ends up between the C10- (2.3 Å) and the Y136- (2.5 Å) OH groups (Fig. 4e, f). The presumed requirement for conformational changes to accommodate substrate binding and catalysis may explain the surprising presence of substrate in the TamL active site. A 10-fold molar excess of TirE over the enzyme could subsequently displace TirD in the active site whose FAD co-factor is stalled in its reduced form in the crystal.
Discussion
Our in vitro characterization of the complete tirandamycin oxidative cascade has revealed a remarkably versatile TamI P450 enzyme that catalyzes at least two hydroxylations and one epoxidation at three distinct sites including a primary allylic C–H bond (C-18), a secondary allylic C–H bond (C-10), and an olefin (C-11/C-12), which to our knowledge is the first reported example of a bacterial biosynthetic P450 with such versatile activity. By comparison, non-heme iron dependent oxygenases are well known for such catalytic versatility23,24, which is thought to be the result of a relatively flexible coordination geometry that might be difficult to achieve through the heme-based coordination of a cytochrome P450. The TamI/TamL reactions occur in a precisely defined order with no detected promiscuity during the individual steps (Figure 3b). Following the initial oxidation of TirC by TamI, TamL must oxidize the C-10 hydroxyl group to the ketone of TirD, which is the exclusively observed substrate for sequential epoxidation to TirA and hydroxylation to TirB. The repeated exchange of substrates between these two enzymes creates a unique tailoring pathway in which a P450 catalyzes multiple oxidations co-dependently with a second biosynthetic enzyme, in contrast to numerous examples of multi-step P450 reactions in which no intervening step by an alternate enzyme is required7,25–27. This intriguing enzymatic interplay parallels that observed in clavaminate biosynthesis, in which a non-heme iron oxidase catalyzes three non-consecutive reactions separated by the action of a separate hydrolase enzyme23. Perhaps the recruitment of TamL to the evolving secondary metabolic pathway was prompted by the inability of TamI to efficiently transform TirE to TirD. Oxidative modification in the tirandamycin pathway contributes significantly to antibiotic potency1, which might have provided evolutionary pressure for the TamI/TamL series of tailoring steps. It appears that this process is facilitated in TamL through bicovalent attachment of FAD, which has been shown to increase redox potential in related enzymes GOOX28 and BBE29 to remarkably high values of ~130 mV. This cofactor configuration accelerates flavin reduction during turnover to the point where its reoxidation by molecular oxygen becomes the rate-limiting step29. It remains to be determined if the nocamycin pathway utilizes a TamL homolog for oxidation of the C-10 hydroxyl of nocamycin II (9) to the ketone of nocamycin, or simply proceeds through P450-mediated geminal-diol formation and equilibration identified as a minor aspect of the TirE → TirD conversion (Fig. 3b).
Conservation of the catalytically essential tyrosine residues in TamL suggests its close mechanistic similarities with GOOX, AknOx and Dbv299,14,15 (Fig. S10). In TamL, four tyrosine residues are situated in the vicinity of the N-5 nitrogen atom of FAD (Fig. 4e): Y136 and Y447 are invariant between TamL, GOOX, AknOx and Dbv29, whereas Y444 is absent from AknOx, and Y64 is present only in TamL and GOOX (Fig. S10). Thus, TamL-mediated oxidation is likely to proceed via proton abstraction from the C-10 hydroxyl group by Tyr447 and concomitant hydride ion transfer from the C-10 carbon atom to the N-5 nitrogen atom of FAD (Fig. 4f). Tyr136 may participate in the proton transfer network to initiate H+ abstraction by Y447. The reduced flavin is likely reoxidized by molecular oxygen with formation of hydrogen peroxide. The oxidative half of the reaction in the flavin-containing oxidases is believed to proceed by a two-step mechanism via transient formation of the red anionic semiquinone20,29. Although it has not been confirmed experimentally for TamL, H130 would be a candidate residue to stabilize the negative charge should it accumulate on the N1-C2=O2 locus in FAD (Fig. 4f). The exact catalytic mechanism and the actual roles of the catalytically important residues require further analysis, which is currently ongoing in our laboratories.
Based on our recent structure-activity relationship analysis of the tirandamycins1, the potency of the early intermediate TirC (MIC 110 μM) is far below that of TirA (MIC 2.25 μM). This makes the above-described oxidative tailoring steps biologically significant, assuming that the observed anti-microbial (e.g. anti-VRE) activity reflects potency against relevant natural targets of the tirandamycins (the molecular target of TirA and TirB is RNA polymerase2). Our in vitro enzymatic characterization and analysis of fermentation broth metabolite profiles all suggest that this biosynthetic pathway is tuned towards the production of TirA, the most potent of the tirandamycin antibiotics. The in vitro conversion of TirC to TirA (passing through TirE and TirD) proceeds efficiently, and the early intermediates TirC and TirD are only observed from wild type culture fermentations if an adsorbent resin is included in the broth to sequester these compounds during production1. Thus, the TirA metabolite that accumulates predominantly in the culture media is the most potent antibiotic. Conversion to the most highly modified, but less potent TirB (MIC 100 μM) occurs with low efficiency in vitro, and TirB only accumulates appreciably in fermentation broths using extended culture times. These observations suggest that the TamI P450 has evolved to efficiently accommodate TirC and TirD, resulting in a substantial increase in potency of the product, but then stalls on the final hydroxylation that converts TirA to the less bioactive TirB.
Methods
General Experimental Procedures
NMR spectra were acquired on a Varian INOVA 400 MHz and a Varian INOVA 600 MHz spectrometer at the Center for Chemical Genomics, University of Michigan. NMR spectra were processed using MestReNova software. High-resolution ESI-MS spectra were measured at the University of Michigan core facility in the Department of Chemistry using a Waters Micromass AutoSpec Ultima. RP-HPLC purification was performed using Waters XBridgeTM 5 μm C18 columns and a solvent system of acetonitrile and H2O supplemented with 0.1% TFA. LC-MS analysis was performed on a Shimadzu 2010 EV ESI spectrometer using an XBridgeTM C18 3.5 μm 50 mm column with the same solvent system supplemented with 0.1% formic acid. DNA sequencing was performed at the University of Michigan DNA Sequencing Core Facility using the dideoxy chain termination method.
Crystallization and structure determination
Crystallization of TamL was sensitive to the presence of Mg2+. Substrate-free crystals obtained from 15% PEG4000, 0.2 M MgCl2 and 0.1 M Tris-HCl, pH 7.0 belong to the space group P212121 with cell dimensions a=64.4, b=129.6, c=134.6. Two molecules in the asymmetric unit related by the two-fold non-crystallographic symmetry form a dimer with the ~1680 Å2 interface. Tirandamycin-bound crystals were obtained in three distinct forms from similar conditions, all of them featuring the same dimeric structure packed in the crystals in different ways (Table S2). Diffraction data were collected at 100–110 K at Beamline 8.3.1, Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, CA. Consistent with the high redox potential of the bicovalently attached FAD is bleaching of the brilliant yellow color of the oxidized flavin, which was observed during the first few seconds of data collection, suggesting conversion into the fully reduced, virtually colorless, hydroquinoid form by the flux of the x-ray electrons. Despite that observation, fully oxidized FAD was fitted into the electron density during the structure refinement. In none of the structures isoalloxazine ring deviated from the exact planarity. Data were processed using ELVES30 to resolutions ranging from ~1.7 to 2.0 Å, depending on the structure. Molecular replacement and refinement routines were carried out using the CCP4 software suite31 with the hexose oxidase atomic coordinates (PDB ID 2WDX) as an initial search model. Data collection and refinement statistics are shown in Table S2.
Enzyme Assays and Preparation of Substrates
Isolation of TirA, TirB, TirC, and TirD have been described previously1. TirE was obtained by preparative scale enzymatic conversion in 40 × 500 μl parallel reactions (overnight, 30°C), each containing approximately 10 μM purified TamI-RhFRED, 0.15 mg TirC, 1 mM NADPH, and 5 μM glucose-6-phosphate and 2 Unit/ml glucose-6-phosphate dehydrogenase for NADPH regeneration. The reaction mixtures were combined and extracted by 3 × 40 mL of CH2Cl2, after which the extract was dried, redissolved in 2 mL of methanol, and purified by RP-HPLC to yield TirE. The structural assignment of TirE was based on analysis of 1H, 13C, COSY, HMQC, HMBC, and NOESY data sets. The complete NMR assignment is available in Supplementary Information. HRMS (m/z): [M-H]− calcd. for C22H29NO6, 402.1922; found, 402.1914
The in vitro enzymatic conversions of tirandamycins were performed in a total volume of 100 μl of conversion buffer (50mM NaH2PO4, pH7.3, 1 mM EDTA, 0.2 mM dithioerythritol, 10% glycerol) containing 2 μM TamL (or 1 μM TamI partnered by 3.5 μM spinach ferredoxin, 0.1 Unit/ml spinach ferredoxin-NADP+ reductase, and 500 μM NADPH), and 200 μM substrate. As negative controls, corresponding enzymes were omitted. After incubation at 30°C for 2 h, the reactions were quenched by extraction, using 2 × 200 μl of CHCl3. The resulting organic extract was dried, redissolved in 120 μl of methanol, and analyzed by LC-MS using 354 nm UV detection and positive/negative ion MS detection.
Kinetic Assays
For TamI kinetics, the standard assays contained TamI-RhFRED (3.2 μM for TirA assay, 25 nM for TirC assay, and 50 nM for TirD assay) and various concentrations of substrates in 190 μl of P450 desalting buffer (50 mM NaH2PO4, pH 7.3, 1 mM EDTA, 0.2 mM dithioerythritol, 10% glycerol). After a pre-incubation at 30 °C for 5 min, the reactions were initiated by adding 10 μl of 50 mM NADPH. For TamL kinetics, the standard assays contained 4 nM of TamL in 198 μl of storage buffer (50 mM NaH2PO4, pH 7.3, 20% glycerol). After a pre-incubation at 30 °C for 5 min, the reactions were initiated by adding 2 μl of TirE (in DMSO) at various concentrations. For both assays, reaction termination and analysis were as follows: three aliquots (50 μl) were taken at three time points within the linear range to thoroughly mix with 100 μl of methanol, and the proteins were pelleted by centrifugation at 13,000g, 4 °C for 15 min. The supernatant was subject to HPLC analysis to monitor substrate consumption within the linear range, thereby deducing the initial velocity of the reactions. All measurements were performed in duplicate, and velocities determined under different substrate concentrations were fit into the Michaelis–Menten equation to calculate the kinetic parameters.
Supplementary Material
Acknowledgements
We thank Petrea M. Kells for excellent technical assistance, and staff members of beamline 8.3.1, James Holton, George Meigs and Jane Tanamachi, at the Advanced Light Source at Lawrence Berkeley National Laboratory, for assistance with data collection. This work was funded by NIH grant GM078553 (to D.H.S. and L.M.P.), U01 TW007404 (ICBG) and the Hans W. Vahlteich Professorship (to D.H.S.). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Author contributions. All authors designed experiments and analyzed data. J. C., S. L., S. G., Y. A., and D. B. performed the experiments. J. C., S. L., L. P., and D. S. wrote the manuscript. J. C. and S. L. contributed equally to this study.
Data deposition note: The atomic coordinates and structure factors (PDBID codes 2Y08, 2Y3R, 2Y4G and 2Y3S) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
References
- 1.Carlson JC, Li S, Burr DA, Sherman DH. Isolation and characterization of tirandamycins from a marine-derived Streptomyces sp. J. Nat. Prod. 2009;72:2076–2079. doi: 10.1021/np9005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Royles BJL. Naturally occurring tetramic acids - structure, isolation, and synthesis. Chem. Rev. 1995;95:1981–2001. [Google Scholar]
- 3.Carlson JC, et al. Identification of the tirandamycin biosynthetic gene cluster from Streptomyces sp. 307-9. ChemBioChem. 2010;11:564–572. doi: 10.1002/cbic.200900658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 5.Li S, Podust LM, Sherman DH. In vitro characterization of a self-sufficient biosynthetic cytochrome P450 PikC fused to a heterologous reductase domain RhFRED. J. Am. Chem. Soc. 2007;129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickens ML, Priestley ND, Strohl WR. In vivo and in vitro bioconversion of epsilon-rhodomycinone glycoside to doxorubicin: functions of DauP, DauK, and DoxA. J. Bacteriol. 1997;179:2641–2650. doi: 10.1128/jb.179.8.2641-2650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 8.Kutchan TM, Dittrich H. Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J. Biol. Chem. 1995;270:24475–24481. doi: 10.1074/jbc.270.41.24475. [DOI] [PubMed] [Google Scholar]
- 9.Winkler A, et al. A concerted mechanism for berberine bridge enzyme. Nat. Chem. Biol. 2008;4:739–741. doi: 10.1038/nchembio.123. [DOI] [PubMed] [Google Scholar]
- 10.Huang CH, et al. Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD. J. Biol. Chem. 2005;280:38831–38838. doi: 10.1074/jbc.M506078200. [DOI] [PubMed] [Google Scholar]
- 11.Alexeev I, Sultana A, Mantsala P, Niemi J, Schneider G. Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6170–6175. doi: 10.1073/pnas.0700579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sosio M, Stinchi S, Beltrametti F, Lazzarini A, Donadio S. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by nonomuraea species. Chem. Biol. 2003;10:541–549. doi: 10.1016/s1074-5521(03)00120-0. [DOI] [PubMed] [Google Scholar]
- 13.Rand T, Qvist KB, Walter CP, Poulsen CH. Characterization of the flavin association in hexose oxidase from Chondrus crispus. FEBS J. 2006;273:2693–2703. doi: 10.1111/j.1742-4658.2006.05285.x. [DOI] [PubMed] [Google Scholar]
- 14.Winkler A, et al. Structural and mechanistic studies reveal the functional role of bicovalent flavinylation in berberine bridge enzyme. J. Biol. Chem. 2009;284:19993–20001. doi: 10.1074/jbc.M109.015727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler A, et al. Berberine bridge enzyme catalyzes the six electron oxidation of (S)-reticuline to dehydroscoulerine. Phytochemistry. 2009;70:1092–1097. doi: 10.1016/j.phytochem.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Mathews FS, Chen ZW, Bellamy HD, McIntire WS. Three-dimensional structure of p-cresol methylhydroxylase (flavocytochrome c) from Pseudomonas putida at 3.0-A resolution. Biochemistry. 1991;30:238–247. doi: 10.1021/bi00215a034. [DOI] [PubMed] [Google Scholar]
- 17.Carter CJ, Thornburg RW. Tobacco nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiol. 2004;134:460–469. doi: 10.1104/pp.103.027482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willie A, Edmondson DE, Jorns MS. Sarcosine oxidase contains a novel covalently bound FMN. Biochemistry. 1996;35:5292–5299. doi: 10.1021/bi952995h. [DOI] [PubMed] [Google Scholar]
- 19.Fitzpatrick PF. Carbanion versus hydride transfer mechanisms in flavoprotein-catalyzed dehydrogenations. Bioorg. Chem. 2004;32:125–139. doi: 10.1016/j.bioorg.2003.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends Bioch. Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu YC, et al. Interception of teicoplanin oxidation intermediates yields new antimicrobial scaffolds. Nat. Chem. Biol. 2011;7:304–309. doi: 10.1038/nchembio.556. [DOI] [PubMed] [Google Scholar]
- 22.Ghisla S, Massey V. Mechanisms of flavoprotein-catalyzed reactions. Eur. J. Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, et al. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat. Struct. Biol. 2000;7:127–133. doi: 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- 24.Salowe SP, Marsh EN, Townsend CA. Purification and characterization of clavaminate synthase from Streptomyces clavuligerus: an unusual oxidative enzyme in natural product biosynthesis. Biochemistry. 1990;29:6499–6508. doi: 10.1021/bi00479a023. [DOI] [PubMed] [Google Scholar]
- 25.Sherman DH, et al. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J. Biol. Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokai T, et al. Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem. Biophys. Res. Commun. 2007;353:412–417. doi: 10.1016/j.bbrc.2006.12.033. [DOI] [PubMed] [Google Scholar]
- 27.Anzai Y, et al. Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics. Chem. Biol. 2008;15:950–959. doi: 10.1016/j.chembiol.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang CH, et al. Functional roles of the 6-S-cysteinyl, 8alpha-N1-histidyl FAD in glucooligosaccharide oxidase from Acremonium strictum. J. Biol. Chem. 2008;283:30990–30996. doi: 10.1074/jbc.M804331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler A, Kutchan TM, Macheroux P. 6-S-cysteinylation of bi-covalently attached FAD in berberine bridge enzyme tunes the redox potential for optimal activity. J. Biol. Chem. 2007;282:24437–24443. doi: 10.1074/jbc.M703642200. [DOI] [PubMed] [Google Scholar]
- 30.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project, Number 4. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.