Abstract
Heparan and chondroitin sulfate proteoglycans (HSPGs and CSPGs, respectively) regulate numerous cell surface signaling events, with typically opposite effects on cell function. CSPGs inhibit nerve regeneration through receptor protein tyrosine phosphatase sigma (RPTPs). Here we report that RPTPs acts bimodally in sensory neuron extension, mediating CSPG inhibition and HSPG growth promotion. Crystallographic analyses of a shared HSPG-CSPG binding site reveal a conformational plasticity that can accommodate diverse glycosaminoglycans with comparable affinities. Heparan sulfate and analogs induced RPTPs ectodomain oligomerization in solution, which was inhibited by chondroitin sulfate. RPTPs and HSPGs colocalize in puncta on sensory neurons in culture, whereas CSPGs occupy the extracellular matrix. These results lead to a model where proteoglycans can exert opposing effects on neuronal extension by competing to control the oligomerization of a common receptor.
Type IIa receptor protein tyrosine phosphatases (RPTPs) are cell surface receptors important for nervous system development, function, and repair (1–3). Vertebrate family members [RPTPs, leukocyte common antigen-related (LAR) protein, and RPTPδ] and invertebrate orthologs [e.g., Drosophila LAR (DLAR)] localize to axonal growth cones, regulating neuronal growth and guidance and participating in excitatory synapse formation and maintenance (1, 4–8). RPTPσ–/– mice exhibit neurological and neuroendocrine defects (9, 10), as well as increased nerve regeneration (11–15); RPTPδ-deficient mice show impaired learning and memory (16). RPTPσ and δ double-mutant mice have a developmental loss of motor neurons leading to paralysis (17).
Type IIa RPTP extracellular regions interact with heparan sulfate proteoglycans (HSPGs) and chondroitin sulfate proteoglycans (CSPGs) (5, 7, 12, 18). These proteoglycans modulate neuronal growth, guidance, and connectivity, typically with CSPGs inhibiting and HSPGs promoting axon extension (19–23). Up-regulation of CSPGs in glial scar tissue after neural injury is an important factor limiting central nervous system axon sprouting and regeneration (2, 21, 24, 25). In adult mouse dorsal root ganglion (DRG) sensory axons, this CSPG inhibitory effect is mediated, at least in part, by RPTPσ (12). In contrast, in developing chick retinal ganglion cell axons, RPTPs promotes growth in response to basal lamina, which contains HSPG ligands (18, 26). We sought to resolve the apparent conundrum of CSPG-RPTPσ and HSPG-RPTPσ interactions eliciting opposing effects on neuronal outgrowth with analyses at the cellular and molecular level.
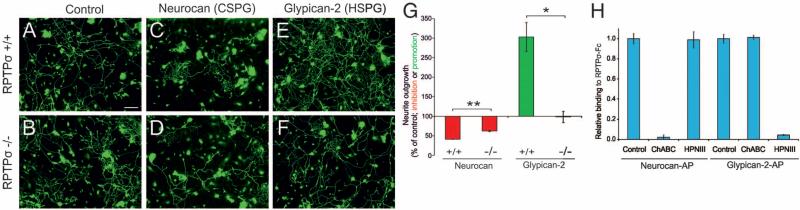
Previous results led to alternative predictions for the potential effects, if any, of HSPGs acting via RPTPσ in DRG neurons: these might inhibit outgrowth, like CSPGs, or promote outgrowth, as observed in retinal ganglion cells. Neurocan, a CSPG, reduced outgrowth of wild-type (WT) DRG neurons by ~60%, as described previously (12), and this inhibitory effect was decreased in RPTPσ–/– neurons (P < 0.001) (Fig. 1, A to D and G). In contrast, glypican-2, an HSPG, strongly promoted outgrowth of WT neurons. This promoting effect was reduced to undetectable levels in RPTPσ–/– neurons, showing a requirement for RPTPσ (P < 0.01) (Fig. 1, E to G). Heparitinase III treatment of the glypican-2 significantly reduced its growth-promoting ability (P < 0.005) (fig. S1), indicating that its glycosaminoglycan (GAG) chains are involved. Chondroitinase ABC and heparitinase III reduced RPTPσ binding by neurocan and glypican-2 respectively, confirming the specific presence of heparan sulfate (HS) and chondroitin sulfate (CS) on these proteoglycans and the role of GAG chains in mediating interactions with RPTPσ (Fig. 1H).
Fig. 1.
RPTPσ-GAG interactions modulate contrasting growth responses. (A, C, E) WT and (B, D, F) RPTPσ–/– P8 mouse DRG neurons grown on poly-d-lysine/laminin alone (control) or supplemented with either neurocan or glypican-2, imaged by GAP-43 immunolabeling. Scale bar in (A), 100 μm. (G) Total neurite outgrowth quantitation reveals neurocan inhibition and glypican-2 promotion. Both effects are reduced in RPTPσ–/– neurons. (H) Pretreatment with chondroitinase ABC (ChABC) or heparitinase III (HPNIII) reduced binding of neurocan and glypican-2, respectively, to RPTPσ in solid-phase binding assays. n = 3 to 4 mice for each genotype. **P < 0.001; *P < 0.01; Student's t test. Error bars indicate SEM. AP, alkaline phosphatase tag; Fc, immunoglobulin fragment crystallizable tag.
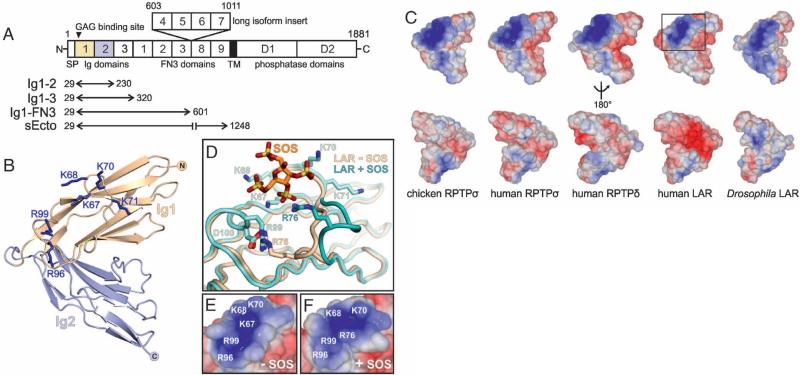
The observed dichotomy in CSPG/HSPG function, apparently mediated through a common receptor, RPTPσ, is intriguing given that mutagenesis studies indicate a common binding site for the two proteoglycan classes (12, 18). To investigate the structural basis of proteoglycan recognition in RPTPσ and type IIa RPTPs in general, we undertook crystallographic studies. The type IIa RPTP ectodomain is predicted to consist of three I-set immunoglobulin (Ig)–like domains followed by either five or nine fibronectin (FN) type III repeats (Fig. 2A). We produced a series of RPTPσ, RPTPδ, and LAR deletion constructs for structural and functional assays (27), all including the N-terminal Ig domain harboring the putative GAG binding site (Fig. 2A) (12, 18). We determined crystal structures of the two N-terminal Ig domains (Ig1-2), which formed the minimal stable unit, for examples across family members and species (chicken and human RPTPσ, human RPTPδ and LAR, Drosophila DLAR) [see (28), fig. S2 and tables S1 and S2]. A V-shape arrangement of Ig1 and Ig2 is stabilized by conserved interactions, irrespective of the crystallization conditions and packing (Fig. 2B and figs. S3 to S5).
Fig. 2.
Structure of the proteoglycan binding site. (A) Type IIa RPTP domain organization. N, amino-terminus (extracellular); SP, secretion signal peptide; TM, transmembrane; C, C terminus (intracellular); Ig, immunoglobulin-like; FN3, fibronectin type-III. The ectodomain may be remodeled by alternative splicing (1). Numbering corresponds to chicken RPTPσ. (B) Ribbon diagram of chicken RPTPσ Ig1-2. Blue sticks represent residues important for proteoglycan binding (12, 18). (C) Solvent-accessible surfaces of Ig1-2 crystal structures, colored by electrostatic potential (±5kT/e, where k is the Boltzmann constant, T is temperature, and e is the charge on an electron). Red, acidic; blue, basic. (D) SOS induces movement of the proteoglycan binding Lys-loop in human LAR (green) relative to the apo-protein (beige). Surface representations of the boxed region in (C) for apo-LAR (E) and SOS-bound LAR (F) highlight the malleability of the proteoglycan binding surface.
RPTPσ residues previously shown to mediate GAG binding (K67, K68, K70, K71, R96, and R99) (29) lie on loops between Ig1 β strands C-D and E-F, forming an extended positively charged surface (Fig. 2, B and C). Our crystal structures, supplemented by sequence comparisons, show that this site is highly conserved across family members and species (Fig. 2C and fig. S2), suggesting a common GAG binding mode. A 2.05 Å resolution crystal structure of human LAR Ig1-2 in complex with sucrose octasulfate (SOS), a synthetic heparin-mimic, confirmed the GAG binding site location and revealed a conformational plasticity of the C-D (“Lys”) loop (residues K67-F77) (Fig. 2D, figs. S6 to S8, and table S2). SOS binding triggered an outwards movement of residues V72 to F77, following rupture of the R76-D100 salt bridge. K67, K68, and R76 form electrostatic interactions with SOS, whereas R99 interacts with D100, resulting in a modified topology of the GAG binding surface but maintaining the overall positive charge (Fig. 2, E and F). Thus, the combination of basic side chains deployed by the type IIa RPTP GAG-binding site may vary to match the sulfate chemistry of a particular disaccharide unit, conferring the ability to accommodate chemically diverse GAGs.
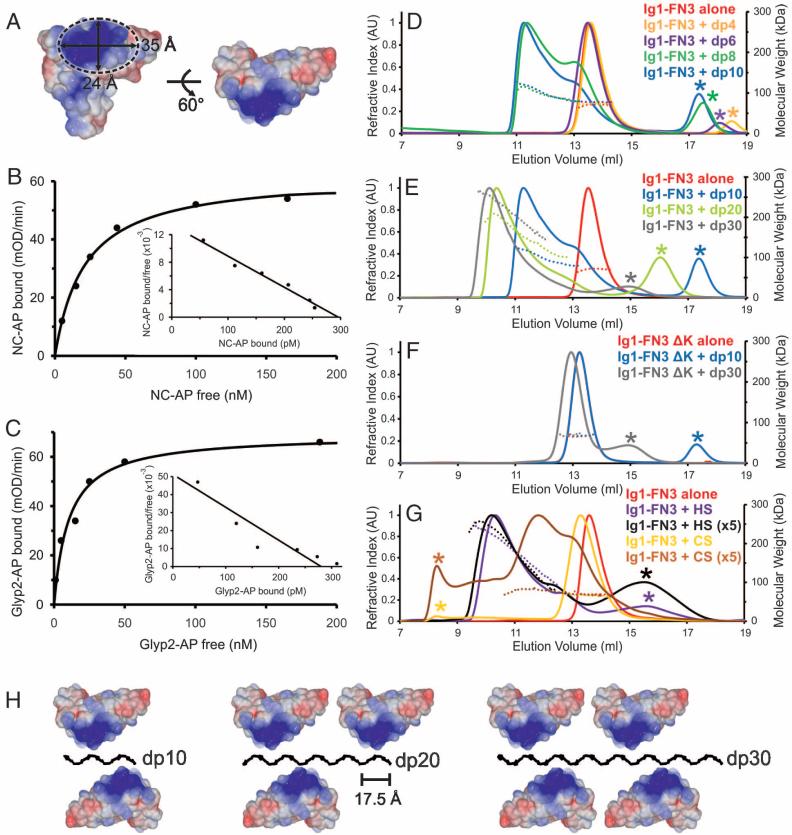
Interaction affinities of RPTPσ sEcto with neurocan and glypican-2, in solid phase binding assays, were in the same range [10 to 20 nM dissociation constant (Kd)] (Fig. 3, F and G), similar to those determined in previous studies of CSPGs or HSPGs binding to type IIa RPTPs (7, 12), although weaker than measured previously for heparin conjugated to bovine serum albumin (0.3 nM) (18). The glycan binding surface on Ig1 forms an elliptical area of ~35 by 24 Å (Fig. 3A). Comparisons with a compact heparin structure, a helix with a four-saccharide pitch of 17.5 Å (30), suggested that GAG chains could assemble RPTPσ oligomers. We therefore incubated a series of size-defined heparin fragments with a RPTPσ construct containing the six N-terminal domains (Ig1-FN3) (Fig. 2A). Heparin fragments containing four [degree of polymerization 4 (dp4)] or six (dp6) saccharide residues did not alter the Ig1-FN3 oligomeric state, as assessed by size-exclusion chromatography coupled with multi-angle light scattering (MALS) (Fig. 3D and table S3). However, fragments containing eight (dp8) or ten (dp10) saccharides induced a shift toward a dimeric Ig1-FN3 species (Fig. 3D and table S3). This trend continued as heparin fragments of increasing length were tested; Ig1-FN3 mixed with dp20 and dp30 formed tri- and tetrameric clusters, respectively (Fig. 3E and table S3). A quadruple K67, K68, K70, K71 mutation to alanine, previously shown to impair binding to both CSPGs and HSPGs (12, 18), abolished the heparin-induced clustering effects (Fig. 3F and table S3). The clustering behavior was reproduced in MALS measurements for an RPTPσ domain deletion series (Fig. 2A and fig. S9) and validated for Ig1-2 by analytical ultracentrifugation and native mass spectrometry (fig. S10). These data provide compelling evidence that the Ig1 GAG-binding site is necessary and sufficient for receptor clustering dependent on heparin fragment length (figs. S9 and S10 and table S3). We demonstrated similar heparin-induced oligomerization characteristics for other type IIa RPTPσ, albeit with some variation (figs. S11 and S12 and table S3).
Fig. 3.
GAG-induced oligomerization of RPTPσ. (A) Dimensions of the proteoglycan binding surface from chicken RPTPσ Ig1-2 crystal structure. (B) and (C) Neurocan-alkaline phosphatase (NC-AP) and glypican-2-AP (Glyp2-AP) bind to immobilized mouse RPTPσ sEcto-Fc with comparable affinities. Kd values were ~21 nM for RPTPσ-neurocan and ~10 nM for RPTPσ–glypican-2 interactions, assuming one-to-one binding. mOD, optical density units × 10–3. (D to G) Size-exclusion chromatography coupled to MALS was used to investigate the oligomerization state of human RPTPσ Ig1-FN3 in solution with an excess of varying-length GAGs. Protein was incubated alone (red), with dp4 (orange), dp6 (purple), dp8 (green), dp10 (blue), dp20 (light green), or dp30 (gray). Heparin dp8 was the minimum length of heparin oligosaccharide to promote oligomerization (D); longer heparin oligosaccharides resulted in larger RPTP oligomers (E). The oligomerization state of a quadruple K67A/ K68A/K70A/K71A mutant of RPTPσ (Ig1-FN3 ΔK) was insensitive to heparin addition (F). HS but not CS induced oligomerization of RPTPσ Ig1-FN3; “×5” indicates increased GAG amounts (G) (28). Refractive index traces (scaled within each panel) and measured molecular weights are represented by bold and dashed lines, respectively. Refractive index peaks indicated by an asterisk correspond to excess glycan ligand. AU, arbitrary units; kDa, kilodaltons. (H) Model for RPTPσ Ig1-2 clustering along the highly sulfated domains of HS based on an unperturbed helical heparin structure (Protein Data Bank accession code 1HPN).
We then used MALS to compare the ability of CS and HS GAGs to induce oligomerization. HS (30 to 150 saccharide units) induced tetrameric clustering of RPTPσ Ig1-FN3, analogous to dp30 heparin fragments (Fig. 3G and table S3). In marked contrast to heparin or HS, comparable CS (30 to 150 saccharide units) quantities did not induce clustering of any type IIa RPTP construct (Fig. 3E and fig. S12). Using fivefold higher CS concentrations, we were able to detect evidence of binding to RPTPσ Ig1-FN3, but the molecular mass did not shift to the levels seen for stable GAG-induced oligomers (Fig. 3G and table S3). Because CSPGs and HSPGs had shown comparable binding affinities to RPTPσ in our solid state assay, we tested whether CS could compete with HS in the MALS assay. Excess CS inhibited both HS- and heparin dp10–induced clustering (fig. S12 and table S3). Thus, differences in GAG chemical structure must be responsible for the contrasting effects of HS and CS on RPTPσ oligomerization (Fig. 3H).
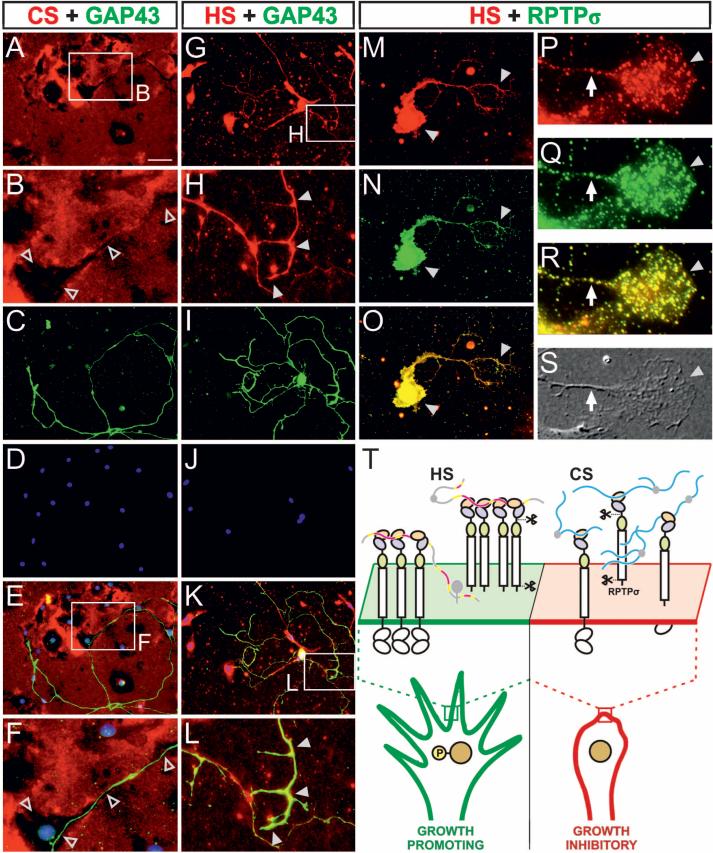
The competing effects of CS and HS on RPTPσ oligomerization suggest that axon outgrowth may be determined by relative amounts, rather than independent effects, of HSPG and CSPG acting on RPTPσ. To investigate this model, we assessed DRG neuron cultures for expression of endogenous HS and CS. Immunofluorescence showed that both CS and HS are endogenously produced by these cultures. CS labeling was seen primarily over the extracellular matrix (ECM), where it was highest adjacent to non-neuronal cells, suggesting production of CSPG by these cells, whereas cell bodies and GAP43-labeled neurites showed little or no evidence of CS labeling and were typically aligned with dark areas in the immunofluorescence (Fig. 4, A to F, and fig. S14). HS labeling, in contrast, was highest over cell bodies and neurites, where RPTPσ labeling was also seen (Fig. 4, G to O, and fig. S14). At higher magnification, RPTPσ labeling revealed a punctate distribution over the growth cone, as described previously (15), and was similar (although not identical) to the HS pattern (Fig. 4, P to S). Thus, these results support a model in which endogeneous HSPGs in these cultures act predominantly in cis on the cell surface, whereas CSPGs are presented in trans by the ECM (Fig. 4T). Exogenous HSPG or CSPG addition (Fig. 1) would shift the HSPG:CSPG ratio. Consistent with this interpretation and with the ability of CS to compete with HS-mediated RPTPσ oligomerization (fig. S12), the addition of CSPG to DRG cultures could block the outgrowth-promoting effect of exogenous HSPG (P < 0.001) (fig. S15).
Fig. 4.
Immunolocalization of endogenous HS, CS, and RPTPσ in DRG neuron cultures. (A to F) Immunolocalization of CS (red) with GAP43 neuronal marker (green) and 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain (blue). Colors are merged in (E) and (F). Boxed areas in (A) and (E) are enlarged in (B) and (F), respectively. Open arrowheads denote dark areas of low CS labeling that overlap with cell bodies and axons. GAP43 labeled axon in (F) grows over a non-neuronal cell at lower left. (G to L) HS (red) with GAP43 (green) and DAPI (blue). Colors are merged in (K) and (L). Boxed areas in (G) and (K) are enlarged in (H) and (L), respectively. Solid arrowheads indicate HS labeling over GAP43-labeled neuronal processes. (M to S) Immunolocalization of HS (red) with RPTPσ (green). (M to O) Labeling of a neuron, including cell body and neurites (arrowheads). (P to S) A growth cone (arrowhead) with axon shaft (arrow) at higher magnification. (S) Differential interference contrast image. Both HS and RPTPσ show punctate labeling, in similar although not identical patterns. Scale bar, 60 μm [(A), (C) to (E), (G), (I) to (K), and (M) to (O)] and 6 μm [(P) to (S)]. (T) Model for type IIa RPTP-proteoglycan interactions and their distinct functional consequences. Islands of high/intermediate sulfation on HS chains (shown in pink/yellow) stabilize receptor oligomers, causing an uneven distribution of tyrosine phosphatase activity and formation of microdomains with high phosphotyrosine levels and supporting neuronal extension. Conversely, secreted CS (blue chains), present in glial scar tissue, is unable to induce tight RPTPσ oligomerization, competing with HS and inhibiting axon growth. Regulatory mechanisms might include shedding (1); crystal structure of human RPTPσ Ig1-3 reveals an exposed furinlike protease cleavage site in the Ig2-3 linker (scissors) (fig. S13).
Mechanistic parallels can be drawn for proteoglycan-specific regulation of other cell surface receptor systems. HS is known to play an essential role in fibroblast growth factor (FGF) signaling (31), and the number of FGF–FGF receptor protomers in a supramolecular assembly directly correlates with the size of the GAG chain (32). Opposing HSPG and CSPG effects have been reported in semaphorin-mediated axon guidance (22). HSPGs and CSPGs differ in the chemical composition of their GAG chains. The distribution of sulfate groups (typically one to two per disaccharide) along CS chains is relatively uniform, whereas HS has a distinct modular composition, with high-sulfation regions (three groups per disaccharide) flanked by intermediately modified transition zones and variably spaced by largely unmodified sections almost devoid of sulfation (33). Our observations suggest a model in which islands of high sulfation present in HS, but not CS, may promote close packing of RPTPσ molecules.
RPTPσ clustering would translate into an uneven distribution of phosphatase activity on the cell surface, consistent with localization-based models for receptor action (34). Small regions depleted in phosphatase activity could enhance the extent and duration of a phosphorylated state for proteins stimulating neuronal extension (1, 35, 36). Our solution and cellular data are consistent with a model in which increasing the CSPG:HSPG ratio shifts the balance away from growth-promoting RPTPσ clusters, stalling neuronal growth cones (Fig. 4T). This model predicts that molecules able to promote RPTPσ oligomerization may prove beneficial in strategies to facilitate plasticity and regeneration after nervous system injury. More generally, proteoglycan-binding is a common property of many cell surface signaling systems involved in normal biology and disease. Our results point to a mechanism by which differences in the structure of GAG chains can serve as a stop/go molecular switch for cell motility and may provide a general paradigm in the biology of cell surface signaling.
Supplementary Material
Acknowledgments
Coordinates and structure factors are deposited in the Protein Data Bank (see table S2). We thank K. Harlos, T. Walter, and staff of the European Synchrotron Radiation Facility and Diamond Light Source for assistance; the Harvard Medical School Nikon Imaging Center for resources and advice; C. Serra-Pagès and A. W. Stoker for RPTPσ cDNAs; M. Tremblay and N. Uetani (McGill Univ.) for the RPTPσ–/– mouse; R. J. Gilbert and R. T. Aplin for analytical ultracentrifugation and mass spectrometry; and J. Silver, B. Lang, and A. W. Stoker for advice and discussions. This work was funded by the Wellcome Trust, the European Research Community Fund (MRTN-CT-2006-035830), the NIH (grants EY11559 and HD29417), Cancer Research UK, and the UK Medical Research Council. C.H.C. was the recipient of a Wellcome Trust D.Phil. studentship, C.S. is a Wellcome Trust Research Career Development Fellow, E.Y.J. is a Cancer Research UK Principal Research Fellow, and A.R.A. is a UK Medical Research Council Career Development Award Fellow. Current patent applications related to this work have been filed by Harvard Univ. A provisional patent filed by the University College London, related to the discovery of HSPGs as RPTPσ ligands, expired in 2001. J.T.G. is the founder, director, and majority shareholder of Iduron, Paterson Institute for Cancer Research, Univ. of Manchester (Manchester, UK).
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/science.1200840/DC1
Materials and Methods
Figs. S1 to S15
Tables S1 to S3
References
References and Notes
- 1.Johnson KG, Van Vactor D. Physiol. Rev. 2003;83:1. doi: 10.1152/physrev.00016.2002. [DOI] [PubMed] [Google Scholar]
- 2.Duan Y, Giger RJ. Sci. Signal. 2010;3:pe6. doi: 10.1126/scisignal.3110pe6. [DOI] [PubMed] [Google Scholar]
- 3.Tonks NK. Nat. Rev. Mol. Cell Biol. 2006;7:833. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 4.Rashid-Doubell F, McKinnell I, Aricescu AR, Sajnani G, Stoker AW. J. Neurosci. 2002;22:5024. doi: 10.1523/JNEUROSCI.22-12-05024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox AN, Zinn K. Curr. Biol. 2005;15:1701. doi: 10.1016/j.cub.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 6.Dunah AW, et al. Nat. Neurosci. 2005;8:458. doi: 10.1038/nn1416. [DOI] [PubMed] [Google Scholar]
- 7.Johnson KG, et al. Neuron. 2006;49:517. doi: 10.1016/j.neuron.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 8.Woo J, et al. Nat. Neurosci. 2009;12:428. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 9.Wallace MJ, et al. Nat. Genet. 1999;21:334. doi: 10.1038/6866. [DOI] [PubMed] [Google Scholar]
- 10.Elchebly M, et al. Nat. Genet. 1999;21:330. doi: 10.1038/6859. [DOI] [PubMed] [Google Scholar]
- 11.McLean J, Batt J, Doering LC, Rotin D, Bain JR. J. Neurosci. 2002;22:5481. doi: 10.1523/JNEUROSCI.22-13-05481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y, et al. Science. 2009;326:592. doi: 10.1126/science.1178310. 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fry EJ, Chagnon MJ, López-Vales R, Tremblay ML, David S. Glia. 2010;58:423. doi: 10.1002/glia.20934. [DOI] [PubMed] [Google Scholar]
- 14.Sapieha PS, et al. Mol. Cell. Neurosci. 2005;28:625. doi: 10.1016/j.mcn.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Thompson KM, et al. Mol. Cell. Neurosci. 2003;23:681. doi: 10.1016/s1044-7431(03)00120-9. [DOI] [PubMed] [Google Scholar]
- 16.Uetani N, et al. EMBO J. 2000;19:2775. doi: 10.1093/emboj/19.12.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uetani N, Chagnon MJ, Kennedy TE, Iwakura Y, Tremblay ML. J. Neurosci. 2006;26:5872. doi: 10.1523/JNEUROSCI.0386-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aricescu AR, McKinnell IW, Halfter W, Stoker AW. Mol. Cell. Biol. 2002;22:1881. doi: 10.1128/MCB.22.6.1881-1892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandtlow CE, Zimmermann DR. Physiol. Rev. 2000;80:1267. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 20.Van Vactor D, Wall DP, Johnson KG. Curr. Opin. Neurobiol. 2006;16:40. doi: 10.1016/j.conb.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Silver J, Miller JH. Nat. Rev. Neurosci. 2004;5:146. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 22.Kantor DB, et al. Neuron. 2004;44:961. doi: 10.1016/j.neuron.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto Y, Irie F, Inatani M, Tessier-Lavigne M, Yamaguchi Y. J. Neurosci. 2007;27:4342. doi: 10.1523/JNEUROSCI.0700-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galtrey CM, Fawcett JW. Brain Res. Brain Res. Rev. 2007;54:1. doi: 10.1016/j.brainresrev.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Case LC, Tessier-Lavigne M. Curr. Biol. 2005;15:R749. doi: 10.1016/j.cub.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Ledig MM, Haj F, Bixby JL, Stoker AW, Mueller BK. J. Cell Biol. 1999;147:375. doi: 10.1083/jcb.147.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aricescu AR, Lu W, Jones EY. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 28.Materials and methods are available as supporting material on Science Online.
- 29.Single-letter abbreviations for the amino acid residues are as follows: D, Asp; F, Phe; K, Lys; R, Arg; and V, Val.
- 30.Mulloy B, Forster MJ, Jones C, Davies DB. Biochem. J. 1993;293:849. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spivak-Kroizman T, et al. Cell. 1994;79:1015. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 32.Harmer NJ, et al. Biochem. J. 2006;393:741. doi: 10.1042/BJ20050985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy KJ, et al. J. Biol. Chem. 2004;279:27239. doi: 10.1074/jbc.M401774200. [DOI] [PubMed] [Google Scholar]
- 34.Groves JT, Kuriyan J. Nat. Struct. Mol. Biol. 2010;17:659. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu DY, Goldberg DJ. J. Cell Biol. 1993;123:653. doi: 10.1083/jcb.123.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robles E, Woo S, Gomez TM. J. Neurosci. 2005;25:7669. doi: 10.1523/JNEUROSCI.2680-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.