Abstract
Small RNA molecules, such as microRNA and siRNA, have emerged as master regulators of gene expression through their ability to suppress target genes in a phenomenon collectively called RNA interference (RNAi). There is growing evidence that small RNAs can also serve as activators of gene expression by targeting gene regulatory sequences. This novel mechanism, known as RNA activation (RNAa), appears to be conserved in at least mammalian cells and triggered by both endogenous and artificially designed small RNAs. RNAa depends on Argonaute proteins, but possesses kinetics distinct from that of RNAi. Epigenetic changes are associated with RNAa and may contribute to transcriptional activation of target genes, but the underlying mechanism remains elusive. Given the potential of RNAa as a molecular tool for studying gene function and as a therapeutic for disease, further research is needed to elucidate fully its molecular mechanism in order to refine the rules for target selection and improve strategies for exploiting it therapeutically.
The quest to manipulate gene expression at will has been a fascinating pursuit since the establishment of the central dogma of gene expression. The discovery of posttranscriptional RNA interference (RNAi) has partially fulfilled this goal—that is the ability to suppress the expression of any gene using small double-stranded RNAs (dsRNAs). Since the initial discovery of RNAi (1, 2), several other related mechanisms of gene silencing have been identified that occur at the levels of chromatin, DNA, transcription, mRNA, and translation, each triggered by small RNA (3–6). It has been proposed that RNAi is an evolutionally conserved defense mechanism to suppress foreign sequences (i.e. viral infection); however, such examples are rare in higher eukaryotes despite the presence of intact RNAi machinery. Therefore, it is now believed instead that evolution has adapted this innate defense mechanism as a means to regulate gene expression. In this regard, it is reasonable to suggest that RNA-mediated gene regulation may have evolved the capability to regulate target sequences both negatively and positively, thereby composing the ‘yin and yang’ of the RNA-mediated gene-regulation network.
Recently, several classes of small RNA have been shown to upregulate gene expression at the transcriptional and/or epigenetic level (7–11). To describe such phenomena, the term RNAa (RNA activation) has been coined to distinguish it from RNAi (7). In this review, we discuss (i) the observations made so far on RNAa; (ii) our current understanding of its mechanism of action; (iii) its potential application both as a tool to study gene function and as a therapy for disease. We also speculate on the possibility of RNAa being mediated by endogenous small non-coding RNA (ncRNA).
1. Small dsRNA-mediated transcriptional activation (RNAa)
The discovery of RNAa came as a surprise. In early 2004, our group was interested in how aberrant DNA methylation of promoter sequences was regulated in cancer cells. It was speculated that ncRNA could induce sequence-specific DNA methylation, a phenomenon that had been known to occur in plants for over 10 years (12). At the time, our laboratory was investigating epigenetic mechanisms of gene silencing, including DNA hypermethylation in gene promoters. One particular gene of interest was E-cadherin, a tumor suppressor gene silenced in several types of cancers. The E-cadherin promoter contains a typical CpG island surrounding the transcription start site that, upon methylation, silences E-cadherin expression (13, 14). We sought to examine whether DNA methylation could be induced at the E-cadherin promoter by exposing cells to synthetic small dsRNAs, the known trigger for RNAi. By implementing rational siRNA (small interfering RNA) design rules (15) against the E-cadherin promoter sequence, two high-scoring targets at sites just outside the CpG island were selected for testing. When dsRNA for either target was transfected into the prostate cancer PC-3 cell line, E-cadherin expression was surprisingly robustly upregulated instead of being downregulated (7). This sparked subsequent identification of additional examples of RNAa (e.g. p21 and VEGF genes) (7) (Table 1). Shortly thereafter, another group reported activation of the progesterone receptor (PR) and major vault protein (MVP) genes by dsRNA (8), implying that the phenomenon could be a general mechanism of gene regulation.
Table 1.
RNAa examples.
| Gene symbol (alias) | Full name | Species | Target location | Trigger RNA | References |
|---|---|---|---|---|---|
| Ccnb1 | Cyclin B1 | Mouse | Promoter | saRNA/agRNA | (11) |
| CDH1 (E-cadherin) | Cadherin 1, type 1, E-cadherin (epithelial) | Human and non-human primates | Promoter | saRNA/agRNA and miRNA | (7, 9, 11, 78–80) |
| CDKN1A (p21) | Cyclin-dependent kinase inhibitor 1A | Human and non-human primates | Promoter | saRNA/agRNA | (7, 11, 75–77, 82) |
| CSDC2 | Cold shock domain-containing protein C2 | Human | Promoter | miRNA | (9) |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | Rat | Promoter | saRNA/agRNA | (11) |
| KLF4 | Kruppel-like factor 4 | Human | Promoter | saRNA/agRNA | (64) |
| LDLR | Low-density lipoprotein receptor | Human | Promoter | saRNA/agRNA | (24) |
| MVP | Major vault protein | Human | Promoter | saRNA/agRNA | (8) |
| NKX3-1 | NK3 homeobox 1 | Non-human primates | Promoter | saRNA/agRNA | (11) |
| PAWR (PAR4) | PRKC, apoptosis, WT1, regulator | Non-human primates | Promoter | saRNA/agRNA | (11) |
| PGR (PR) | Progesterone receptor | Human | Promoter and 3′ terminus | saRNA/agRNA | (8, 25) |
| TP53 (p53) | Tumor protein 53 | Non-human primates | Promoter | saRNA/agRNA | (11) |
| VEGFA (VEGF) | Vascular endothelial growth factor A | Human and non-human primates | Promoter | saRNA/agRNA | (7, 11) |
| Vegfa (Vegf) | Vascular endothelial growth factor A | Mouse | Promoter | shRNA | (10) |
| WT1 | Wilms tumor 1 | Non-human primates | Promoter | saRNA/agRNA | (11) |
1.1. Definition, observations, and features of RNAa
1.1.1. Definition
The term RNAa was originally used to describe gene activation at the transcriptional level mediated by small dsRNA designed to target gene promoter sequences (7). These dsRNAs, termed ‘small activating RNAs’ (saRNAs) to distinguish them from siRNAs, exert an effect opposite to that of RNAi. Similar to the term RNAi, whose definition has been expanded to describe both transcriptional and posttranscriptional gene silencing mechanisms, RNAa may also be used to describe other related mechanisms by which small RNAs positively regulate gene expression and the epigenome, including transcriptional activation via targeting the 3′ flanking regions of genes with small dsRNAs (16), piwi-interacting RNA (piRNA)-mediated epigenetic activation (17), miRNA-mediated translational activation (18, 19), and additional gene activation mechanisms yet to be discovered.
1.1.2. Promoter-targeting RNAa
In our initial studies, by testing 6 genes and designing 1 or 2 saRNA targets on the promoter of each gene, 3 of the tested genes (E-cadherin, p21, and VEGF) were activated by their respective saRNAs (7). These saRNAs have a target size of 19 nt with 3′ dTdT overhangs – an identical structure to standard siRNAs. They targeted locations on gene promoters ranging from −200 to −700 relative to the transcription start site. Soon after, the Corey group reported that synthetic dsRNAs, termed antigene RNAs (agRNAs) to distinguish them from siRNAs that target mRNA, targeting the promoter regions of PR and MVP activated the expression of their respective target genes. These agRNAs also have a target size of 19 nt but they target sequences located on or near the transcription start site, ranging from −56 to −2. Interestingly, in the example of the PR gene, the agRNA PR11, which targets sequence −11/+8, robustly induced PR gene expression in the MCF7 breast cancer cell line, but suppressed PR expression in the T47D cell line which has higher basal expression of PR compared to MCF7 cells (8). In these studies, RNAa was observed when the saRNA/agRNA was introduced into the cells at concentrations ranging from 5 to 50 nM (7, 8). Since these initial reports, additional examples of promoter-targeting RNAa have been identified (Table 1).
1.1.3. Non-promoter-targeting RNAa
Targeting genomic regions outside promoter sequences may also affect gene transcription both negatively and positively. Yue et al. found that agRNAs targeting the 3′ terminal region of the PR gene caused either transcriptional gene silencing or activation by interacting with an overlapping noncoding sense transcript (16). It has been proposed that the agRNAs trigger a looping mechanism bringing the 3′ terminus and promoter into close proximity to allow recruitment of additional proteins and modulation of promoter activity (16). Likewise, targeting enhancer sequences may also exert some regulatory effect on gene activity as enhancers use a similar looping mechanism to activate transcription (20).
1.1.4. Distinct kinetics of RNAa
An interesting feature associated with RNAa is its kinetics, which diverges from traditional RNAi. It is well known that RNAi can be induced within hours in mammalian cells, lasting for a period of ~5–7 days and subsequently disappearing when the exogenous siRNA is exhausted (21, 22). In contrast, RNAa takes a very different time course. Gene activation by transfecting promoter-targeting saRNA has been reported to be delayed by ~24–48 hours in comparison to RNAi. For example, in a side-by-side comparison, p21 activation by saRNA did not emerge until ~48 hours following saRNA transfection, while knockdown of two genes (MOF or E2F1) by siRNA was detectable as early as 6 hours (22). Perhaps, the delay in RNAa activity reflects a more complex mechanism with additional rate-limiting steps. Because RNAa is a nuclear process acting on gene transcription, acquiring access to the nucleus may be one of the rate-limiting steps. Changes in chromatin structure associated with RNAa may also contribute to the delayed kinetics.
Another peculiar feature of RNAa is its prolonged effect. After a single saRNA transfection, gene activation can last for nearly two weeks in certain cell lines (7, 22). The duration of PR gene activation demonstrated by the Corey group (8) and of E-cadherin and p21 induction observed by our group (7, 22) is remarkably similar, suggesting prolonged activation is a general feature of RNAa. It is reasonable to speculate that such long-lasting gene activation is due to epigenetic changes, which may persist well beyond the life of saRNA molecules. In support, several types of epigenetic changes have been associated with RNAa (see section 1.3.3).
1.1.5. Complementarity requirement of guide-target sequences for RNAa
Classic RNAi mediated by siRNA requires perfect complementarity between the siRNA and its cognate mRNA sequence. This complementarity is necessary for the RNA-induced silencing complex (RISC) to cleave the mRNA transcript at the siRNA recognition site. In contrast, translational repression mediated by miRNA can tolerate mismatches between non-seed sequence and its cognate mRNA (23). Interestingly, promoter-targeting saRNAs share similar recognition features as miRNA: mismatches in regions outside the seed sequence are tolerable and retain partial RNAa activity (7, 24). Based on this observation, it is possible that RNAa may be triggered by endogenous miRNA with imperfect matches to promoter sequences.
1.2. Conservation of RNAa
RNAa was initially discovered in human cells (7–9). By targeting the mouse VEGF promoter using short hairpin RNAs (shRNAs) via lentiviral-mediated overexpresion, Turunen et al. demonstrated RNAa in mouse cells in vivo and in vitro (10). RNAa has also been reported in several other mammalian species, including rat and non-human primates (11). Because non-human primates share almost identical genome sequences with human, most saRNA targets designed on the promoters of human genes are well-conserved in non-human primates (11) and these saRNAs can also activate their cognate genes in non-human primate cells (11). The conservation of saRNA targets in primates suggests the possibility of validating RNAa-based therapeutics in non-human primate disease models, whereas human and rodent promoter sequences diverge significantly, saRNAs that are functional in rodents may not be active in human cells.
1.3. Mechanism of RNAa
Although the mechanism of RNAa remains largely elusive, recent studies have provided insight into several important aspects of RNAa.
1.3.1. Target molecules of RNAa
Unlike targeting single-stranded mRNA sequences, saRNAs/agRNAs designed for promoter regions may have different modes of strand preference: sense or antisense. At present, a universal model for RNAa is lacking and different mechanisms may exist in context with different promoters.
Sense or antisense target?
Whereas the trigger for RNAa is small duplex RNAs, evidence suggests that only one strand is required to guide activity. In a manner seemingly analogous to strand selection in the case of siRNAs, duplex saRNA is recruited by endogenous machinery in which one strand is discarded and the remaining strand facilitates target recognition via complementary base-pairing to initiate RNAa. The question remains: which strand guides RNAa activity when targeting noncoding sequence? It has been demonstrated that activation of the PR and low-density lipoprotein receptor (LDLR) genes by targeting promoter sequences with agRNA is facilitated through interacting with a noncoding antisense transcript (i.e. synthesized in the opposite direction as target gene transcription), which runs through the promoter (24, 25). In these cases, the antisense transcript serves as a docking site for target recognition, implying that RNAa activity is mediated by the sense strand in the agRNA duplex. In another system, in which PR expression was activated by targeting the 3′ terminal region with agRNA, it was revealed that a noncoding sense transcript (i.e. synthesized in the same direction as target gene transcription) recruits agRNA (16). Such evidence would imply that the antisense strand in this agRNA duplex guides gene activation. Overlapping noncoding RNAs transcribed in the sense and antisense orientations have already been shown to serve as docking sites for transcriptional gene silencing mediated by small duplex RNAs (26–28). Likewise, such sense and antisense transcripts may serve as the targets for RNAa as well.
Though PR gene activation by targeting promoter sequence was facilitated by the sense strand of its agRNA duplex, other target genes may not have the same requirement. For instance, using chemical modifications to inactivate strand function selectively, we showed that the antisense strand in saRNA duplexes was responsible for RNAa activity in two examples targeting the p21 and E-cadherin promoters (22). It is likely that strand function in saRNA/agRNA duplexes is dependent on the context of the gene and/or orientation of the targeted molecule. Utilizing modified saRNA/agRNAs with inactivated strands can define strand activity and assist in determining orientation of putative target noncoding transcripts.
RNA or DNA as target?
Recent studies have revealed that both sense and antisense noncoding transcripts are pervasive in the human genome (29, 30). At promoter sites, transcripts are also abundant, with some as short as ~20 nucleotides (31). Some, referred to as nascent transcripts, have been found to be usually tethered to gene promoters (31–33). Long noncoding RNAs (lncRNAs), as large as a few thousand nucleotides, can also overlap promoter regions. These promoter-overlapping transcripts may serve as binding sites of Ago-loaded agRNAs. For example, long noncoding antisense transcripts overlapping the PR or LDLR promoter are considered to be the target of agRNAs (24, 25) because their depletion by an RNase H-based mechanism renders the agRNAs nonfunctional (25). The transcript–agRNA–Ago complex is thought to act as a scaffold for recruiting other proteins [e.g. hnRNP-k (heterogenous nuclear ribonucleoprotein-k), HP1 (heterochromatin protein 1γ), etc.] to effect gene regulation by enhancing or reducing association of RNA polymerase II (RNAP II) (25). Interestingly, the abundance of these promoter-overlapping transcripts is not affected by agRNA targeting, implying that an RNAi cleavage mechanism is not involved (34).
RNA has long been known to hybridize to double-stranded DNA (35, 36) with high stability comparable to that of protein-DNA complexes (37). It has been hypothesized that RNA hybridization with the non-template strand of the promoter could expose the template strand for RNAP II binding (36). In plants, RNA-directed DNA methylation (RdDM) only occurs to the cytosines along RNA-DNA duplexes, indicating a direct RNA-DNA interaction that provides a strong and specific signal for de novo DNA methylation (38, 39). Thus, it is possible that, with the help of an Ago protein, saRNA interacts directly with its DNA target without the need of a third ncRNA as a docking molecule. This viewpoint is further supported by recent observations that many ncRNAs can guide chromatin-modifying complexes to specific genomic sites in mammalian cells (40, 41).
1.3.2. Dependence of RNAa on Argonaute proteins
Argonaute (Ago) proteins belong to a large family of proteins containing the PIWI and PAZ domains. Humans have a total of four closely related Ago proteins (Ago1-4) that function in small RNA-mediated gene regulation. During conventional RNAi, the Ago2 protein recruits siRNAs to form the RISC complex, which cleaves target mRNA by virtue of Ago2’s catalytic activity (42). Both Ago1 and Ago2 have been implicated in transcriptional RNAi, frequently referred to as transcriptional gene silencing (TGS) (43, 44), although the details of the mechanism remain elusive. The requirement of Ago proteins in RNAa has also been examined by RNAi knockdown and chromatin immunoprecipitation (ChIP) experiments. In knockdown experiments, RNAa-mediated induction of E-cadherin and p21 was abolished by Ago2 depletion, whereas knockdown of the other Ago members (Ago1, 3, and 4) did not significantly impair RNAa (7, 22). Using ChIP assays, an agRNA-dependent association of Ago2 has been demonstrated with the targeted antisense transcripts in the PR and LDLR promoter (24, 34). Turunen et al. also showed that Ago2 is recruited to the mouse VEGF promoter by activator shRNAs (10). The dependence of RNAa on Ago2 is surprising because target cleavage is not observed in RNAa (34). A reasonable explanation is that Ago2 is required for initial processing of saRNA/agRNA duplexes in a manner similar to siRNA maturation. The endonuclease activity of Ago2 cleaves and discards the passenger strand to form an active RNA-Ago complex capable of recognizing complementary sequences (45, 46). This view is supported by the observation that 2′-OMe modification at the passenger-strand cleavage site inhibits RNAa activity (22). In addition, Ago proteins were found to be as abundant in the nucleus as in the cytoplasm in a number of studies (47–49). However, it is unclear whether Ago2 is directly involved in subsequent transcriptional activation at the targeted gene or if additional proteins are essential for RNAa. Moreover, other Ago proteins may participate in RNAa when it is triggered by small RNA with an imperfect duplex structure (i.e. miRNA), in a manner similar to conventional RNAi.
1.3.3. Transcriptional activation is associated with epigenetic changes
Most examples of RNAa were demonstrated by an increase in target gene expression at the mRNA and/or protein level. Enhanced mRNA stability may also lead to increased mRNA and protein levels and represents an alternative mechanism for RNAa. However, this possibility does not appear to be true when assessing mRNA stability after saRNA transfection. Determination of both p21 and E-cadherin mRNA levels across a period of time after actinomycin D or α-amanitin treatment revealed that saRNA treatment does not increase the stability of their corresponding mRNAs compared to control treatments (Portnoy et al., unpublished data). Instead, several studies have shown an increase in RNAP II association with the core promoter, indicating RNAa is a transcriptional mechanism (9, 16, 24, 34).
Various types of chromatin modifications have also been identified at different promoters following RNAa. For example, loss of di- and tri-methylation at histone H3K9 has been associated with RNAa at the E-cadherin promoter in PC-3 cells (7). In another study, reduced acetylation at H3K9 and H3K4 together with increased di- and tri-methylation at H4K4 has been associated with RNAa at the PR promoter (8). Also, activation of the mouse VEGF promoter by shRNAs has been linked to increased H3K4me2 and H3K4me3 levels, as well as decreased H3K9me2, leading to a more accessible chromatin structure (10). It is plausible to hypothesize that Ago2 guides saRNA molecules to complementary target sequence where they serve as a scaffold to recruit histone-modifying enzymes and activate transcription. The diversity in chromatin modifications may reflect gene-specific epigenetic changes associated with RNAa (8). Furthermore, the question still remains as to whether the changes in chromatin are the cause or consequence of RNAa. Regardless, successful mapping of epigenetic changes at target-gene promoters has caught RNAa in action and indirectly provides evidence toward target specificity.
2. Examples of endogenous RNAa
The fact that synthetic saRNA/agRNA duplexes are able to elicit RNAa in a variety of mammalian species suggests that RNAa is an endogenous mechanism of gene regulation. Given the similarity between miRNA and saRNA/agRNA in structure and chemistry, it is tempting to speculate that naturally occurring miRNA may serve as an endogenous trigger of RNAa or TGS. In support, mutation experiments have revealed that RNAa can tolerate mismatches between synthetic saRNAs and target promoter sites at nucleotides outside the “seed” sequence; features indicative of miRNA target recognition (7, 22).
Initial evidence that miRNA positively regulates target sequences came from a study by Jopling et al. (50). In this study, miR-122 expressed in human liver cells enhanced hepatitis C viral (HCV) gene replication by targeting the 5′ non-coding elements in the HCV genome (50), whereas sequestering miR-122 resulted in loss of autonomous replication of HCV RNAs. In two other studies, miR-369-3 was shown to positively regulate mRNA translation by targeting AU-rich elements in 3′UTRs (untranslated regions) in stressed cells (18, 19). Recently, miR-10a also has been shown to positively regulate gene expression by interacting with the 5′UTR of ribosomal protein mRNAs to enhance translation (51).
2.1. miRNA target prediction on promoter sequences
miRNA target prediction in 3′UTRs is facilitated by examining target conservation across different species. Otherwise, computationally predicted targets with good homology may not necessarily be bona fide targets (52, 53). Predicting miRNA targets in gene promoters is even more challenging due to a lack of relevant experimental data and general poorer conservation across species. The first attempt to identify miRNA targets on promoter sequences came from a study by Place et al. (9) in which the E-cadherin promoter was scanned for putative target sites highly complementary to miRNA. This study led to the identification and verification of miR-373 target sites in the promoters of E-cadherin and cold shock domain containing protein C2 (CSDC2) (9). Other groups have subsequently used/designed other algorithms to scan 200-bp portions of all proximal promoter regions in the human genome. It was found that promoter sequences complementary to miRNAs are as common as those within 3′UTRs, some of which possess unusually high complementarity (54, 55).
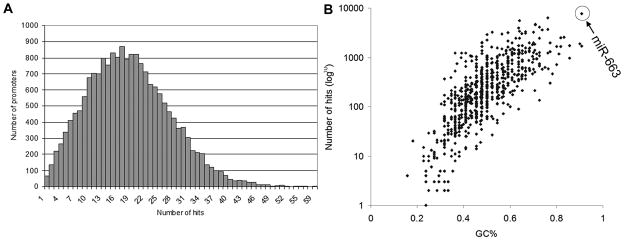
Considering that miRNA target sites may not be restricted to only the 200-bp proximal promoter region, we used the miRanda algorithm to scan 1 kb of all human promoter sequences for putative target sites (56). By setting the prediction stringency to medium-high, the prediction identified a total of 345,259 targets on 18,687 promoters for 701 human miRNAs, with an average of 18.5 miRNA targets per promoter (1 kb) (Fig. 1A). Only a few miRNAs target promoter sequences with 100% complementarity. Six of these miRNAs (miR-611, miR-191, miR-484, miR-320a, miR-34b, and let-7i) are directly transcribed from the promoter regions of protein-coding genes. Among them, miR-320a, transcribed from the promoter of POLR3D gene, has already been shown to modulate in cis the expression of POLR3D (54). Two additional miRNAs (miR-574-5p and miR-548c-5p) have perfect complementarity with multiple promoters. As it turns out, these miRNAs are transcribed from repetitive sequences in the genome and share perfect complementarity with similar repetitive sequences found in the promoters of multiple genes. The impact of these miRNAs on gene transcription deserves further investigation.
Figure 1. Whole-genome promoter miRNA target prediction.
A. Histogram depicting the number of promoters (1 kb) in the human genome versus the number of sites predicted to be complementary to known human miRNAs. B. The number of miRNA hits in gene promoters positively correlates to the GC content of miRNAs. Shown is a semi-log plot with the number of promoter hits in logarithmic scale on the y-axis and GC content (GC%) of miRNAs in linear scale on the x-axis.
One intriguing observation from whole-genome target prediction is that the number of potential targets in gene promoters positively correlates to the GC content of the miRNA (Fig. 1B). For example, miR-663 has the highest number of targets (19,263 hits, Fig. 1B) followed by miR-638, -661 and -608. A feature common to these miRNAs is that they have relatively high GC content and contain several CpG sites within their sequence; miR-663 has a GC content of 90.9%, the highest among all human miRNAs, and contains 5 CpG sites. It may come as no surprise that these miRNAs would have such high numbers of hits in gene promoters because 60% of all human genes have CpG islands within their promoters, which are characterized by high GC content and a high ratio of observed-to-expected CpG frequency (57). Interestingly, GC-rich miRNAs are not well conserved across species. This raises an interesting possibility that such miRNAs may preferentially target gene-promoters, which are also poorly conserved compared to protein-coding sequences. Perhaps, such miRNAs rapidly evolve in parallel with targeted promoter sequences.
2.2. miRNA-mediated RNAa
Based on target-prediction algorithms, miR-373 possesses a target site in the E-cadherin promoter at position -645 relative to the transcription start site, with 87% sequence complementarity. Transfection of either mature miR-373 or precursor (pre-miR-373) mimics into PC-3 cells readily activates E-cadherin expression (9). E-cadherin induction by pre-miR-373 requires the Dicer protein for maturation and activation is specific to the miR-373 sequence. Mechanistically, miR-373 enhanced the transcription of its target genes by recruiting RNAP II (9). By analyzing additional putative miR-373 targets, CSDC2 also was confirmed to be activated by miR-373 in PC-3 cells. However, not all genes containing promoter sequences complementary to miR-373 were susceptible to gene activation. This suggests that promoter environment, chromatin accessibility and so on may play a role in determining susceptibility to miRNA-mediated RNAa.
The miR-373 study provides the first evidence that promoter-targeting miRNAs can elicit RNAa in manner similar to saRNA/agRNA duplexes. However, several questions still remain. For instance, what is the physiological relevance of miRNA-mediated RNAa via targeting promoter sequence? Examples of endogenous miRNAs positively regulating gene expression in a physiological context, including phenotypic consequences and functional significance, need to be examined. Given that miRNA can have oncogenic or tumor suppressor-like function, one might envision endogenous miRNA influencing tumorigenesis via RNAa. However, the involvement of RNAa in cancer remains unclear.
2.3. piRNA-mediated RNAa
piRNAs comprise a class of 26- to 31-nt small RNAs that interact with Piwi subfamily members of the Argonaute proteins and suppress transposable elements, thus playing roles in germline development and transposon silencing (58–61). By deep sequencing of Piwi-bound small RNA of Drosophila melanogaster, Yin et al. identified a 20-nt piRNA that uniquely maps to a region of the telomere-associated sequence (TAS) on the right arm of chromosome 3 (3R-TAS). This piRNA, called 3R-TAS1 piRNA, is enriched in the nucleus and expressed in both the germline and soma. In wild-type flies, 3R-TAS is associated with both euchromatic chromatin markers, including H3K4me2, H3K4me3 and H3K9ac, and heterochromatic markers such as H3K9m2, H3K9m3, H4K12ac and HP1. Depletion of the 3R-TAS1 piRNA by Piwi mutation results in a dramatic shift of the TAS chromatin markers toward heterochromatin markers, whereas restoration of 3R-TAS1 piRNA reinstates the euchromatic features of TAS (17). Such epigenetic activation by piRNA provides an additional example that an otherwise largely suppressing small RNA can have dual functions by acting as an activator.
3. Proposed model for RNAa
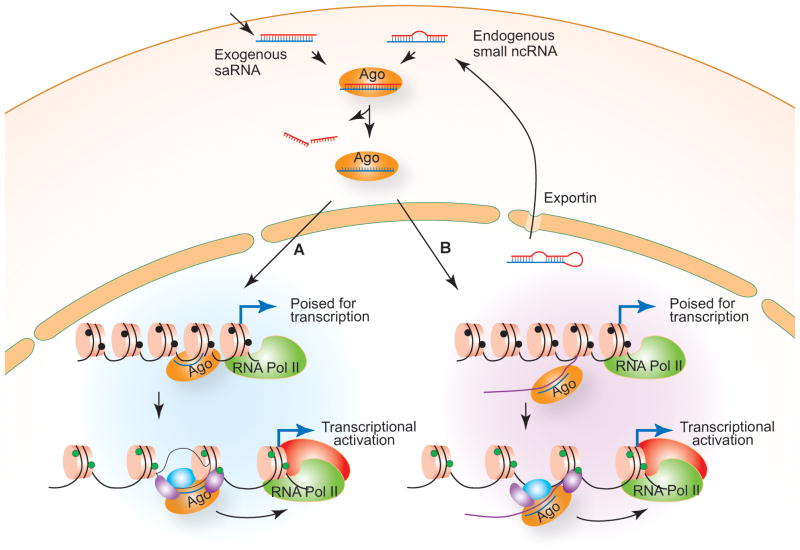
Evidence accumulated so far points to the possibility that there exists an endogenous cellular mechanism by which endogenous ncRNA is used to positively regulate gene expression by modulating chromatin and epigenetic states. This mechanism can be exploited by using artificially designed saRNA/agRNA molecules that target gene promoters to increase gene transcriptional activity. Based on our understanding of the current literature, we propose a model for RNAa induced by promoter-targeted saRNA/agRNA (Fig. 2). In this model, an exogenously introduced saRNA/agRNA or an endogenously transcribed and processed small RNA such as miRNA is loaded onto an Ago protein in the cytoplasm. The Ago protein processes the saRNA/agRNA to form an active Ago-RNA complex by cleaving and discarding one of the RNA strands. The Ago-RNA complex then enters the nucleus through active transport or passive diffusion when the nuclear envelope breaks down during cell division. It is also possible that saRNA/agRNA is first imported into the nucleus where it programs an active Ago-RNA complex. If complementary targets for the RNA guider exist in genomic DNA sequences (Fig. 2A) or in ncRNA sequences that are tethered to the DNA (Fig. 2B), then the Ago-RNA complex would initiate a process that diverges from RNAi to modify chromatin structure and epigenetic states at the target location via two potential mechanistic models. In model A, the RNA guider guides the Ago protein to its DNA target by forming an RNA-DNA duplex or triplex structure and the Ago protein then serves as a recruiting platform to attract histone-modifying activities, leading to an open chromatin structure and active transcription. Alternatively, as depicted in model B (Fig. 2B), the RNA guider binds to cognate promoter transcripts and the Ago protein recruits histone-modifying enzymes to institute active chromatin marks on the local chromosome, resulting in transcriptional activation.
Figure 2. Mechanism for RNAa.
An exogenously introduced or naturally occurring saRNA/agRNA is loaded into an Ago protein (e.g. Ago2) where the passenger strand is cleaved and discarded, resulting in an active Ago-RNA complex. This complex gains access to the nuclear compartment by either passive transport when the nuclear envelope disappears during mitosis or active transport mechanisms. The complex may then bind to (A) complementary DNA sequences or (B) nascent cognate transcripts in promoters or 3′ flanking regions and further recruit histone modifiers, leading to an open chromatin structure and active transcription.
4. Exploiting RNAa
RNAa has been shown to be generally potent and long-lasting (7, 8), making it a promising new tool for gene overexpression studies and a novel therapeutic strategy for treating disease by stimulating gene expression.
4.1. Technical considerations for RNAa
Several technical issues should be taken into consideration when designing and performing RNAa experiments.
4.1.1. saRNA target selection
Design of saRNAs has largely been a hit-or-miss process due to a lack of complete understanding of the underlying mechanism and the heterogeneous nature of different target regions in promoters with regard to local chromatin structure and its impact on gene transcription. Shifting a targeting sequence by just a few bases may change a potent saRNA/agRNA into a less effective activator (8). The basis for this spatial sensitivity is unclear, but it may be dependent on the context of duplex sequence and/or target site. Different target locations may result in saRNA sequences with varying thermodynamic properties, and guide-strand selection and effective processing by Ago proteins are dependent on the thermodynamic characteristics within dsRNAs. As such, improper selection of the guide strand may sequester RNAa activity. Certain small RNA sequences may also be restricted to the cytoplasm and have limited access to the nuclear compartment thereby limiting their opportunity to facilitate RNAa. In support, the sequences in some miRNAs have been shown to specify compartment localization (62). It has been reported that some miRNAs are restricted to only the cytoplasm or nucleus (63). Additional parameters associated with a particular region of a gene promoter such as chromatin/DNA accessibility, distance from the transcription start site, presence or absence of nascent transcripts, and so on may also determine whether the region is a suitable target for RNAa. Efficient saRNA design will be greatly improved as our understanding of the mechanism(s) of RNAa develops. To increase the likelihood of identifying a functional saRNA for a particular gene, generally, at least 5 saRNAs should be designed and tested.
4.1.2. Off-target effects
Short dsRNA is known to be able to induce off-target effects and change the expression of many unrelated genes. These effects could be sequence-specific through the miRNA-like regulatory mechanism or non-sequence-specific through an interferon response or immunostimulation. To avoid off-target effects, saRNA targets should be carefully selected to ensure that the target sequences do not have significant homology with any other region in the genome, including coding and non-coding regions. Induced expression of the intended target gene following saRNA transfection should also be carefully interpreted so as to exclude any potential off-target effect. This can be done by correlating the expression induction with phenotypic changes of the transfected cells and expression changes of downstream genes known to be regulated by the target gene (64). Validation of RNAa-induced phenotypic and downstream gene changes by vector-based overexpression of the RNAa target gene may also help in determining specificity (64).
4.1.3. Maximizing the RNAa effect
To achieve maximal gene activation by RNAa in cultured cells, we consider the two most important aspects to be saRNA transfection concentration and duration. Because RNAa is a nuclear process and transfected small dsRNAs largely localize in the cytoplasm due to an exportin-5-mediated dsRNA nuclear exclusion mechanism (65), a higher concentration of saRNA is needed to transfect cells compared to siRNA transfection for RNAi experiments. Since there is an ~48-hour delay in the onset of RNAa activity, and peak gene-activation does not appear until 4–5 days post-transfection (7), it is advised that the optimal window for observing gene activation is between 3–5 days (22). In addition, some general rules for cell-transfection experiments should be followed in RNAa experiments, including: using healthy and uncontaminated cells at their exponential growth phase, and transfecting the cells at an optimal density so that the cells are not over-confluent at harvest. A reverse-transfection protocol generally yields better results than forward transfection for RNAa.
4.2. RNAa as a tool for studying gene function
Conventionally, gain-of-function studies are often carried out by vector-based systems. However, ectopic expression vectors do not typically resemble natural genes (66, 67). They are frequently created from cDNA libraries or amplicons, which do not contain introns or UTR elements. Such regions can have pivotal effects on endogenous gene function (66, 68). For instance, intronic regions allow for multiple splicing variants that can have unique functions in different cell or cancer types (69, 70). RNAa has the unique ability to enhance endogenous transcription of a targeted gene and its potential splicing variants. Such ability has been demonstrated by the simultaneous upregulation of multiple splicing variants of the VEGF (7) or PR (8, 16) gene using a single promoter-targeted saRNA. Very recently, our group has used RNAa as a molecular tool to interrogate the function of KLF4, a member of the Krüppel-like family of transcription factors, in prostate cancer cells in which its expression is silenced and its function had never been studied (64). RNAa-mediated KLF4 restoration in prostate cancer cells caused modulation of its regulated genes, including genes that regulate the cell cycle and cell motility (64), and led to inhibition of both cell proliferation/survival and invasion/migration, suggesting a tumor-suppressor role for KLF4 in prostate cancer cells. These results could be fully recapitulated by vector-mediated KLF4 overexpression (64). RNAa thus offers a promising new approach to interrogating gene function (e.g. tumor suppressor genes) and may serve as a surrogate tool for traditional vector-based overexpression systems.
4.3. Potential therapeutic use of RNAa
RNAi-based therapies are in development for the treatment of a variety of diseases (e.g. cancer, etc.). RNAi offers high specificity and efficacy with generally minimal toxicity. RNAi is ideal for the treatment of disorders that are caused by the expression of mutant genes encoding dominant-negative proteins, aberrant splicing isoforms, or overexpressed genes leading to gain-of-function effects (71). However, RNAi can only offer antagonism of a specific molecular target for disease treatment. Strategies that can lead to activation of a molecular target (e.g. tumor suppressor genes) are equally crucial for effective disease control. Although traditional gene therapy has the capacity to correct abnormal gene copies or augment the expression of normal genes, it has inherent drawbacks, including the tedious process of constructing expression vectors and potential detrimental effects on host genome integrity.
By utilizing saRNAs as therapeutic compounds, RNAa offers similar benefits as RNAi, while facilitating the exact opposite response – gene activation. This approach addresses a missing element in RNA-based gene therapies and offers a novel solution to provide greater efficacy in disease control. Several studies have begun to explore the therapeutic use of RNAa in suppressing tumor growth by activating tumor suppressor genes. For instance, p21 and E-cadherin are tumor suppressor genes readily susceptible to RNAa. E-cadherin is a key functional component at adherens junctions in all epithelial cells and serves as a potent inhibitor of invasion and growth (72, 73). p21 is a cyclin-dependent kinase (CDK) inhibitor that functions as a key mediator of cell-cycle arrest (74). Restoration of p21 expression in different cancer cells, including prostate, bladder, and lung cancer, has been shown to inhibit proliferation (7, 75–77) and sensitizes lung cancer cells to chemotherapeutic agents (77). Similarly, activation of E-cadherin via RNAa inhibits growth of bladder cancer and prostate cancer cells in vitro (78, 79). Recently, Junxia et al. reported that transfection of E-cadherin saRNA into breast cancer cells restores endogenous E-cadherin expression, induces apoptosis, and inhibits proliferation in vitro and in vivo (80).
The VEGF gene plays an important role in angiogenesis. Turunen et al. used lentivirus to deliver a mouse Vegf saRNA in the form of shRNA into ischemic mouse hindlimb and found that such treatment improved vascularity and blood flow (10). This study opens a potential new avenue for the treatment of cardiovascular diseases. Similarly, activation of the LDLR gene by RNAa may provide a new therapeutic option for hypercholesterolemia because increased LDLR expression can lead to reduced cholesterol levels (24).
Identifying chemical modifications is also necessary to improve the medicinal properties of saRNAs. For instance, blocking the 5′-OH of the passenger strand in synthetic saRNAs has been shown to reduce its off-target potential (22). Furthermore, incorporating intentional mismatches opposite the 5′-most nucleotide in the guide strand has been shown to enhance target-gene induction in the case of E-cadherin and p21 (22). Modifications to the 2′ backbone (i.e. 2′-OMe and 2′-Fluoro), as well as locked nucleic acid substitutions, are also tolerated by saRNA/agRNA duplexes (22, 24, 81). These modifications may improve therapeutic application by increasing endonuclease resistance and serum stability, much as they are utilized to stabilize siRNAs. Exploiting these chemical modifications alone or in combination, as well as identifying new features of saRNAs, are ultimately needed to improve the medicinal properties of saRNAs for in vivo application.
5. Conclusion
Evidence suggests that RNAa is an endogenous mechanism of gene regulation guided by small RNA molecules and mediated, in part, by Ago proteins. In parallel to RNAi mechanisms, RNAa may serve as a countervailing component in gene regulatory networks. As such, saRNAs, much like siRNAs, provide a new approach for interrogating and modulating endogenous gene function in living cells. Augmenting gene expression with saRNAs also has potential for treating diseases (e.g. cancer, etc.) and application in stem cell research. However, our understanding of RNAa is still limited and many questions remain unanswered. For example, how is transcriptional activation achieved? What are the molecular machines involved? Is RNAa naturally exploited by cells for physiological function? On a practical level, several challenges with RNAa still remain. For instance, multiple targets need to be screened in order to activate a particular promoter. Regardless, RNAa offers a new approach to enhance endogenous gene expression, which may be manipulated to target a variety of genes for therapeutics or functional studies.
Acknowledgments
This work was supported by grants from the National Cancer Institute at the National Institutes of Health (1R21CA131774-01 to L.C.L.), the National Institutes of Health (1R01GM090293-0109 to L.C.L.), Department of Defense (W81XWH-08-1-0260 to L.C.L.), and California Institute for Regenerative Medicine (RL1-00660-1 to L.C.L.).
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305(5688):1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 4.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 5.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216(2):671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 6.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Li LC, Okino ST, Zhao H, Pookot D, Place RF, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103(46):17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3(3):166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 9.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, et al. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ Res. 2009;105(6):604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- 11.Huang V, Qin Y, Wang J, Wang X, Place RF, et al. RNAa is conserved in mammalian cells. PLoS ONE. 2010;5(1):e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76(3):567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 13.Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55(22):5195–5199. [PubMed] [Google Scholar]
- 14.Li LC, Zhao H, Nakajima K, Oh BR, Filho LA, et al. Methylation of the E-cadherin gene promoter correlates with progression of prostate cancer. J Urol. 2001;166(2):705–709. [PubMed] [Google Scholar]
- 15.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 16.Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, et al. Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat Chem Biol. 2010;6(8):621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450(7167):304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 18.Vasudevan S, Steitz JA. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell. 2007;128(6):1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science (New York, NY. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 20.Petrascheck M, Escher D, Mahmoudi T, Verrijzer CP, Schaffner W, Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2010;33(12):3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4(6):457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 22.Place RF, Noonan EJ, Foldes-Papp Z, Li LC. Defining features and exploring chemical modifications to manipulate RNAa activity. Curr Pharm Biotechnol. 2010;11(5):518–526. doi: 10.2174/138920110791591463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novina CD, Sharp PA. The RNAi revolution. Nature. 2010;430(6996):161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 24.Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem Biol. 2010;17(12):1344–1355. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15(8):842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A. 2007;104(30):12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33(4):462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7(16):2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 29.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. The transcriptional landscape of the mammalian genome. Science. 2010;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 30.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. Antisense transcription in the mammalian transcriptome. Science. 2010;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 31.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 32.Goodrich JA, Kugel JF. From bacteria to humans, chromatin to elongation, and activation to repression: The expanding roles of noncoding RNAs in regulating transcription. Crit Rev Biochem Mol Biol. 2009;44(1):3–15. doi: 10.1080/10409230802593995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 34.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bekhor I, Bonner J, Dahmus GK. Hybridization of chromosomal RNA to native DNA. Proc Natl Acad Sci U S A. 1969;62(1):271–277. doi: 10.1073/pnas.62.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frenster JH. Selective control of DNA helix openings during gene regulation. Cancer Res. 1976;36(9 PT 2):3394–3398. [PubMed] [Google Scholar]
- 37.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258(5087):1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 38.Pelissier T, Thalmeir S, Kempe D, Sanger HL, Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27(7):1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, et al. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta. 2004;1677(1–3):129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 40.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert SL, Pehrson JR, Sharp PA. XIST RNA associates with specific regions of the inactive X chromatin. J Biol Chem. 2000;275(47):36491–36494. doi: 10.1074/jbc.C000409200. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 43.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13(9):787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 44.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13(9):793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 45.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123(4):621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 46.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 47.Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, et al. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36(20):6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA (New York, NY) 2008;14(6):1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, et al. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 51.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Saito T, Saetrom P. MicroRNAs - targeting and target prediction. N Biotechnol. 2010;27(3):243–249. doi: 10.1016/j.nbt.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Verbeek FJ. Comparison and Integration of target prediction algorithms for microRNA studies. J Integr Bioinform. 2010;7(3):1–13. doi: 10.2390/biecoll-jib-2010-127. [DOI] [PubMed] [Google Scholar]
- 54.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Younger ST, Pertsemlidis A, Corey DR. Predicting potential miRNA target sites within gene promoters. Bioorg Med Chem Lett. 2009;19(14):3791–3794. doi: 10.1016/j.bmcl.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60(8):1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 59.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 60.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 62.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 63.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE. 2010;5(5):e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Place RF, Huang V, Wang X, Noonan EJ, et al. Prognostic Value and Function of KLF4 in Prostate Cancer: RNAa and Vector-Mediated Overexpression Identify KLF4 as an Inhibitor of Tumor Cell Growth and Migration. Cancer Res. 2010;70(24):10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34(5):1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark AJ, Archibald AL, McClenaghan M, Simons JP, Wallace R, Whitelaw CB. Enhancing the efficiency of transgene expression. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences. 1993;339(1288):225–232. doi: 10.1098/rstb.1993.0020. [DOI] [PubMed] [Google Scholar]
- 68.Stemmler MP, Hecht A, Kemler R. E-cadherin intron 2 contains cis-regulatory elements essential for gene expression. Development. 2005;132(5):965–976. doi: 10.1242/dev.01662. [DOI] [PubMed] [Google Scholar]
- 69.Breitbart RE, Nguyen HT, Medford RM, Destree AT, Mahdavi V, Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated troponin T isoforms from a single gene. Cell. 1985;41(1):67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- 70.Leff SE, Rosenfeld MG, Evans RM. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- 71.Aigner A. Gene silencing through RNA interference (RNAi) in vivo: strategies based on the direct application of siRNAs. J Biotechnol. 2006;124(1):12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 72.Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113(1):173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richards FM, McKee SA, Rajpar MH, Cole TR, Evans DG, et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum Mol Genet. 1999;8(4):607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 74.Fang L, Igarashi M, Leung J, Sugrue MM, Lee SW, Aaronson SA. p21Waf1/Cip1/Sdi1 induces permanent growth arrest with markers of replicative senescence in human tumor cells lacking functional p53. Oncogene. 1999;18(18):2789–2797. doi: 10.1038/sj.onc.1202615. [DOI] [PubMed] [Google Scholar]
- 75.Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya R, Li LC. Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene activation in human bladder cancer cells. Mol Cancer Ther. 2008;7(3):698–703. doi: 10.1158/1535-7163.MCT-07-2312. [DOI] [PubMed] [Google Scholar]
- 76.Yang K, Zheng XY, Qin J, Wang YB, Bai Y, et al. Up-regulation of p21(WAF1/Cip1) by saRNA induces G1-phase arrest and apoptosis in T24 human bladder cancer cells. Cancer Lett. 2008;265(2):206–214. doi: 10.1016/j.canlet.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 77.Wei J, Zhao J, Long M, Han Y, Wang X, et al. p21WAF1/CIP1 gene transcriptional activation exerts cell growth inhibition and enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC Cancer. 2010;10:632. doi: 10.1186/1471-2407-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mao Q, Li Y, Zheng X, Yang K, Shen H, et al. Up-regulation of E-cadherin by small activating RNA inhibits cell invasion and migration in 5637 human bladder cancer cells. Biochem Biophys Res Commun. 2008;375(4):566–570. doi: 10.1016/j.bbrc.2008.08.059. [DOI] [PubMed] [Google Scholar]
- 79.Mao Q, Zheng X, Yang K, Qin J, Bai Y, et al. Suppression of Migration and Invasion of PC3 Prostate Cancer Cell Line via Activating E-Cadherin Expression by Small Activating RNA. Cancer Invest. 2010 doi: 10.3109/07357900802620844. early online. [DOI] [PubMed] [Google Scholar]
- 80.Junxia W, Ping G, Yuan H, Lijun Z, Jihong R, et al. Double strand RNA-guided endogeneous E-cadherin up-regulation induces the apoptosis and inhibits proliferation of breast carcinoma cells in vitro and in vivo. Cancer Sci. 2010;101(8):1790–1796. doi: 10.1111/j.1349-7006.2010.01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watts JK, Yu D, Charisse K, Montaillier C, Potier P, et al. Effect of chemical modifications on modulation of gene expression by duplex antigene RNAs that are complementary to non-coding transcripts at gene promoters. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whitson JM, Noonan EJ, Pookot D, Place RF, Dahiya R. Double stranded-RNA-mediated activation of P21 gene induced apoptosis and cell cycle arrest in renal cell carcinoma. Int J Cancer. 2009;125(2):446–452. doi: 10.1002/ijc.24370. [DOI] [PMC free article] [PubMed] [Google Scholar]