Abstract
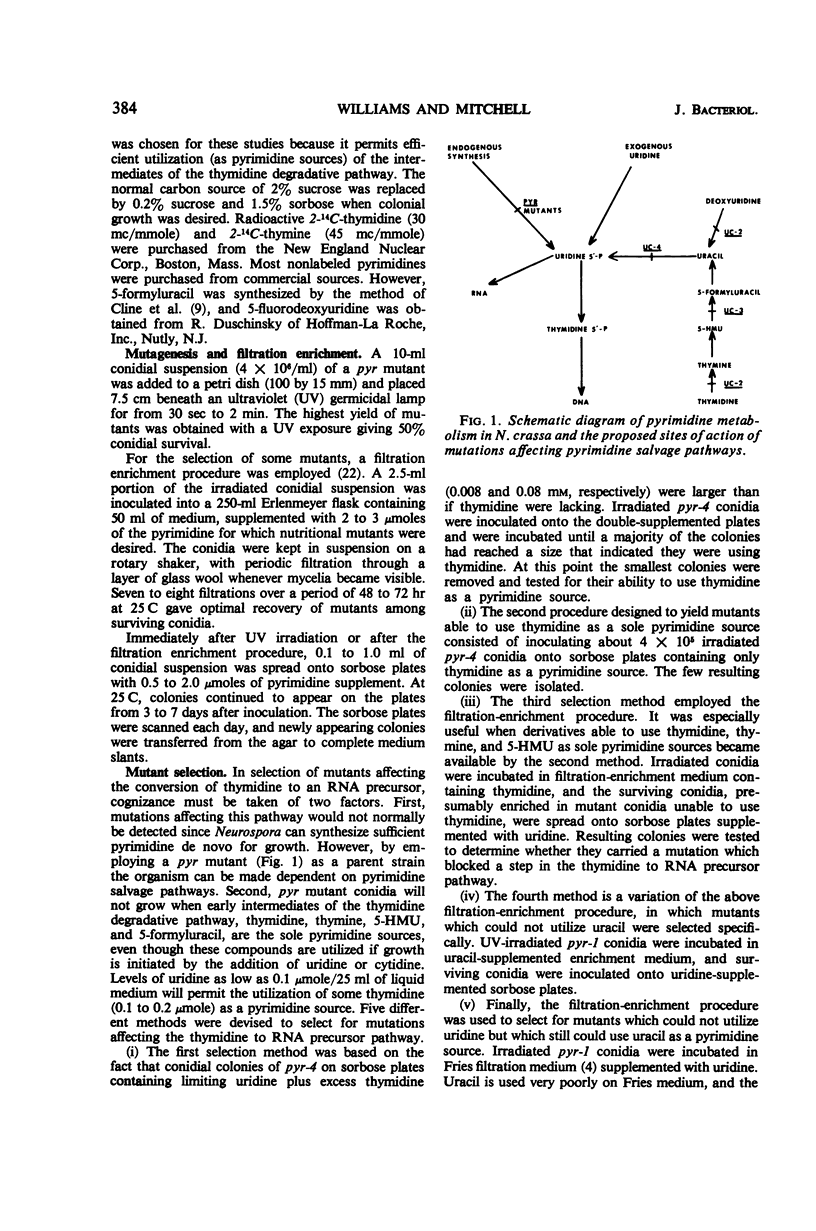
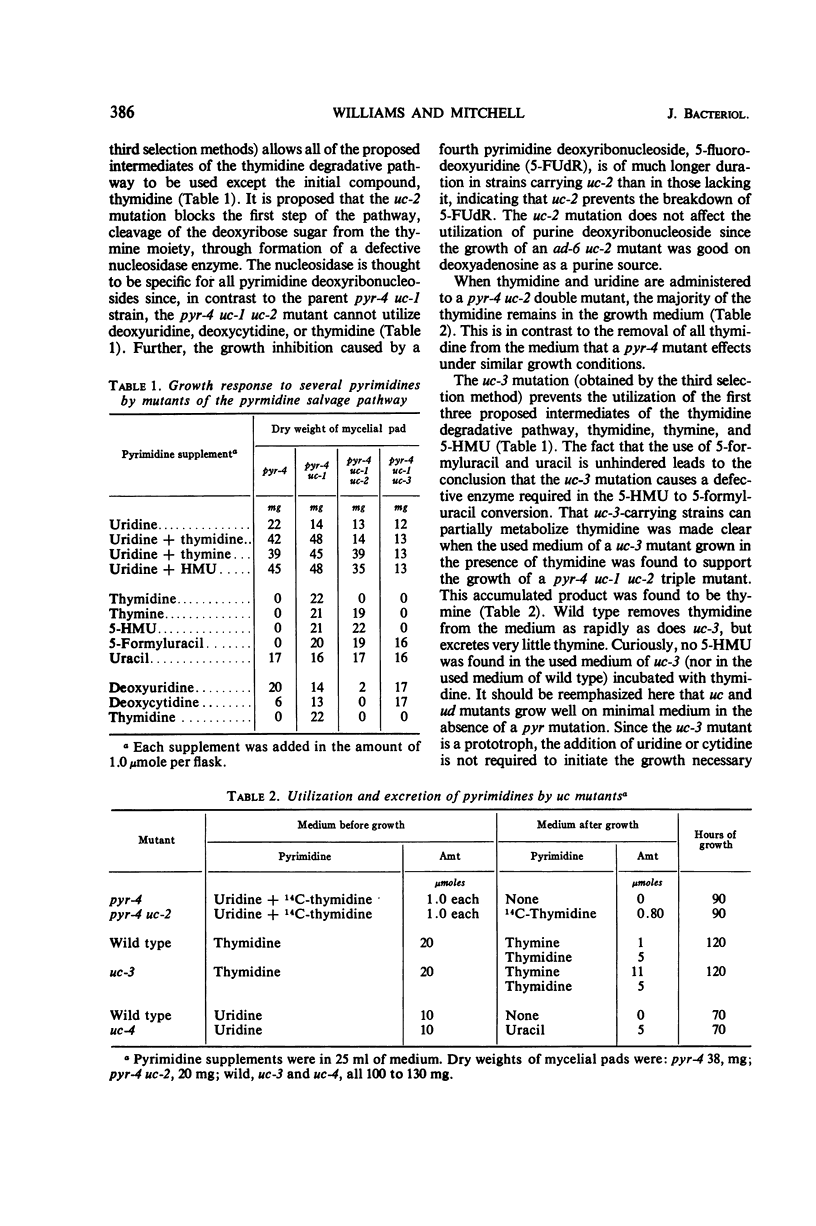
When 14C-thymidine labeled only in the ring is administered to Neurospora crassa, the majority of the recovered label is found in the ribonucleic acid (RNA). Three mutants were isolated in which different steps are blocked in the pathway that converts the pyrimidine ring of thymidine to an RNA precursor. Evidence from genetic, nutritional, and accumulation studies with the three mutants shows the pathway to proceed as follows: thymidine → thymine → 5-hydroxymethyluracil → 5-formyluracil → uracil → uridylic acid. A mutant strain in which the thymidine to thymine conversion is blocked is unable to metabolize thymidine appreciably by any route, including entry into nucleic acids. This suggests that Neurospora lacks a thymidine phosphorylating enzyme. A second mutation blocks the pathway at the 5-hydroxymethyluracil to 5-formyluracil step, whereas a third prevents utilization of uracil and all compounds preceding it in the pathway. The mutant isolation procedures yielded three other classes of mutations which are proposed to be affecting, respectively, regulation of the thymidine degradative pathway, transport of pyrimidine free bases, and transport of pyrimidine nucleosides.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abbott M. T., Dragila T. A., McCroskey R. P. The formation of 5-formyluracil by cell-free preparations from Neurospora crassa. Biochim Biophys Acta. 1968 Nov 20;169(1):1–6. doi: 10.1016/0005-2787(68)90002-6. [DOI] [PubMed] [Google Scholar]
- Abbott M. T., Schandl E. K., Lee R. F., Parker T. S., Midgett R. J. Cofactor requirements of thymine 7-hydroxylase. Biochim Biophys Acta. 1967 Mar 15;132(2):525–528. doi: 10.1016/0005-2744(67)90177-5. [DOI] [PubMed] [Google Scholar]
- BERECH J., Jr, VAN WAGTENDONK W. J. An autoradiographic study of the macronuclear changes occurring in Paramecium aurelia during autogamy. Exp Cell Res. 1962 Mar;26:360–372. doi: 10.1016/0014-4827(62)90188-x. [DOI] [PubMed] [Google Scholar]
- BRACHET J. The effects of various metabolites and anti-metabolites on the regeneration of fragments of Acetábularia mediterranea. Exp Cell Res. 1958 Jun;14(3):650–651. doi: 10.1016/0014-4827(58)90178-2. [DOI] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANELLAKIS E. S. Pyrimidine metabolism. II. Enzymatic pathways of uracil anabolism. J Biol Chem. 1957 Jul;227(1):329–338. [PubMed] [Google Scholar]
- CRAWFORD I., KORNBERG A., SIMMS E. S. Conversion of uracil and orotate to uridine 5'-phosphate by enzymes in lactobacilli. J Biol Chem. 1957 Jun;226(2):1093–1101. [PubMed] [Google Scholar]
- FINK R. M., FINK K. Biosynthesis of radioactive RNA and DNA pyrimidines from thymidine-2-C-14. Biochem Biophys Res Commun. 1961 Oct 23;6:7–10. doi: 10.1016/0006-291x(61)90174-7. [DOI] [PubMed] [Google Scholar]
- FINK R. M., FINK K. Relative retention of H3 and C14 labels of nucleosides incorporated into nucleic acids of Neurospora. J Biol Chem. 1962 Sep;237:2889–2891. [PubMed] [Google Scholar]
- FINK R. M., FINK K. Utilization of radiocarbon from thymidine and other precursors of ribonucleic acid in Neurospora crassa. J Biol Chem. 1962 Jul;237:2289–2290. [PubMed] [Google Scholar]
- Fangman W. L., Novick A. Mutant bacteria showing efficient utilization of thymidine. J Bacteriol. 1966 Jun;91(6):2390–2391. doi: 10.1128/jb.91.6.2390-2391.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivell A. R., Jackson J. F. Thymidine kinase: evidence for its absence from Neurospora crassa and some other micro-organisms, and the relevance of this to the specific labelling of deoxyribonucleic acid. J Gen Microbiol. 1968 Dec;54(2):307–317. doi: 10.1099/00221287-54-2-307. [DOI] [PubMed] [Google Scholar]
- LUBIN M., KESSEL D. H., BUDREAU A., GROSS J. D. The isolation of bacterial mutants defective in amino acid transport. Biochim Biophys Acta. 1960 Aug 26;42:535–538. doi: 10.1016/0006-3002(60)90836-2. [DOI] [PubMed] [Google Scholar]
- OGUR M., ROSEN G. The nucleic acids of plant tissues; the extraction and estimation of desoxypentose nucleic acid and pentose nucleic acid. Arch Biochem. 1950 Feb;25(2):262–276. [PubMed] [Google Scholar]
- Woodward V. W., De Zeeuw J. R., Srb A. M. THE SEPARATION AND ISOLATION OF PARTICULAR BIOCHEMICAL MUTANTS OF NEUROSPORA BY DIFFERENTIAL GERMINATION OF CONIDIA, FOLLOWED BY FILTRATION AND SELECTIVE PLATING. Proc Natl Acad Sci U S A. 1954 Mar;40(3):192–200. doi: 10.1073/pnas.40.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]



