Abstract
The pathogenesis of schizophrenia is thought to involve aberrant immune and inflammatory responses. Nuclear factor kappa B (NF-κB) has important roles in the immune and inflammatory responses. The v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA) gene encodes the major component of the NF-κB complex. We genotyped four single-nucleotide polymorphisms (SNPs) in the RELA gene and performed a gene-based association analysis using 1224 patients with schizophrenia and 1663 controls. We found significant associations of three SNPs (rs11820062: p=0.00011, rs2306365: p=0.0031, and rs7119750: p=0.0080) with schizophrenia and stronger evidence for association in a multi-marker sliding window haplotype analysis (the lowest p=0.00006). The association between this gene and schizophrenia was evident in male subjects but not in female subjects, when separately analyzed by gender. In silico genotype-gene expression analysis using web database and the WGAViewer software revealed that these three schizophrenia-associated SNPs might be related to RELA mRNA expression in immortalized B-lymphocytes. In silico analysis also suggested the putative promoter SNP, rs11820062, might disrupt the consensus transcription factor binding sequence of the androgen receptor. The impact of four RELA polymorphisms on pre-pulse inhibition (PPI) was investigated in 53 patients with schizophrenia. We provided evidence that at risk genotypes of three SNPs were associated with deficits in PPI; however, there was no effect of the one non-risk SNP on PPI. These findings suggest that variants of the RELA gene are associated with risk for schizophrenia and PPI deficits in a Japanese population.
Keywords: schizophrenia, v-rel reticuloendotheliosis viral oncogene homolog A (avian) (RELA), NF-κB, pre-pulse inhibition (PPI), single-nucleotide polymorphism, gene expression
INTRODUCTION
Schizophrenia is a common and complex psychiatric disease. The lifetime morbidity rate is 0.5–1.0% across distinct populations. Family, twin, and adoption studies of schizophrenia have indicated that there are strong genetic factors with an estimated heritability of 80% (Cardno and Gottesman, 2000; Tsuang, 2000). Although genes implicated in the pathogenesis of schizophrenia have been found through intense research efforts, eg association studies of candidate gene approach, genomewide association studies (GWAS), copy number variation (CNV) studies, and pedigree studies (Harrison and Weinberger, 2005; Cichon et al, 2009), the exact genetic factors of this complex disease remain to be explained. Recent polygenic component analysis in GWAS studies demonstrated the less effect between different populations, eg Europeans and African–Americans, Europeans, and Japanese, compared with the effects between two European populations (Purcell et al, 2009; Ikeda et al, 2010). As it may be due to aggregate differences in allele frequencies and patterns of linkage disequilibrium and/or population specific risk for schizophrenia, association studies using multiple populations are expected.
Alterations in cytokine expression in schizophrenia have been extensively studied. The levels of cytokines, such as interleukin-1β (IL-1β), IL-1 receptor antagonist (IL-1RA), IL-6, and tumor necrosis factor-α (TNF-α), are increased in the plasma and cerebrospinal fluid of patients with schizophrenia (Naudin et al, 1997; Potvin et al, 2008). Nuclear factor-kappa B (NF-κB), a prototypical transcription factor, regulates the expression of cytokines. Conversely, NF-κB can be activated by pro-inflammatory cytokines, including IL-1β and TNF-α, and in response to various cell stressors. Many genes have been shown to be responsive to NF-κB (Baeuerle, 1991), including genes involved in survival/apoptosis, immune and inflammatory responses, and cell differentiation. In addition, the NF-κB-responsive genes IL2, IL6, TNF-α, major histocompatibility complex (MHC), Bcl-2 family members, Calbindin, and ICAM-1 have been reported to be associated with schizophrenia (Jarskog et al, 2000, 2004; Schwarz et al, 2000; Benes et al, 2001, 2003; Potvin et al, 2008; Woo et al, 2008; Purcell et al, 2009; Shi et al, 2009; Song et al, 2009; Stefansson et al, 2009).
NF-κB is present in synaptic terminals and serves as a regulator of neuronal plasticity, which is activated by the activity of neuronal circuits (Mattson et al, 2000). The NF-κB complex is inhibited by the IκB complex, which inactivates NF-κB by sequestering it in the cytoplasm (Huxford and Ghosh, 2009). After the phosphorylation of serine residues on the IκB proteins, IκB dissociates from and activates the NF-κB complex. The activated NF-κB complex translocates into the nucleus and binds to regulatory elements in target genes. Constitutively, activated NF-κB is detected mostly in glutamatergic neurons, whereas NF-κB in glia has a lower basal activity and is heavily inducible (Kaltschmidt and Kaltschmidt, 2009). Knockout of a subunit of NF-κB or inhibition of NF-κB by super-repressor IκB in neuron of mice resulted in defects in learning and memory and the loss of neuroprotection (Kaltschmidt and Kaltschmidt, 2009). It has been shown that activation of NF-κB prevents neuronal apoptosis in various cell types. H2O2 increased NF-κB activation and dopamine D2 receptor expression (Larouche et al, 2008), suggesting that NF-κB may participate in the psychopathology of schizophrenia through its effect on the neurotransmitter system. The association between cytokine expression and NF-κB activation has been reported in schizophrenia (Song et al, 2009). These findings support the hypothesis that alterations in cytokines and NF-κB, which cause abnormal inflammatory responses, might contribute to the pathogenesis of schizophrenia.
Although three components of NF-κB, NFKB1, NFKB2, and NFKB3, were not in major locus in schizophrenia (OMIM181500: http://www.ncbi.nlm.nih.gov/omim), NFKB3 located on chromosome 11q13 showed a suggestive linkage to schizophrenia in a family-based linkage disequilibrium analysis in a Japanese population (Yamada et al, 2004). The v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA) gene (OMIM164014; alternative names include nuclear factor kappa-B, subunit 3 (NFKB3), transcription factor NFKB3, NFKB, p65 subunit, and nuclear factor of kappa light chain gene enhancer in B cells 3) encodes the major subunit of the NF-κB protein complex, are abundantly expressed in neurons and glia (Kaltschmidt and Kaltschmidt, 2009). Mice lacking RELA showed a learning deficit in the spatial version of the radial arm maze (Meffert et al, 2003), indicating the critical involvement of the RELA gene in memory function, which may be related to pathophysiology of memory dysfunction in schizophrenia. Microarray Expression Data in UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) showed moderate mRNA expression of the RELA gene and the components of IκB (NFKBIA and NFKBIB) in human brain and prominent expression in prefrontal cortex. Altered expression of these genes in postmortem brain of patients with schizophrenia was not found in the pooled gene expression data in the Stanley Genomic Medical Research Institute Online Genomics Database (https://www.stanleygenomics.org). In this study, we first investigated the association between the RELA gene and schizophrenia in a Japanese population, and then performed in silico genotype–gene expression analysis.
Impaired sensorimotor gating is considered to be a common psychophysiological feature of schizophrenia that could theoretically lead to a variety of severe defects in perception, attention, and thinking (Braff and Geyer, 1990). Pre-pulse inhibition (PPI) of the acoustic startle reflex (ASR) is the most common psychophysiological index of sensorimotor gating. PPI is emerging as an important intermediate phenotype for schizophrenia (Braff and Light, 2005), because it has high heritability (Anokhin et al, 2003) and PPI deficits have been found in high-risk subjects (Cadenhead et al, 2000). It has been hypothesized that the maternal immune response to infection may influence fetal brain development and lead to schizophrenia (Brown and Derkits, 2010). Prenatal immune challenge by bacterial endotoxin lipopolysaccharide (LPS) or polyriboinosinic–polyribocytidylic acid (poly I : C) resulted in deficits in PPI (Cardon et al, 2010; Romero et al, 2010). Thus, it has been hypothesized that there are genetic variants that are related to PPI deficits in patients with schizophrenia. Genetic variations in the serotonin-2A receptor, Catechol O-methyltransferase and neuregulin-1 genes have been associated with PPI in schizophrenia (Quednow et al, 2008, 2010; Hong et al, 2008). Thus, we also analyzed the association between the identified SNPs in the RELA gene and PPI in patients with schizophrenia.
MATERIALS AND METHODS
Subjects
The subjects of our genetic association study consisted of 1224 patients with schizophrenia (50. 9% male (623/601), mean age±SD: 46.2±15.0 years) and 1663 healthy controls (46.5% male (773/890), mean age±SD: 46.9±20.7 years). The mean age did not differ significantly between the groups (Z=−1.10, p=0.27), while the sex ratio differed significantly between the groups (χ2=5.51, p=0.019). All the subjects were biologically unrelated Japanese individuals. Patients were recruited at the National Center Hospital of Neurology and Psychiatry, Showa University School of Medicine, Fujita Health University School of Medicine and Osaka University Graduate School of Medicine. Cases were recruited from both outpatients and inpatients at the hospitals. Each patient with schizophrenia had been diagnosed by at least two trained psychiatrists according to the criteria of the Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV), on the basis of unstructured clinical interviews. When the diagnosis of two trained psychiatrists was discordant, they started to discuss the diagnosis. When the disputes about the diagnosis were resolved and the patient was diagnosed as schizophrenia, we included the patients. When the disputes were not resolved by discussion or the patients was not diagnosed as schizophrenia, we excluded the patients. Controls, including hospital and institutional staff, were recruited through local advertisements. Psychiatrically healthy controls were evaluated using unstructured interviews to exclude individuals who had current or past contact with psychiatric services, who had received psychiatric medication or who were not Japanese. We did not assess the controls for the family history of mental disorders, such as schizophrenia, bipolar disorder, or major depressive disorder. The ethnicity was determined by the self-report and it was not confirmed by genetic analyses.
Data for the PPI analysis were available for 53 patients with schizophrenia (56.6% males (30/23), mean age±SD: 39.1±13.2 years). The subjects included in the PPI analysis met additional criteria. All subjects were recruited at Osaka University, and subjects were excluded from this study if they had neurological or medical conditions that could potentially affect the central nervous system, such as atypical headaches, head trauma with loss of consciousness, chronic lung disease, kidney disease, chronic hepatic disease, thyroid disease, active cancer, cerebrovascular disease, epilepsy, seizures, substance-related disorders, or mental retardation. None of the subjects reported history of any known hearing impairment and all participants were able to clearly detect 70 dB noise. Written informed consent was obtained from all subjects after the procedures had been fully explained. This study was performed in accordance with the World Medical Association's Declaration of Helsinki and approved by the institutions' ethical committees (National Center Hospital of Neurology and Psychiatry, Showa University School of Medicine, Fujita Health University School of Medicine and Osaka University Graduate School of Medicine).
SNP Selection and Genotyping
Venous blood was drawn from the subjects, and genomic DNA was extracted from whole blood according to standard procedures. We used the Tagger program (Paul de Bakker, http://www.broad.mit.edu/mpg/tagger) of the Haploview software (ver. 4.1) (Barrett et al, 2005) to select SNPs in the HapMap database (release #22/phase II, April 2007, www.hapmap.org, population: Han Chinese in Beijing (CHB) + Japanese in Tokyo (JPT); minor allele frequencies (MAFs) of more than 0.05) that covered the RELA gene spanning 8.5 Kb (5′-flanking regions including approximately 2 kb from the first exon and approximately 0.5 kb downstream (3′) from the last exon; HapMap database contig number chr11: 65178000.65189000). The criterion for detecting tag SNPs was an r2 threshold greater than 0.80 in ‘pair-wise tagging only' mode. Two tag SNPs were selected (rs2306365 and rs11820062) by the Tagger program among five SNPs in the HapMap database in the RELA gene region. We also searched putative functional SNPs, which are located in exons, exon-intron boundaries and putative promoter regions (5′-flanking region including approximately 2 kb from the first exon and 3′ region approximately 1 kb from first exon). We only find one SNP (rs11820062) fulfilled the criteria, which was already selected by Tagger program. Because these two SNPs were located in the 5′ region, we added two SNPs (rs11568300 and rs7119750) on the 3′ region in this gene for better coverage (2.2 kb per SNP) (Figure 1). The four selected SNPs (rs7119750 (SNP1), rs11568300 (SNP2), rs2306365 (SNP3), and rs11820062 (SNP4)) in the RELA gene were genotyped using the TaqMan 5′-exonuclease allelic discrimination assay (Applied Biosystems, Foster City, CA) as described previously (Hashimoto et al, 2006, 2007; Ohi et al, 2009). The positions of the four SNPs analyzed in the present study are indicated in Figure 1. Primer and probe sequences for detection of the SNPs are as follows: SNP1: forward primer 5′-GCTCAGGCTCAATCCCTCTCTA-3′, reverse primer 5′-CCTACAGGCTGGGTCAGATG-3′, probe 1 VIC-ACTGCCAACACCC, probe 2 FAM-CACTGCTAACACCC; SNP2: forward primer 5′-GGTGTGCGCAGAGAAGCA-3′, reverse primer 5′-CCTTCTCCATGCAGCTGTCT-3′, probe 1 VIC-CACACTGGCCTCCG, probe 2 FAM-CACAGTGGCCTCCG; SNP3: forward primer 5′-GCCAAGAAAACAGGCGATCAG-3′, reverse primer 5′-CCTCCTCTAGGACTTGTGTTTCAC-3′, probe 1 VIC-CCCTCCCAGTGCAGAG, probe 2 FAM-CCTCCCAGCGCAGAG; SNP4: forward primer 5′-CGCATCTGATTCAGTTTCCTCTCT-3′, reverse primer 5′-AATCAGGGCCTGTTGTACTTTCTT-3′, probe 1 VIC-CTCCCTCAATTTTCCT, probe 2 FAM-TCCCTCAGTTTTCCT.
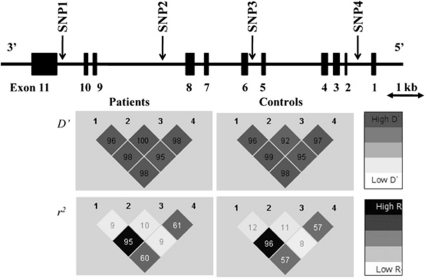
Figure 1.
Genomic structure of RELA, including locations of the four SNPs studied and linkage disequilibrium of these four SNPs in the patient and control groups. The genomic structure of RELA is based on an entry in the Entrez Gene database (National Center for Biotechnology Information). The locations of the SNPs analyzed in this study are indicated with arrows. The distances of the exons–introns and intermarkers are drawn to scale. The linkage disequilibrium (LD) between pairwise SNPs, using D′ and r2 values, are shown at the bottom of the gene structure map separately for 1224 cases and 1663 controls. High levels of LD are represented by increasing gray scale intensity from 0 to 100, as shown by the bars.
In Silico Analysis to Identify SNPs Associated with RELA Expression (eQTLs)
To identify whether the SNPs in the RELA gene might be expression quantitative trait loci (eQTLs), we used GeneVar (http://www.sanger.ac.uk/humgen/genevar/). Genotype and gene lymphoblastoid expression data from multiple HapMap ethnic samples such as Japanese, Han Chinese, Utah residents with Northern and Western European ancestry from the CEPH collection, and Yoruban in Ibadan, Nigeria are deposited in GeneVar. Users could not access the original genotype, gene expression and demographic data in each individual; however, users are able to analyze the association between genotype and gene expression by the WGAViewer software as described by the group that developed GeneVar (Stranger et al, 2007). WGAViewer software is able to perform correlational analysis with number of allele as a continuous variable (allele dose effect: 1/1=0, 1/2=1, 2/2=2). However, any other statistical models such as categorical analysis such as dominant and recessive are not able to perform by the software. We searched for potential transcription factor binding sites in the sequence that included SNP4 with the Patch 1.0 pattern search program and the TRANSFAC 6.0 public site (http://www.gene-regulation.com/cgi-bin/pub/programs/patch/bin/patch.cgi).
Startle Response Measurement
A computerized human startle response monitoring system (Startle Eyeblink Reflex Analysis System Map1155SYS, NIHONSANTEKU, Osaka, Japan) was used to measure PPI. The methods for the startle paradigm, eyeblink acquisition, scoring parameters, and the procedure are described in detail elsewhere (Takahashi et al, 2008, 2010; Moriwaki et al, 2009). The startle paradigm was a total of 44 trials that consisted of three blocks with a continuous 70 dB sound pressure level (SPL) background white noise. Pulse stimuli consisted of broadband white noises at 115 dB SPL with an instantaneous rise/fall time of 40 ms. The pre-pulse stimuli were also broadband white noises with an instantaneous rise/fall time of 20 ms presented at three different intensities (82, 86, and 90 dB SPL). The lead interval (from pre-pulse onset to pulse onset) was 120 ms. In block 1, the startle response for the pulse alone trial (PA trial) was recorded six times. Block 2 consisted of PA trials or trials of pulse with pre-pulse at the three different intensities (PP trials) performed eight times for each condition. Block 3 was the same as block 1 to observe the habituation phenomenon. All trials were presented in a fixed pseudorandom order and were separated by inter-trial intervals of 15–25 s (20 s on average). The session lasted approximately 20 min, including 5 min of acclimation to the background noise. The following startle measures were calculated: (i) for the acoustic startle reflex, the average eyeblink amplitude of startle response to PA trials in block 1; (ii) habituation of the startle response during the session, calculated as the percentage of amplitude reduction between blocks 1 and 3 with the formula ((1−average eyeblink amplitude of the startle response in block 3/average eyeblink amplitude of the startle response in block 1) × 100); and (iii) PPI82, PPI86, PPI90, the pre-pulse inhibitions at intensities of 82 dB, 86 dB, and 90 dB SPL, respectively. The PPI for each intensity level was calculated as the percentage of the amplitude reduction between the PA and PP trials in block 2 with the following formula: (1−average eyeblink amplitude of the startle response in the PP trials in block 2/average eyeblink amplitude of the startle response in the PA trials in block 2) × 100.
Statistical Analyses
Differences in clinical characteristics between patients and controls or between genotype groups were analyzed using the χ2-test for categorical variables and the Mann–Whitney U-test for continuous variables using the PASW Statistics 18.0. software (SPSS Japan, Tokyo, Japan). Statistical analyses for genetic association were performed using the SNPAlyze v5.1.1 Pro software (DYNACOM, Yokohama, Japan). A logistic regression analysis (forced entry method) was conducted to examine the independent association of the sex (1: male, 2: female) and each genotype (0:M/M, 1:M/m, 2:m/m; M:major allele, m:minor allele) on the categorical diagnosis of schizophrenia (0: control, 1: patient). Sex and genotype statuses were included in the model as independent variables, and diagnosis was included as dependent variables. Deviation from the Hardy-Weinberg equilibrium (HWE) was tested separately in cases and controls using χ2-tests for goodness of fit. The allelic and genotypic distributions of RELA polymorphisms between patients and controls were analyzed using χ2-tests. The number of effective independent SNPs assayed was estimated to correct for multiple testing by the spectral decomposition method of Nyholt using the SNPSpD software (Nyholt, 2004). Haplotype frequencies were estimated by the maximum likelihood method using the genotyping data and the expectation-maximization algorithm. Rare haplotypes found in less than 3% of both patients and controls were excluded from the association analysis. We performed 10 000 permutations for the most significant tests to determine the empirical significance. We used a 2–4-window fashion analysis. Pairwise linkage disequilibrium (LD) analyses, expressed by D′ and r2, were applied to detect the intermarker relationships in each group using the Haploview 4.1 software (http://www.broad.mit.edu/mpg/haploview/contact.php). We performed post hoc power calculations using the Power Calculator for Two Stage Association Studies (http://www.sph.umich.edu/csg/abecasis/CaTS (Skol et al, 2006)). Power estimates were based on the allele frequencies in patients, which ranged from 0.43 (SNP1) to 0.45 (SNP4); the odds ratios, which ranged from 1.16 (SNP1) to 1.23 (SNP4), for each associated SNP in this study and an alpha level of 0.05. Power was calculated under a prevalence of 0.01 using a multiplicative model, which assumed varying degrees of marker allele frequency and odds ratios. The effects of the RELA genotypes on the PPI in patients with schizophrenia were analyzed by one-way analysis of covariance (ANCOVA) to adjust for possible confounding factors (gender, current smoking status, and age) using the PASW Statistics 18.0 software (Swerdlow et al, 2008). Standardized effect sizes were calculated using Cohen's d method (http://www.uccs.edu/faculty/lbecker). All p-values reported are two tailed. Statistical significance was defined as p<0.05.
RESULTS
Genetic Association Analysis
The genotype and allele frequencies of four SNPs located in the RELA gene are summarized in Table 1. The genotyping call rates were 96.4% (SNP1), 99.3% (SNP2), 96.7% (SNP3), and 97.5% (SNP4). No deviation from HWE was detected in the cases or controls (p>0.05). Significant differences in the genotype and allele frequencies between patients and controls were observed for SNP1, SNP3, and SNP4 (SNP1: genotype χ2=7.1, p=0.028, allele χ2=7.0, p=0.0080; SNP3: genotype χ2=8.6, p=0.014, allele χ2=8.8, p=0.0031; SNP4: genotype χ2=14.7, p=0.00064, allele χ2=14.9, p=0.00011). These associations remained significant even after the SNPSpD correction for multiple SNP tests, except for the genotypic association for SNP1 (the effective number of independent marker loci: 2.76: corrected p values, SNP1: genotype 0.078, allele 0.0021; SNP3: genotype 0.039, allele 0.0086; SNP4: genotype 0.0018, allele 0.00030). The frequencies of the C allele of SNP1, the G allele of SNP3 and the T allele of SNP4 were higher in patients than in controls. There was no allelic or genotypic association between SNP2 and schizophrenia. We additionally performed association analyses in males and females, separately. The association between the RELA gene and schizophrenia was found in males but not in females (Table 2). As the gender ratio was not matched in this sample, a logistic regression analysis (dependent variable: diagnosis, independent variables: genotype and gender) was performed for four SNPs. This analysis showed that the sex and each genotype were significant predictors for diagnosis of schizophrenia (all p<0.05), except for genotype of SNP2 (p=0.19) (Supplementary Table S1). Diagnosis of schizophrenia was significantly predicted by sex and genotype (SNP1, SNP3, and SNP4), respectively.
Table 1. Genotypic and Allelic Distributions for SNPs in the RELA Gene Between Patients with Schizophrenia and Controls.
|
Marker |
M/m | Location |
SCZ |
CON |
Genotypic p value (df=2) |
SCZ |
CON |
Allelic p value (df=1) | OR (95%CI) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Positiona | M/M | M/m | m/m | M/M | M/m | m/m | MAF | ||||||
| SNP1 | 10728386 | C/T | Intron 10 | 0.32 | 0.48 | 0.19 | 0.29 | 0.48 | 0.23 | 0.028 | 0.43 | 0.47 | 0.008 | 0.87 (0.78–0.96) |
| SNP2 | 10730962 | G/C | Intron 8 | 0.78 | 0.21 | 0.0091 | 0.76 | 0.23 | 0.01 | 0.45 | 0.12 | 0.13 | 0.22 | 0.90 (0.77–1.06) |
| SNP3 | 10733141 | G/A | Intron 5 | 0.32 | 0.48 | 0.20 | 0.28 | 0.48 | 0.24 | 0.014 | 0.44 | 0.48 | 0.0031 | 0.85 (0.77–0.95) |
| SNP4 | 10735731 | C/T | Intron 1 | 0.31 | 0.48 | 0.21 | 0.36 | 0.47 | 0.16 | 0.00064 | 0.45 | 0.40 | 0.00011 | 1.23 (1.11–1.37) |
Abbreviations: CI, confidence interval; CON, controls; M, major allele; m, minor allele; MAF, minor allele frequency; OR, odds ratio; SNP, single-nucleotide polymorphism; SCZ, patients with schizophrenia.
All the alleles are represented according to the minus strand DNA sequence.
P values <0.05 are in bold and underlined.
db SNP build 129.
Table 2. Genotypic and Allelic Distributions for SNPs in the RELA Gene Between Patients in Schizophrenia and Controls.
| (a) In males | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Marker |
M/m | Location |
SCZ (N=623) |
CON (N=773) |
Genotypic p value (df=2) |
SCZ |
CON |
Allelic p value (df=1) | OR (95%CI) | |||||
| SNP ID | Positiona | M/M | M/m | m/m | M/M | M/m | m/m | MAF | ||||||
| SNP1 | 10728386 | C/T | Intron 10 | 0.35 | 0.47 | 0.18 | 0.28 | 0.48 | 0.23 | 0.014 | 0.42 | 0.48 | 0.0030 | 0.79 (0.68–0.92) |
| SNP2 | 10730962 | G/C | Intron 8 | 0.76 | 0.23 | 0.01 | 0.75 | 0.24 | 0.01 | 0.88 | 0.12 | 0.13 | 0.69 | 0.95 (0.76–1.20) |
| SNP3 | 10733141 | G/A | Intron 5 | 0.34 | 0.47 | 0.19 | 0.28 | 0.48 | 0.24 | 0.0080 | 0.42 | 0.48 | 0.0016 | 0.78 (0.67–0.91) |
| SNP4 | 10735731 | C/T | Intron 1 | 0.30 | 0.48 | 0.22 | 0.38 | 0.45 | 0.16 | 0.0022 | 0.46 | 0.39 | 0.00037 | 1.32 (1.13–1.54) |
| (b) In females | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Marker |
M/m | Location |
SCZ (N=601) |
CON (N=890) |
Genotypic p value (df=2) |
SCZ |
CON |
Allelic p value (df=1) | OR (95%CI) | |||||
| SNP ID | Positiona | M/M | M/m | m/m | M/M | M/m | m/m | MAF | ||||||
| SNP1 | 10728386 | C/T | Intron 10 | 0.30 | 0.50 | 0.20 | 0.30 | 0.48 | 0.23 | 0.52 | 0.45 | 0.47 | 0.45 | 0.94 (0.81–1.10) |
| SNP2 | 10730962 | G/C | Intron 8 | 0.80 | 0.19 | 0.01 | 0.77 | 0.22 | 0.01 | 0.35 | 0.11 | 0.12 | 0.16 | 0.84 (0.67–1.07) |
| SNP3 | 10733141 | G/A | Intron 5 | 0.30 | 0.49 | 0.21 | 0.29 | 0.47 | 0.24 | 0.52 | 0.45 | 0.47 | 0.32 | 0.93 (0.80–1.08) |
| SNP4 | 10735731 | C/T | Intron 1 | 0.31 | 0.49 | 0.20 | 0.34 | 0.49 | 0.16 | 0.13 | 0.45 | 0.41 | 0.054 | 1.16 (1.00–1.35) |
CI, confidence interval; CON, controls; M, major allele; m, minor allele; MAF, minor allele frequency; OR, odds ratio; SNP, single-nucleotide polymorphism; SCZ, patients with schizophrenia.
All the alleles are represented according to the minus strand DNA sequence.
P values <0.05 are in bold and underlined.
db SNP build 129.
Two-four SNP sliding window haplotype analysis revealed significant association of this gene with schizophrenia (the lowest global p=0.00040, SNP3-SNP4) (Table 3). The differences in the detailed haplotype frequencies between cases and controls are shown in Table 3 (lowest p=0.00006, G-C haplotype compared with other haplotypes of SNP3-SNP4). The LD relationships between markers are provided in Figure 1. As the strong LD pattern observed in patients with schizophrenia was nearly identical to that among our controls and the JPT, CEU, and YRI HapMap samples, the strong LD patterns in this gene were likely to be common among ethnic groups.
Table 3. Haplotype Analysis of the RELA Gene Between Patients and Controls.
| Haplotypea |
Frequency |
Individual p (χ2) | Global p (χ2) | |
|---|---|---|---|---|
| Patients | Controls | |||
| SNP1-SNP2 | 0.0019 (12.13) | |||
| 1–0 | 0.43 | 0.47 | 0.012 (6.88) | |
| 0–0 | 0.45 | 0.40 | 0.00060 (12.10) | |
| 0–1 | 0.12 | 0.13 | 0.19 (1.59) | |
| SNP2-SNP3 | 0.00080 (13.24) | |||
| 0–1 | 0.44 | 0.48 | 0.0053 (8.03) | |
| 0–0 | 0.45 | 0.40 | 0.00030 (13.24) | |
| 1–0 | 0.12 | 0.13 | 0.23 (1.38) | |
| SNP3-SNP4 | 0.00040 (15.14) | |||
| 1–0 | 0.43 | 0.48 | 0.0031 (9.19) | |
| 0–1 | 0.45 | 0.40 | 0.00006 (15.14) | |
| 0–0 | 0.12 | 0.13 | 0.21 (1.56) | |
| SNP1-SNP2-SNP3 | 0.0022 (12.03) | |||
| 1–0–1 | 0.43 | 0.47 | 0.0071 (7.07) | |
| 0–0–0 | 0.45 | 0.40 | 0.00070 (12.02) | |
| 0–1–0 | 0.12 | 0.13 | 0.22 (1.40) | |
| SNP2-SNP3-SNP4 | 0.00090 (13.66) | |||
| 0–1–0 | 0.44 | 0.48 | 0.0028 (8.68) | |
| 0–0–1 | 0.45 | 0.40 | 0.00040 (13.65) | |
| 1–0–0 | 0.11 | 0.12 | 0.26 (1.20) | |
| SNP1-SNP2-SNP3-SNP4 | 0.0022 (13.11) | |||
| 1–0–1–0 | 0.43 | 0.47 | 0.0058 (8.32) | |
| 0–0–0–1 | 0.45 | 0.40 | 0.00060 (13.11) | |
| 0–1–0–0 | 0.12 | 0.12 | 0.27 (1.17) | |
0, major allele; 1, minor allele. Haplotypes with frequencies of <3% in each group were excluded. Significant p values <0.05 are represented by bold faces and underlines.
In Silico Genotype-Expression Analysis
We analyzed the associations between the SNPs and the expression levels of the RELA gene in lymphoblasts in GeneVar database (Table 4). Unfortunately, data concerning SNP2 was not available in this database. We found that there were significant correlations between the RELA gene expression and all three SNPs associated with schizophrenia only in the Japanese population (SNP1: r2=0.27, p=0.0003; SNP3: r2=0.25, p=0.0005; SNP4: r2=0.15, p=0.0089). Interestingly, the risk alleles in all SNPs were associated with lower gene expression (SNP1: C allele, SNP3: G allele, SNP4: T allele).
Table 4. Association Between the RELA Gene SNPs and mRNA Expression in a Japanese Population.
| SNP | Location | Population | r | Beta | SE | t | p |
|---|---|---|---|---|---|---|---|
| SNP1 | Intron 10 | JPT | 0.52 | 0.24 | 0.06 | 3.91 | 0.0003 |
| CHB | −0.05 | −0.02 | 0.06 | −0.30 | 0.76 | ||
| CEU | −0.07 | −0.03 | 0.05 | −0.52 | 0.61 | ||
| YRI | 0.15 | 0.05 | 0.05 | 1.16 | 0.25 | ||
| SNP3 | Intron 5 | JPT | −0.50 | −0.23 | 0.06 | −3.74 | 0.00050 |
| CHB | 0.05 | 0.02 | 0.06 | 0.30 | 0.76 | ||
| CEU | 0.07 | 0.03 | 0.05 | 0.51 | 0.61 | ||
| YRI | −0.13 | −0.05 | 0.05 | −1.01 | 0.32 | ||
| SNP4 | Intron 1 | JPT | −0.39 | −0.17 | 0.06 | −2.74 | 0.0089 |
| CHB | 0.11 | 0.04 | 0.06 | 0.74 | 0.47 | ||
| CEU | −0.01 | 0.00 | 0.05 | −0.06 | 0.95 | ||
| YRI | −0.16 | −0.05 | 0.04 | −1.20 | 0.24 |
JPT, Japanese in Tokyo, Japan; CHB, Han Chinese in Beijing, China; CEU, Utah residents with Northern and Western European ancestry from the CEPH collection (Parent); YRI, Yoruban in Ibadan, Nigeria (Parent). P values <0.05 are in bold and underlined.
Because SNP4 is located in intron 1, this SNP is possibly related to the regulation of gene transcription. An in silico search for potential transcription factor binding sites in the sequences surrounding SNP4 by the Patch 1.0 program showed that SNP4 could potentially alter androgen receptor (AR) binding. The consensus sequence of the AR binding site is AAAACT (C allele of SNP4 is a non-risk allele). Thus, the mismatch in the AR consensus sequence created by the single nucleotide change of SNP4 (C allele to risk T allele: AAAATT) could lead to lower transcriptional activity of the RELA gene, which is consistent with in silico expression data and genetic association results in male.
The Effect of RELA Genotypes on PPI
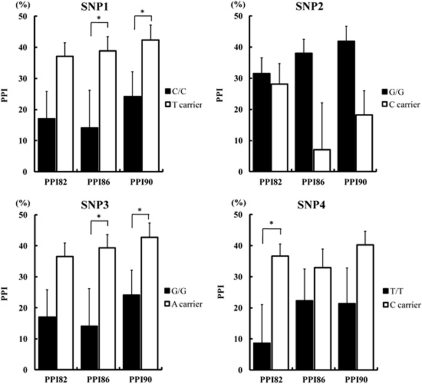
We examined possible associations between the genotype of four SNPs and PPI in patients with schizophrenia, as PPI is one of physiological phenotypes of schizophrenia. There was no difference in the demographic variables, including age, sex, current smoking status, years of education, chlorpromazine equivalents of total anti-psychotics, age at onset, duration of illness, medicated years with anti-psychotics, acoustic startle reflex, and habituation, between genotype groups, except for the chlorpromazine equivalents of total anti-psychotics of SNP1 (p=0.028) and SNP3 (p=0.043) (Supplementary Table S2). One-way ANCOVA showed significant effects of the four genotypes on at least one of the PPI conditions (Table 5). The SNPSpD correction for multiple SNPs tested, revealed the association between three at risk genotypes (SNP1, SNP3, and SNP4) and PPI (SNP1 PPI86: corrected p=0.033, PPI90: corrected p=0.044; SNP3 PPI86: corrected p=0.036, PPI90: corrected p=0.044; SNP4 PPI82: corrected p=0.019) (Table 5, Figure 2). However, the effects of the non-risk SNP (SNP2) on PPI was no longer significant after the SNPSpD correction (PPI86: corrected p=0.083) (Table 5, Figure 2). The patients with the C/C at risk genotype of SNP1, patients with the G/G at risk genotype of SNP3 and patients with the T/T at risk genotype of SNP4 showed significant deficits in PPI.
Table 5. Association Between PPI and RELA Genotypes in Patients with Schizophrenia.
| SNP ID | PPI | P value (F value) | Corrected P | Cohen's d | |
|---|---|---|---|---|---|
| SNP1 | C/C | T carrier | |||
| PPI82 | 17.0±8.8 | 37.1±4.4 | 0.024 (5.39) | 0.066 | −0.63 |
| PPI86 | 14.1±12.1 | 38.9±4.6 | 0.012 (6.75) | 0.033 | −0.61 |
| PPI90 | 24.2±8.0 | 42.4±4.8 | 0.016 (6.25) | 0.044 | −0.58 |
| SNP2 | G/G | C carrier | |||
| PPI82 | 31.5±5.1 | 28.1±6.6 | 0.95 (<0.01) | >0.99 | 0.12 |
| PPI86 | 38.0±4.5 | 7.0±15.1 | 0.030 (4.97) | 0.083 | 0.73 |
| PPI90 | 41.9±4.8 | 18.2±7.7 | 0.072 (3.38) | 0.20 | 0.82 |
| SNP3 | G/G | A carrier | |||
| PPI82 | 17.0±8.8 | 36.5±4.4 | 0.029 (5.06) | 0.080 | −0.62 |
| PPI86 | 14.1±12.1 | 39.3±4.3 | 0.013 (6.73) | 0.036 | −0.64 |
| PPI90 | 24.2±8.0 | 42.7±4.7 | 0.016 (6.30) | 0.044 | −0.61 |
| SNP4 | T/T | C carrier | |||
| PPI82 | 8.6±12.5 | 36.6±4.0 | 0.0068 (8.00) | 0.019 | −0.81 |
| PPI86 | 22.3±10.2 | 32.9±6.0 | 0.35 (0.89) | 0.97 | −0.29 |
| PPI90 | 21.4±11.5 | 40.2±4.5 | 0.041 (4.40) | 0.11 | −0.56 |
SNP1: C/C, patients with C/C genotype (n=17); T carrier, patients with C/T or T/T genotype (n=37). SNP2: C carrier, patients with C/C or C/G genotype (n=12); G/G, patients with G/G genotype (n=42). SNP3: G/G, patients with G/G genotype (n=17); A carrier, patients with G/A or A/A genotype (n=35). SNP4: T/T, patients with the T/T genotype (n=11); C carrier, patients with the T/C or C/C genotypes (n=42). Means±SE are shown. The effects of the RELA genotypes on PPI were analyzed by ANCOVA with age, sex, and current smoking status as covariates. P values <0.05 are in bold face and underline. SNPSpD correction was applied to correct for multiple SNPs tested (the effective number of independent marker loci: 2.76).
Figure 2.
Effects of the genotypes in the RELA gene on PPI. PPI82, PPI86, and PPI90: pre-pulse inhibition of acoustic startle reflex in pre-pulse intensities of 82 dB, 86 dB, and 90 dB, respectively. Error bars represent the standard error of the mean. *p<0.05. **p<0.01.
DISCUSSION
In this study, we first provided evidence that genetic variants of the RELA gene are associated with the risk for schizophrenia. Next, in silico analysis suggested that the risk SNPs in the RELA gene might be associated with gene expression differences in lymphoblasts. Finally, we measured the effects of the RELA genotypes on PPIs in patients with schizophrenia. Our results indicated that the risk alleles were associated with reduced PPIs.
As the association of the RELA gene and schizophrenia was supported by a number of statistical analyses such as genotypic and allelic associations for four SNPs (total 28) and haplotype analysis (total 24), the correction for multiple testing should be considered. In this study, overall genetic association tests were 52; however, all tests were not independent and multiple hypotheses were included. Thus, Bonferroni correction, a method to correct for multiple independent tests for one hypothesis, might not be appropriate. The consensus how to correct such multiple testing has not been reached in this research field. Thus, we only applied SNPSpD correction for genotypic and allelic association analysis for four SNPs, because the number of effective independent SNPs could be calculated by the SNPSpD method.
The reason why we obtained such low p values in our association analysis could be due to a relatively large sample size and high frequency of minor allele of the SNP4. Indeed, power analysis showed that our subjects had sufficient power (>0.95) to detect an effect of the odds ratios for SNP4, 1.23 in total subjects and 1.32 in male subjects. Although the strong association between the RELA gene and schizophrenia has been observed, the biological significance of this gene in susceptibility for schizophrenia might not be large, because 22% of the patients with schizophrenia have homozygous of risk allele in SNP4, but 16% of the controls also are homozygous of risk allele in SNP4. However, the association between the RELA gene and schizophrenia might explain, at least in part, the relation between immune system and schizophrenia.
It is of interest to study how genetic variation affects RELA function/expression. There is no experimental-based evidence that any of the SNPs or haplotypes is functional. Very little is known about the potential function of specific intronic sequences with regard to protein binding, stability, and splicing efficacy. However, the genotype-expression analysis showed that the risk alleles are associated with lower RELA gene expression in Japanese lymphoblasts. On the other hand, no association was found between other SNPs and gene expression in other ethnicities. No data could explain these results. A possible explanation for this ethnic difference is that the allele frequencies of some target genes in the signal transduction pathway of NF-κB, such as interleukins, chemokines, and interferons, and molecules involved in apoptosis or adhesion (Li and Verma, 2002), which could be associated with the pathophysiology of schizophrenia, might be different among populations. For example, a GWAS study in Caucasian population showed significant association with schizophrenia were in a region of LD on chromosome 6p22.1, including several immunity related genes other than the RELA gene (Stefansson et al, 2009). There were four SNPs associated with schizophrenia in the immunity related genes. Three SNPs out of four SNPs were not polymorphic in Japanese population and the minor allele frequency of the remained one SNP is 0.01 in Japanese population (0.19 in Caucasian population). This difference of allele frequency of SNP could alter the effect of the presumably functional risk alleles in the RELA gene in other ethnicities. A risk SNP of the RELA gene was found in the possible transcription factor binding sequence of AR. Indeed, it was reported that AR activation decreased the expression of RELA and reduced its nuclear localization and transcriptional activity (Nelius et al, 2007). As the risk allele destroyed the consensus sequence for AR binding, this SNP could be functional. Several studies have examined the association between AR and schizophrenia, and there are clinical differences between males and females, such as greater lifetime risk, earlier age of onset and poorer outcome in males (Tandon et al, 2008). Crow et al. reported the association of AR with schizophrenia in males (Crow et al, 1993); however, negative results have also been reported (Arranz et al, 1995; Tsai et al, 2006). When we examined the association between the RELA gene and schizophrenia by gender, a significant association was observed in males but not in females. Our results suggest that the T allele of SNP4 in the RELA gene, which might be functional for AR binding and transcription, could be a risk-associated allele for male schizophrenia.
There are numerous genes in neurons that are regulated by NF-κB (Kaltschmidt et al, 2005). Among these genes are molecules related to neurotransmission, including subunits of N-methyl-D-aspartate receptors, voltage-dependent calcium channels and the calcium-binding protein calbindin; cell survival factors, including Bcl-2, Mn-SOD, and inhibitor of apoptosis proteins (IAPs); and cell death factors, including Bcl-x(S) and Bax (Mattson, 2005). It is noteworthy that the expression levels of some of these downstream molecules have been reported to be altered in postmortem brains of patients with schizophrenia, for example, increased: Bax, calbindin, and NR2B; decreased Bcl-2 (Gao et al, 2000; Jarskog et al, 2000, 2004; Fung et al, 2010).
PPI of the acoustic startle response has been demonstrated from mice to humans, and is considered to be a measure of ‘sensorimotor gating,' whereby pre-pulses reduce the effect of subsequent sensory stimuli to protect the brain from sensory overload (Braff and Geyer, 1990). A deficit in PPI is a reliable feature of schizophrenia, where reduced gating is thought to be one possible neurobiological mechanism that underlies the basic cognitive abnormalities associated with this disorder (Braff et al, 2001). We observed associations between risk SNPs in the RELA gene and some pre-pulse intensities in patients with schizophrenia. Although we cannot explain the differences in the associated pre-pulse intensities among SNPs, our findings suggest that the RELA gene might modulate PPI in patients with schizophrenia.
The PPI deficits observed in schizophrenia can be mimicked in animals by the administration of dopamine agonizts and NMDA antagonists, such as phencyclidine (PCP), and reversed by anti-psychotic drugs (Mansbach et al, 1988; Geyer et al, 2001; Wang et al, 2001). Typical and atypical anti-psychotic drugs also reverse the PPI deficits observed in schizophrenic patients (Kumari and Sharma, 2002). Consistent with our data, PCP administration to rats that showed deficits in PPI elicited the abnormal nuclear translocation of NF-κB in the frontal cortex (Wang et al, 2001), which is indicative of a functional correlation between the RELA gene and PPI in a potential animal model for schizophrenia.
There are several limitations to interpret our results. Our study size of 1224 cases and 1663 controls had sufficient power (>0.80) to detect the effects of odds ratios of 1.16 or greater as indicated in recent genome-wide association studies for each SNP (O'Donovan et al, 2009). However, the possibility of false positive results due to type I errors could not be excluded. Our positive results might be derived from sample bias due to population stratification and non-sex-matched samples, although the Japanese are a relatively homogeneous population and the logistic regression study revealed genotype effects on the diagnosis independent of sex. We did not perform a systematic mutation search using these Japanese schizophrenia samples. In silico analysis of gene expression in lymphoblasts and AR binding site in SNP4, raised a possibility that the SNP4 might be a functional SNP. However, further biological study of the function of SNP4 is required to verify these in silico results. As the sample size we used for the PPI analysis was small, gender effect on the association between PPI and the SNPs in the RELA gene was not able to be analyzed. An increased sample size for schizophrenia and control subjects is needed before a firm conclusion can be drawn. We used PPI as a main phenotype of interest. Other phenotypes such as neurocognitive dysfunction that has larger effect size than PPI and brain morphology, which is more stable overtime were not tested in this study; however, PPI is a physiological phenotype, which is reliable, easily measured, and relatively specific for schizophrenia. In the present study, we propose RELA as a new candidate gene for susceptibility to schizophrenia.
Acknowledgments
We thank all individuals who participated in this study. This work was funded in part by Grants-in-aid from the Japanese Ministry of Health, Labor and Welfare (H19-kokoro-002, H22-rinken-ippan-002), the Japanese Ministry of Education, Culture, Sports, Science and Technology (18689030, 22390225), CREST of JST, Grant-in-aid for Scientific Research on Priority Areas -Research on the Pathomechanisms of Brain Disorders- from the MEXT (18023045) and Innovative Areas (Comprehensive Brain Science Network) and the Japan Foundation for Neuroscience and Mental Health.
The authors RH, KO, YY, MF, HY, HT, MI, TO, HK, OS, MT, NI, NO, KK, HK and MT declare that no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Anokhin AP, Heath AC, Myers E, Ralano A, Wood S. Genetic influences on prepulse inhibition of startle reflex in humans. Neurosci Lett. 2003;353:45–48. doi: 10.1016/j.neulet.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Arranz M, Sharma T, Sham P, Kerwin R, Nanko S, Owen M, et al. Schizophrenia and the androgen receptor gene: report of a sibship showing co-segregation with Reifenstein syndrome but no evidence for linkage in 23 multiply affected families. Am J Med Genet. 1995;60:377–381. doi: 10.1002/ajmg.1320600506. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA. The inducible transcription activator NF-kappa B: regulation by distinct protein subunits. Biochim Biophys Acta. 1991;1072:63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benes FM, Vincent SL, Todtenkopf M. The density of pyramidal and nonpyramidal neurons in anterior cingulate cortex of schizophrenic and bipolar subjects. Biol Psychiatry. 2001;50:395–406. doi: 10.1016/s0006-3223(01)01084-8. [DOI] [PubMed] [Google Scholar]
- Benes FM, Walsh J, Bhattacharyya S, Sheth A, Berretta S. DNA fragmentation decreased in schizophrenia but not bipolar disorder. Arch Gen Psychiatry. 2003;60:359–364. doi: 10.1001/archpsyc.60.4.359. [DOI] [PubMed] [Google Scholar]
- Braff DL, Geyer MA. Sensorimotor gating and schizophrenia. Human and animal model studies. Arch Gen Psychiatry. 1990;47:181–188. doi: 10.1001/archpsyc.1990.01810140081011. [DOI] [PubMed] [Google Scholar]
- Braff DL, Geyer MA, Swerdlow NR. Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- Braff DL, Light GA. The use of neurophysiological endophenotypes to understand the genetic basis of schizophrenia. Dialogues Clin Neurosci. 2005;7:125–135. doi: 10.31887/DCNS.2005.7.2/dlbraff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Derkits EJ. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am J Psychiatry. 2010;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenhead KS, Swerdlow NR, Shafer KM, Diaz M, Braff DL. Modulation of the startle response and startle laterality in relatives of schizophrenic patients and in subjects with schizotypal personality disorder: evidence of inhibitory deficits. Am J Psychiatry. 2000;157:1660–1668. doi: 10.1176/appi.ajp.157.10.1660. [DOI] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- Cardon M, Ron-Harel N, Cohen H, Lewitus GM, Schwartz M. Dysregulation of kisspeptin and neurogenesis at adolescence link inborn immune deficits to the late onset of abnormal sensorimotor gating in congenital psychological disorders. Mol Psychiatry. 2010;15:415–425. doi: 10.1038/mp.2009.66. [DOI] [PubMed] [Google Scholar]
- Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, Kelsoe J, et al. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow TJ, Poulter M, Lofthouse R, Chen G, Shah T, Bass N, et al. Male siblings with schizophrenia share alleles at the androgen receptor above chance expectation. Am J Med Genet. 1993;48:159–160. doi: 10.1002/ajmg.1320480309. [DOI] [PubMed] [Google Scholar]
- Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2010;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- Gao XM, Sakai K, Roberts RC, Conley RR, Dean B, Tamminga CA. Ionotropic glutamate receptors and expression of N-methyl-D-aspartate receptor subunits in subregions of human hippocampus: effects of schizophrenia. Am J Psychiatry. 2000;157:1141–1149. doi: 10.1176/appi.ajp.157.7.1141. [DOI] [PubMed] [Google Scholar]
- Geyer MA, Krebs-Thomson K, Braff DL, Swerdlow NR. Pharmacological studies of prepulse inhibition models of sensorimotor gating deficits in schizophrenia: a decade in review. Psychopharmacology (Berl) 2001;156:117–154. doi: 10.1007/s002130100811. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR.2005Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence Mol Psychiatry 1040–68.image 45. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Hashimoto H, Shintani N, Chiba S, Hattori S, Okada T, et al. Pituitary adenylate cyclase-activating polypeptide is associated with schizophrenia. Mol Psychiatry. 2007;12:1026–1032. doi: 10.1038/sj.mp.4001982. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, Ishimoto T, et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet. 2006;15:3024–3033. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- Hong LE, Wonodi I, Stine OC, Mitchell BD, Thaker GK. Evidence of missense mutations on the neuregulin 1 gene affecting function of prepulse inhibition. Biol Psychiatry. 2008;63:17–23. doi: 10.1016/j.biopsych.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxford T, Ghosh G. A structural guide to proteins of the NF-kappaB signaling module. Cold Spring Harb Perspect Biol. 2009;1:a000075. doi: 10.1101/cshperspect.a000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Kinoshita Y, Okochi T, Kawashima K, Kushima I, et al. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2010;69:472–478. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Jarskog LF, Gilmore JH, Selinger ES, Lieberman JA. Cortical bcl-2 protein expression and apoptotic regulation in schizophrenia. Biol Psychiatry. 2000;48:641–650. doi: 10.1016/s0006-3223(00)00988-4. [DOI] [PubMed] [Google Scholar]
- Jarskog LF, Selinger ES, Lieberman JA, Gilmore JH. Apoptotic proteins in the temporal cortex in schizophrenia: high Bax/Bcl-2 ratio without caspase-3 activation. Am J Psychiatry. 2004;161:109–115. doi: 10.1176/appi.ajp.161.1.109. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt B, Kaltschmidt C. NF-kappaB in the nervous system. Cold Spring Harb Perspect Biol. 2009;1:a001271. doi: 10.1101/cshperspect.a001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt B, Widera D, Kaltschmidt C. Signaling via NF-kappaB in the nervous system. Biochim Biophys Acta. 2005;1745:287–299. doi: 10.1016/j.bbamcr.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Kumari V, Sharma T. Effects of typical and atypical antipsychotics on prepulse inhibition in schizophrenia: a critical evaluation of current evidence and directions for future research. Psychopharmacology (Berl) 2002;162:97–101. doi: 10.1007/s00213-002-1099-x. [DOI] [PubMed] [Google Scholar]
- Larouche A, Berube P, Sarret P, Grignon S. Subacute H2O2, but not poly(IC), upregulates dopamine D2 receptors in retinoic acid differentiated SH-SY5Y neuroblastoma. Synapse. 2008;62:70–73. doi: 10.1002/syn.20458. [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- Mansbach RS, Geyer MA, Braff DL. Dopaminergic stimulation disrupts sensorimotor gating in the rat. Psychopharmacology (Berl) 1988;94:507–514. doi: 10.1007/BF00212846. [DOI] [PubMed] [Google Scholar]
- Mattson MP. NF-kappaB in the survival and plasticity of neurons. Neurochem Res. 2005;30:883–893. doi: 10.1007/s11064-005-6961-x. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Culmsee C, Yu Z, Camandola S. Roles of nuclear factor kappaB in neuronal survival and plasticity. J Neurochem. 2000;74:443–456. doi: 10.1046/j.1471-4159.2000.740443.x. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Moriwaki M, Kishi T, Takahashi H, Hashimoto R, Kawashima K, Okochi T, et al. Prepulse inhibition of the startle response with chronic schizophrenia: a replication study. Neurosci Res. 2009;65:259–262. doi: 10.1016/j.neures.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Naudin J, Capo C, Giusano B, Mege JL, Azorin JM. A differential role for interleukin-6 and tumor necrosis factor-alpha in schizophrenia. Schizophr Res. 1997;26:227–233. doi: 10.1016/s0920-9964(97)00059-5. [DOI] [PubMed] [Google Scholar]
- Nelius T, Filleur S, Yemelyanov A, Budunova I, Shroff E, Mirochnik Y, et al. Androgen receptor targets NFkappaB and TSP1 to suppress prostate tumor growth in vivo. Int J Cancer. 2007;121:999–1008. doi: 10.1002/ijc.22802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126:3–12. doi: 10.1007/s00439-009-0703-0. [DOI] [PubMed] [Google Scholar]
- Ohi K, Hashimoto R, Yasuda Y, Yoshida T, Takahashi H, Iike N, et al. Association study of the G72 gene with schizophrenia in a Japanese population: a multicenter study. Schizophr Res. 2009;109:80–85. doi: 10.1016/j.schres.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, Kouassi E. Inflammatory cytokine alterations in schizophrenia: a systematic quantitative review. Biol Psychiatry. 2008;63:801–808. doi: 10.1016/j.biopsych.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quednow BB, Kuhn KU, Mossner R, Schwab SG, Schuhmacher A, Maier W, et al. Sensorimotor gating of schizophrenia patients is influenced by 5-HT2A receptor polymorphisms. Biol Psychiatry. 2008;64:434–437. doi: 10.1016/j.biopsych.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Quednow BB, Wagner M, Mossner R, Maier W, Kuhn KU. Sensorimotor gating of schizophrenia patients depends on Catechol O-methyltransferase Val158Met polymorphism. Schizophr Bull. 2010;36:341–346. doi: 10.1093/schbul/sbn088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero E, Guaza C, Castellano B, Borrell J. Ontogeny of sensorimotor gating and immune impairment induced by prenatal immune challenge in rats: implications for the etiopathology of schizophrenia. Mol Psychiatry. 2010;15:372–383. doi: 10.1038/mp.2008.44. [DOI] [PubMed] [Google Scholar]
- Schwarz MJ, Riedel M, Ackenheil M, Muller N. Decreased levels of soluble intercellular adhesion molecule-1 (sICAM-1) in unmedicated and medicated schizophrenic patients. Biol Psychiatry. 2000;47:29–33. doi: 10.1016/s0006-3223(99)00206-1. [DOI] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Song XQ, Lv LX, Li WQ, Hao YH, Zhao JP. The interaction of nuclear factor-kappa B and cytokines is associated with schizophrenia. Biol Psychiatry. 2009;65:481–488. doi: 10.1016/j.biopsych.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Weber M, Qu Y, Light GA, Braff DL. Realistic expectations of prepulse inhibition in translational models for schizophrenia research. Psychopharmacology (Berl) 2008;199:331–388. doi: 10.1007/s00213-008-1072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwase M, Canuet L, Yasuda Y, Ohi K, Fukumoto M, et al. Relationship between prepulse inhibition of acoustic startle response and schizotypy in healthy Japanese subjects. Psychophysiology. 2010;47:831–837. doi: 10.1111/j.1469-8986.2010.01000.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Iwase M, Ishii R, Ohi K, Fukumoto M, Azechi M, et al. Impaired prepulse inhibition and habituation of acoustic startle response in Japanese patients with schizophrenia. Neurosci Res. 2008;62:187–194. doi: 10.1016/j.neures.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, ‘Just the Facts': what we know in 2008 part 1: overview. Schizophr Res. 2008;100:4–19. doi: 10.1016/j.schres.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Hong CJ, Liao DL, Chiang CH. Distribution of androgen receptor CAG repeat polymorphism in Chinese schizophrenia and its correlation with age at onset. Psychoneuroendocrinology. 2006;31:270–274. doi: 10.1016/j.psyneuen.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Tsuang M. Schizophrenia: genes and environment. Biol Psychiatry. 2000;47:210–220. doi: 10.1016/s0006-3223(99)00289-9. [DOI] [PubMed] [Google Scholar]
- Wang C, McInnis J, Ross-Sanchez M, Shinnick-Gallagher P, Wiley JL, Johnson KM. Long-term behavioral and neurodegenerative effects of perinatal phencyclidine administration: implications for schizophrenia. Neuroscience. 2001;107:535–550. doi: 10.1016/s0306-4522(01)00384-0. [DOI] [PubMed] [Google Scholar]
- Woo TU, Shrestha K, Lamb D, Minns MM, Benes FM. N-methyl-D-aspartate receptor and calbindin-containing neurons in the anterior cingulate cortex in schizophrenia and bipolar disorder. Biol Psychiatry. 2008;64:803–809. doi: 10.1016/j.biopsych.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Iwayama-Shigeno Y, Yoshida Y, Toyota T, Itokawa M, Hattori E, et al. Family-based association study of schizophrenia with 444 markers and analysis of a new susceptibility locus mapped to 11q13.3. Am J Med Genet B Neuropsychiatr Genet. 2004;127B:11–19. doi: 10.1002/ajmg.b.20166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.