Abstract
Innate behaviours are flexible: they change rapidly in response to transient environmental conditions, and are modified slowly by changes in the genome. A classical flexible behaviour is the exploration-exploitation decision, which describes the time at which foraging animals choose to abandon a depleting food supply. Here we use quantitative genetic analysis to examine the decision to leave a food patch in Caenorhabditis elegans. We find that patch-leaving is a multigenic trait regulated in part by naturally-occurring noncoding polymorphisms in tyra-3, which encodes a G protein-coupled catecholamine receptor related to vertebrate adrenergic receptors. tyra-3 acts in sensory neurons that detect food-related cues, suggesting that the internal catecholamines detected by tyra-3 regulate responses to external conditions. These results indicate that genetic variation and environmental cues can converge on common circuits to regulate behaviour, and suggest that catecholamines have an ancient role in regulating behavioural decisions.
Despite abundant evidence for heritability of behavioural traits within and between species, only a few naturally varying traits have been associated with polymorphisms in specific genes 1. Foraging for food is an ecologically relevant, environmentally regulated behaviour that is suitable for genetic analysis, as it can differ between populations of a species that live in different habitats 2. An essential foraging decision is the choice between exploiting existing resources and exploring other options that may provide new resources. This decision can be described by Charnov’s marginal value theorem, which proposes that the optimal time for an animal to leave a foraging ground occurs when local resource levels fall below the average level in the entire habitat3. The marginal value theorem was developed for animals foraging for food in patchy environments, but has analogies with diverse decision-making processes in field biology, cognitive neuroscience, and economics2,4–6.
Studies of patch-leaving behaviour in the nematode C. elegans have revealed innate, environmental, and experience-dependent factors that affect its foraging decisions. C. elegans rarely leaves a dense lawn of high-quality bacterial food 7,8, but more frequently leaves lawns of pathogenic bacteria or lawns that are spiked with chemical repellents 9,10. Males will leave lawns that do not contain potential mates 11, while hermaphrodites leave lawns when animal density is high 12. In addition, wild-type strains vary in their propensity to leave bacterial lawns based on a genetic polymorphism that affects the G protein-coupled neuropeptide receptor NPR-1 12–14. This npr-1 polymorphism affects many foraging behaviours; low-activity npr-1 strains aggregate into social feeding groups, move quickly on food, and have altered responses to oxygen, carbon dioxide, and pheromones compared to the N2 laboratory strain 15–20. The high-activity allele of npr-1 in N2 arose in the laboratory, probably as an adaptation to laboratory conditions 19, so it is not known whether genetic variation affects C. elegans foraging in natural environments.
Natural genetic variation within a species can generate diversity in foraging behaviour, as exemplified by the polymorphic Drosophila melanogaster foraging (for) gene, which encodes a cGMP-dependent protein kinase 21. A low-activity allele of for is present in Drosophila sitter larvae, which move slowly on a food patch; a high-activity allele of for is present in rover larvae, which move quickly and disperse rapidly 22. A for-related cGMP-dependent kinase affects foraging in honeybees, ants, and nematodes, suggesting that diverse animals share molecular mechanisms for behavioural regulation 22,23.
To gain further insight into the genetics and neurobiology of lawn-leaving behaviour in C. elegans, we here use quantitative genetic analysis to examine its genetic architecture in wild-type strains, and show that genetic variation in multiple loci, including a catecholamine receptor, interacts with environmental conditions to regulate the exploitation-exploration decision.
Multiple loci affect leaving behaviour
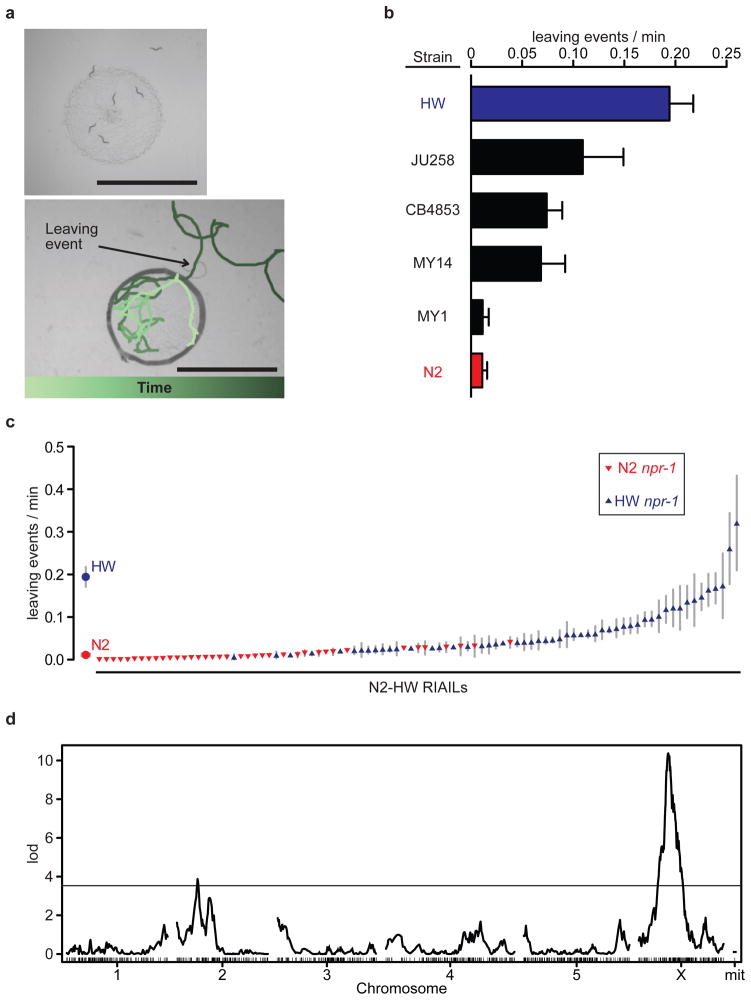
Different wild-type strains of C. elegans vary in their tendency to leave or remain on a standardized small lawn of bacterial food (Fig. 1a). For example, adult hermaphrodites from the laboratory strain N2 leave the lawn only once every 100 minutes, whereas animals from the CB4856 (HW) strain isolated from pineapple fields in Hawaii leave the lawn once every 5–6 minutes (Fig. 1b, Supplementary Movies 1 and 2). To determine the genetic architecture of this behavioural difference between N2 and HW, we quantified leaving rates in 91 N2-HW recombinant inbred advanced intercross lines (RIAILs) 24. 58 of the RIAILs had low leaving rates comparable to N2, only 6–10 had high leaving rates comparable to HW, and 23 had intermediate rates (Fig. 1c). The excess of low leaving rates and the continuous behavioural distribution in RIAILs suggest that leaving is a multigenic quantitative trait.
Figure 1. Lawn-leaving behaviour varies between wild-type C. elegans strains.
a) Lawn-leaving assays. Top: Six adult HW hermaphrodites on a bacterial lawn. One animal has left the lawn and one is leaving. Bottom: Track of a HW animal during 5 min of an assay; colour shows passage of time. The border of the lawn is outlined. Scale bar, 6 mm. b) Leaving rates of six wild-type strains. c) Leaving rates of 91 N2-HW recombinant inbred advanced intercross lines (RIAILs) 24 and parental strains. d) QTL analysis of RIAILs shown in c. The horizontal line denotes the P<0.01 genome-wide significance threshold. Error bars indicate s.e.m.
Quantitative trait locus (QTL) analysis of the RIAILs uncovered two regions with significant effects on leaving rates, one on the X chromosome and one on chromosome II (Fig. 1d). The X chromosome QTL overlapped with the location of the polymorphic G protein-coupled neuropeptide receptor NPR-1, which affects many food-related behaviours 12,15. The npr-1 polymorphism has previously been shown to affect leaving, as well as locomotion speed on food, a behaviour that partially correlates with leaving rate 12,15 (Supplementary Fig. 2). Examining the npr-1 genotype in the RIAILs revealed a strong but asymmetric correlation with leaving rates (Fig. 1c). Every strain with the N2 allele of npr-1 had low leaving rates (≤1 event every 20 minutes), but strains with the HW allele of npr-1 could have either low or high leaving rates (Fig. 1c). The asymmetric distribution is consistent with a role for npr-1 in leaving behaviour, but indicates that npr-1 has epistatic interactions with other loci segregating in the RIAILs.
The involvement of npr-1 in leaving behaviour was confirmed by analyzing near-isogenic lines (NILs) containing the N2 and HW npr-1 alleles in the reciprocal strain background, and by examining npr-1 null mutants (Supplementary Fig. 3). Specific transgenic expression of the N2 npr-1 allele in its essential site of action, the RMG motor neurons 20, sharply reduced the leaving rate of HW animals (Supplementary Fig. 3). Thus npr-1 is a regulator of HW leaving rates, but not the only contributing gene.
tyra-3 affects leaving behaviour
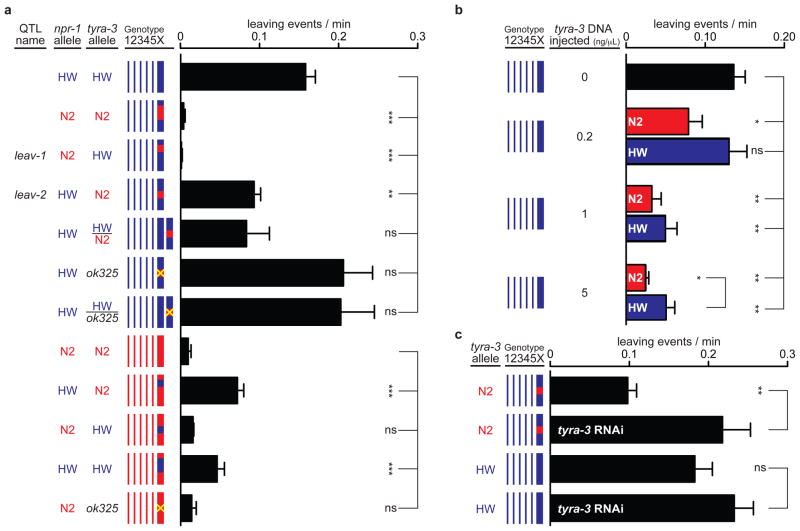
Studies in yeast, flies, mice, and plants have shown that individual QTLs often resolve into several genes that contribute to phenotypic variance 25–28. Similarly, fine-mapping of the ~ 1 Mb QTL that contained npr-1 suggested the existence of multiple loci that affected leaving rates. A NIL with <150 kb of N2 DNA spanning the npr-1 locus introgressed into HW had N2-like leaving rates (leav-1 QTL, Fig 2a and Supplementary Fig. 3). A second NIL with 700 kb of N2 DNA that did not cover npr-1 introgressed into HW also had a low leaving rate, with about half the leaving rate of HW (leav-2 QTL, Fig. 2a). These results suggest the existence of a second X-linked locus that affects leaving rates, which we called leav-2. The leav-2 region did not affect leaving in the N2 genetic background (Fig. 2a), so all subsequent experiments were conducted in the HW background.
Figure 2. N2 and HW tyra-3 alleles differentially affect leaving rates.
a) Dissection of the QTL on X into two loci: leav-1 (4.70–4.78 Mb) and leav-2 (4.78–5.75 Mb). ‘Genotype’ shows chromosomes; thick line is X chromosome. Blue denotes HW DNA, red denotes N2 DNA, and yellow denotes the tyra-3(ok325) null mutant. In heterozygous strains, both X chromosomes are diagrammed. b) tyra-3 genomic fragments (Fig. 3a) reduce HW leaving rates. Blue, HW transgenes; red, N2 transgenes. Two-way ANOVA showed significant effects of both transgene concentration and DNA strain of origin. c) Effect of tyra-3 RNAi. Error bars indicate s.e.m. * P<0.05, ** P<0.01, or *** P<0.001 by t-test or ANOVA with Dunnett test.
A 100 kb minimal region for leav-2 was identified by analyzing the breakpoints of individual RIAILs (Supplementary Fig. 4 and Supplementary Methods). We characterized the genetic properties of leav-2 by crossing the leav-2 NIL strain with HW. The heterozygous F1 progeny had leaving rates similar to the leav-2 NIL (Fig. 2a), indicating that the N2 leav-2 locus was dominant to HW and suggesting that N2 transgenes covering the relevant gene should reduce the leaving rate of HW animals. Therefore, overlapping N2 genomic DNA fragments from the 100 kb minimal leav-2 region were introduced into HW animals by microinjection (Fig. 2b and Supplementary Fig. 5). A single gene in this region reduced leaving rates: tyra-3, which encodes a G protein-coupled receptor for the invertebrate norephinephrine-like neurotransmitters tyramine and octopamine 29. Tyramine and octopamine receptors are related to vertebrate adrenergic receptors, and are thought to carry out analogous functions. tyra-3 genomic fragments from the N2 strain were more active than tyra-3 fragments from the HW strain injected at the same concentration, consistent with the possibility that tyra-3 is a polymorphic gene that differs between N2 and HW (Fig. 2b).
If leav-2 corresponds to tyra-3, a tyra-3 mutation should eliminate its activity 30. To test this prediction genetically, a null allele of tyra-3 in an N2 background was introgressed into a HW background. The N2 region in the resulting NIL covered from 4.9 to 5.4 MB of the X chromosome, the inferred position of leav-2. The tyra-3(ok325) null NIL had high (HW-like) leaving rates, suggesting that N2 leav-2 activity was not present in the strain (Fig. 2a). Heterozygotes between HW and the near-isogenic tyra-3(ok325) null strain also had high leaving rates (Fig. 2a). These results are as expected if the active locus in leav-2 is tyra-3; however, other genes within the introgressed regions could also contribute to the different leaving rates.
To strengthen the connection between tyra-3 and leav-2, RNAi against tyra-3 was performed in the leav-2 NIL that has low leaving rates due to the presence of the N2 QTL. Knockdown of tyra-3 increased the leaving rate of the leav-2 NIL to levels observed in HW animals, the result predicted if the tyra-3 locus from N2 reduces leaving (Fig. 2c). Comparable experiments in a pure HW strain had minimal effects, as expected if tyra-3 activity in HW is already low.
Further confirmation that the HW allele of tyra-3 has reduced biological activity was provided by examining the one phenotype previously associated with tyra-3, avoidance of dilute octanol 29. tyra-3 null mutants avoid octanol more strongly than wild-type N2; the NIL strain with the HW tyra-3 allele had a similar enhanced octanol response, suggesting that the HW tyra-3 allele has reduced tyra-3 function (Supplementary Fig. 6).
Noncoding changes affect tyra-3 activity
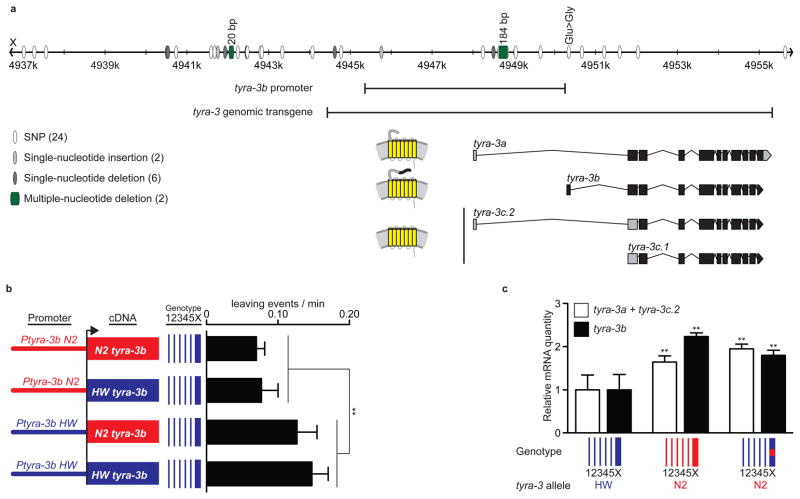
The differential activity of N2 and HW genomic tyra-3 fragments in the leaving assay suggested that N2 and HW alleles are functionally distinct (Fig. 2b). To identify polymorphisms between N2 and HW alleles of tyra-3, we sequenced ~19 kb surrounding the tyra-3 locus in HW. There were 34 differences between HW and the N2 consensus genomic sequence (Fig. 3a): 33 noncoding changes and a single coding difference that changed a glutamate in the tyra-3b isoform to glycine.
Figure 3. Noncoding changes in tyra-3 affect its activity and expression level.
a) HW polymorphisms in the tyra-3 locus relative to N2. tyra-3 encodes three predicted G protein-coupled receptors. The genomic region examined in Fig. 2b and the 4.9 kb promoter used in Figs. 3b and 4a are indicated. b) Leaving rates of transgenic HW animals with tyra-3b promoters fused to tyra-3b cDNAs. Error bars indicate s.e.m. ** P<0.01 by two-way ANOVA; no statistical interaction between the promoter and the cDNA. c) Relative amounts of tyra-3 isoform mRNAs in HW, N2, and leav-2 strains (Fig. 2a). Error bars indicate s.d. ** P<0.01 compared to HW, ANOVA with Dunnett test.
Sequences that contribute to the differential activity of N2 and HW tyra-3 alleles were localized further using transgenic assays. We fused N2 and HW tyra-3b cDNAs to 4.9 kb of noncoding N2 or HW sequence upstream of the tyra-3b start site and introduced each of the four resulting clones into the HW strain. tyra-3 transgenes with the N2 noncoding sequence were significantly more potent than comparable transgenes with the HW sequence, regardless of whether they preceded N2 or HW tyra-3 cDNAs (Fig. 3b), excluding the coding polymorphism and localizing a functional difference between N2 and HW tyra-3 genes to a 4.9 kb region that harbours 5 noncoding SNPs, 1 single nucleotide insertion, and a 184 bp deletion in HW. To narrow the relevant change down further, the 184 bp deletion was engineered into the N2 tyra-3 genomic fragment; this clone was significantly less potent in the leaving assay than the full N2 genomic fragment (Supplementary Fig. 7). These results indicate that the 184 bp deletion represents at least part of the functional difference between N2 and HW tyra-3 alleles.
Sequence variation in tyra-3 noncoding regions could affect the level or location of tyra-3 expression. Quantitative RT-PCR of tyra-3 mRNA levels in mixed-stage animals indicated that N2 expressed approximately twice as much tyra-3 mRNA as HW, consistent with increased tyra-3 activity in the N2 strain (Fig. 3c). The leav-2 NIL with N2 tyra-3 introgressed into HW also had high tyra-3 mRNA levels, suggesting that cis-acting changes affect tyra-3 expression (Fig. 3c).
Since both N2 and HW were cultivated in the laboratory for many years before permanent cultures were frozen, we wished to exclude the possibility that the tyra-3 polymorphisms were laboratory-derived 19. Therefore, 19 kb of the tyra-3 locus was sequenced in all wild strains tested for leaving behaviour in Fig. 1, including three strains that were frozen immediately after their isolation. Each strain represents a different C. elegans haplotype group 24. Both N2-like and HW-like tyra-3 sequences were represented in the wild-caught strains, confirming the wild ancestry of both alleles (Supplementary Table 1 and Supplementary Methods). Notably, the tyra-3 locus of MY1 was identical to N2 and, correspondingly, the leaving rate of MY1 was similar to that of N2.
tyra-3 acts in sensory neurons
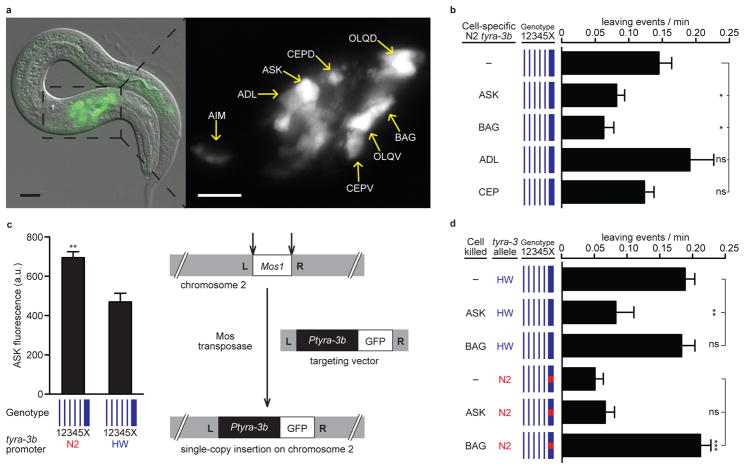
The identification of tyra-3 provided an opportunity to characterize the neuronal basis of the decision to leave or remain on a food patch. The biological activity of a transgene with 4.9 kb upstream of the tyra-3b start site fused to a tyra-3 cDNA (Fig. 3b) implied that it was expressed in cells that regulate leaving behaviour. When this 4.9 kb region was fused to GFP, it drove reliable expression in ASK, ADL, AIM, AUA, BAG, CEP, OLQ, and SDQL neurons, in other unidentified neurons in the ventral ganglion and the tail, occasionally in AFD and AWC neurons, and in two non-neuronal cell types, the spermatheca and the distal tip cell (Fig. 4a and data not shown). The same set of cells was observed with reporter genes bearing either N2 or HW tyra-3 upstream regions, and in both N2 and HW genetic backgrounds. Together with the quantitative RT-PCR data (Fig. 3c), these results suggest that different tyra-3 expression levels, not different sites of expression, distinguish N2 and HW alleles.
Figure 4. tyra-3 acts in ASK and BAG sensory neurons.
a) Expression of 4.9 kb N2 tyra-3b promoter::GFP fusion (Fig. 3a) in HW animal; HW tyra-3b promoter::GFP is expressed in the same cells. Posterior signal is gut autofluorescence. Scale bar = 20μm. b) Leaving rates of HW strains expressing tyra-3b in specific cells. c) Left: GFP fluorescence intensity in ASK of HW animals with a MosSCI insertion of N2 or HW 4.9 kb tyra-3b promoter::GFP. Right: Schematic of MosSCI technique 36. d) Leaving rates after killing ASK or BAG in HW and leav-2 strains (Fig. 2a). Error bars indicate s.e.m. * P<0.05, ** P<0.01, or *** P<0.001 by t-test or ANOVA with Dunnett test.
The neurons whose activity is regulated by tyra-3 were localized further by expressing tyra-3 cDNAs from cell type-specific promoters. tyra-3 expression in ASK or BAG sensory neurons significantly reduced leaving, but expression in the CEP or ADL sensory neurons did not (Fig. 4b). The ASK neurons sense attractive food-derived amino acids 31 and regulate search behaviours after animals are removed from food 32,33. The BAG neurons sense CO2 and O2, two cues associated with bacterial metabolism 34,35. Lowering O2 to levels that activate BAG reduced leaving rates (Supplementary Fig. 8).
To ask whether the tyra-3 noncoding polymorphism affects expression in relevant neurons, single-copy N2 or HW tyra-3b promoters driving GFP were inserted into a single, defined chromosomal location using the MosSCI technique 36. GFP levels in ASK neurons were significantly higher for transgenes containing the N2 promoter compared to those containing the HW promoter (Fig. 4c). These results suggest that the N2 tyra-3 locus is associated with higher tyra-3 expression in ASK, as well as higher tyra-3 mRNA expression at a whole-animal level; expression in BAG was not examined.
The behavioural functions of ASK and BAG, and tyra-3’s effect on those functions, were assessed by killing the neurons in different genetic backgrounds. Killing the ASK neurons reduced the leaving rate of HW animals, indicating that ASK can promote leaving (Fig. 4d). The ablation resembled the effect of the ASK::tyra-3 transgene, suggesting that tyra-3 reduces ASK activity. In agreement with this idea, killing the ASK neurons in a strain with the N2 high-activity tyra-3 allele did not reduce their leaving rates further. The effect of tyra-3 on ASK was selective for this assay; tyra-3 did not reduce lysine chemotaxis, a second ASK-dependent behaviour (Supplementary Fig. 9).
Killing the BAG neurons increased leaving rates in the strain with the N2 tyra-3 allele, demonstrating that BAG neurons prevent leaving (Fig. 4d). However, killing BAG had no effect in the strain with the HW tyra-3 allele, suggesting that BAG activity is already low in this strain under the assay conditions. The ablation and genetic results suggest that the N2 tyra-3 allele decreases ASK activity and increases BAG activity, two changes that act together to prevent leaving (Supplementary Fig. 1).
Gene-gene-environment interactions
Like most natural behaviours, the decision to leave a food patch is regulated by multiple genes and the environment; it responds to genetic variation in tyra-3, npr-1, and additional genes on the autosomes (Fig. 1) as well as food quality and quantity 7,8. Our results suggested that the N2 npr-1 allele was epistatic to tyra-3; animals with the N2 npr-1 allele had low leaving rates regardless of the tyra-3 genotype (Fig. 2a). However, N2 npr-1 reduced the leaving rate to almost zero, making it difficult to detect any further reduction. To make the assay more powerful, leaving was assayed on bacterial lawns of different densities. Leaving rates of all genotypes increased on thinner lawns and decreased on thicker lawns (Supplementary Fig. 10), but the thickness of the lawn changed the genetic interaction between tyra-3 and npr-1. In the standard leaving assay, tyra-3 polymorphisms had different effects only in the presence of the HW npr-1 allele; on a thinner lawn, only in the presence of the N2 npr-1 allele (Supplementary Fig. 10). Thus the epistatic relationship between npr-1 and tyra-3 is defined by the specific environment, not by an intrinsic regulatory relationship between the genes.
Discussion
Our results show that natural variation in tyra-3 affects patch leaving, a behaviour representative of the exploration-exploitation decision. tyra-3 encodes a G protein-coupled receptor activated by the invertebrate transmitters tyramine and octopamine 29, which are structurally related to vertebrate epinephrine and norepinephrine. Catecholamines are known to regulate arousal systems that affect many behaviours and behavioural decisions. In C. elegans, octopamine drives sensory, molecular, and behavioural responses to starvation, and tyramine affects specific aspects of locomotion 37–40. In insects, octopamine affects locomotory activity, arousal, and aggression 41–43. Mammalian norephinephrine is generally implicated in arousal behaviours, and norepinephrine release from the locus coeruleus is associated with switching between different tasks, a cognitive function with analogies to the exploration-exploitation decision 6.
Relatively few natural behavioural variations have been mapped to the single-gene level in any animal, and it is interesting that several of these variations affect G protein-coupled receptor signalling systems 44,45. We speculate that these receptor pathways may serve as common substrates of behavioural variation. All animal genomes encode many G protein-coupled receptors with different expression patterns. These receptors may provide a reservoir for genetic changes, as alteration in an individual receptor could cause relatively discrete effects without disrupting the entire system.
QTL mapping in rodents and in Drosophila indicates that most behavioural traits are polygenic, with widespread epistatic effects 1,46. In agreement with this conclusion, our analysis suggests the existence of epistatic interactions between tyra-3, npr-1, and at least one additional locus. Importantly, the non-additive interactions between tyra-3 and npr-1 are not stable, but vary based on the genetic background and the environment, similar to what has been found with yeast sporulation QTLs 47.
By integrating genetic studies of C. elegans foraging with neuronal analysis, we can provide a first-level description of underlying mechanisms. The sensory neurons that express tyra-3 detect food-related cues; we suggest that they integrate these external cues with internal arousal states detected by tyra-3, and that different tyra-3 alleles confer differential sensitivity to these arousal states (Supplementary Fig. 1). Thus variation in tyra-3 lies at the intersection of many forms of behavioural flexibility: rapid responses to environmental cues, short-term modulation by internal state fluctuations, and long-term genetic changes that lead to adaptive changes in innate behaviours.
Methods summary
Standardized leaving assays were conducted by videotaping seven adult hermaphrodite animals for 30 min on a low-density bacterial lawn compared to their growth lawn. The number of leaving events was recorded manually by examining the video recordings, and further behavioural analysis was conducted with a Matlab code adapted from the Parallel Worm Tracker 48. A leaving event was defined as an episode in which the whole body of an animal left the bacterial lawn and the animal did not reverse immediately to return to the lawn. The leaving rate was calculated as the number of leaving events per worm minute spent inside the bacterial lawn. Experiments on each strain were repeated at least three times.
QTL analysis was performed on the mean leaving rates of N2-HW recombinant inbred advanced intercross lines (RIALs) by nonparametric interval mapping in R/qtl 24,49. Significance levels were estimated from 10,000 permutations of the data. Near-Isogenic Lines were created by backcrossing a chromosomal region or allele into the desired genetic background at least 9 times.
Extrachromosomal transgenes were made by injection of DNA clones into the gonads of young adult hermaphrodites together with a fluorescent coinjection marker 50. To control for variation between transgenes, between two and five independent lines from each injection were characterized.
Single-copy insertion of transgenes was performed using the direct MosSCI transposition technique, targeting the ttTi5605 Mos allele on chromosome II 36.
Methods
Analysis of Behaviour in the Leaving Assay
6 cm NGM agar plates were seeded with 70 μL (conditioning plate) or with 10 μL (assay plate) of a fresh overnight culture of E. coli HB101 diluted in LB to OD600nm=2.0. 90 min after seeding the plates, ten young adult hermaphrodites were picked onto the conditioning plate. 30 min after being placed on the conditioning plates, seven of the animals were transferred onto the lawn of the assay plate. The 30 min leaving assay began 1 hr after placing the seven animals on the assay plate. The number of leaving events was recorded manually by examining the video recordings, and further behavioural analysis was conducted with a Matlab code adapted from the Parallel Worm Tracker 48. A leaving event was defined as an episode in which the whole body of an animal left the bacterial lawn and the animal did not reverse immediately to return to the lawn. The leaving rate was calculated as the number of leaving events per worm minute spent inside the bacterial lawn. Experiments on each strain were repeated at least three times.
Quantitative Trait Locus Analysis
The N2-HW recombinant inbred advanced intercross lines (RIAILs) used in this study represent the terminal generation of a 20-generation pedigree founded by reciprocal crosses between N2 and HW. The lines were constructed through 10 generations of intercrossing followed by 10 generations of selfing 24. They have been genotyped at 1454 nuclear and one mitochondrial markers and have a 5.3-fold expansion of the F2 genetic map 24. QTL analysis was performed on the mean leaving rates of N2-HW recombinant inbred advanced intercross lines (RIALs) by nonparametric interval mapping in R/qtl 49. Significance levels were estimated from 10,000 permutations of the data.
Quantitative RT-PCR
Total RNA from mixed stage worms was isolated with Trizol. 1.5 μg of RNA and oligo-dT were used for reverse transcription using SuperScript III First-Strand Synthesis (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed with Fast SYBR Green Master Mix (Applied Biosystems) on a 7900HT Real-Time PCR System (Applied Biosystems). act-3 was used as the calibrator for relative quantitation. 5′ primers corresponded to upstream exons that distinguished tyra-3 isoforms, and 3′ primers corresponded to shared exon sequence. Primers used were:
tyra-3a&c.2_F, ccacttgcaaatagcagcag
tyra-3b_F, ggctatttggtggtggtttg
tyra-3a & tyra-3b_R, tccttctggcgtcgaaatac
act-3_F, tcacgatcatgagaccattcaaa
act-3_R, gcaaattgtagtggggtcttcttatg
tyra-3 Expression Pattern
The N2 and HW 4.9 kb tyra-3b promoters were amplified using primers: tcaacctaaccactaactaaggg and cGatgaagcaagatgtcaggt, which overlaps the coding region by 4 bp. The ATG start codon is mutated to ATC (mutation is uppercase in primer). These promoters were individually fused by PCR to a fragment containing GFP followed by the unc-54 3′-UTR, as described 51. These PCR products were injected individually into both HW and N2 animals at 20 ng/μL. Cells expressing GFP were identified by Nomarski microscopy in both L1 and adult hermaphrodites. The identification of some cells was aided by injecting Ptyra-3b::GFP-expressing animals with promoter-mCherry fusions with established expression patterns. In this manner, the AIM neurons were identified as Ptyra-3b::GFP-expressing cells based on their position and the absence of co-localization with Pttx-3::mCherry. The BAG neurons co-expressed Ptyra-3b::GFP and Pflp-17::mCherry. The CEP neurons co-expressed Ptyra-3b::GFP and Pdat-1::mCherry. The ASK neurons co-expressed Ptyra-3b::GFP and Psra-9::mCherry. The ADL neurons co-expressed Ptyra-3b::GFP and Psri-51::mCherry.
Extrachromosomal transgenes
Transgenes were made by injection of DNA clones into the gonads of young adult hermaphrodites together with a fluorescent coinjection marker 50. To control for variation between transgenes, between two and five independent lines from each injection were characterized.
Generation of MosSCI Lines and Quantitation of GFP Fluorescence in ASK
Single-copy insertion of transgenes was performed using the direct MosSCI technique targeting the ttTi5605 Mos allele on chromosome II, as described 36. A schematic of the mechanism underlying MosSCI is shown in Fig. 4c.
The pCFJ151 targeting vector was modified by the introduction of an FseI restriction site into the multiple cloning site by site-directed mutagenesis using the primers gtaatacgactcacttaaggccggccctagagggtaccagagctcacc and ggtgagctctggtaccctctagggccggccttaagtgagtcgtattac to make pAB1. An FseI-SpeI fragment from a pSM vector containing N2-Ptyra-3b::N2-tyra-3b::SL2 GFP::unc-54 3′-UTR or HW-Ptyra-3b::N2-tyra-3b::SL2 GFP::unc-54 3′-UTR was cloned into pAB1.
For each tyra-3-containing test plasmid, about fifty EG4322 animals were injected with a mixture of tyra-3 plasmid, pGH8, pCFJ90, pCFJ104, and pJL43.1. After positive and negative selection and full sequencing of the insert, two inserted transgenes each of N2-Ptyra-3b and HW-Ptyra-3b were backcrossed to HW males seven times, selecting GFP-fluorescent hermaphrodites each generation. The transgene-containing chromosome was then homozygosed.
The strains containing the single-copy transgene in a HW background were injected with Psra-9::mCherry to identify ASK. Young adult hermaphrodites were examined on a Zeiss Imager Z.1 with a 60X objective focused on ASK using mCherry to prevent bleaching of GFP signal. Fluorescence signals were acquired with fixed acquisition times (30–50 msec for mCherry, 100 msec for GFP). Background mean fluorescence intensity adjacent to ASK was subtracted from the ASK signal.
RNAi
RNA interference was performed essentially as described 52. A fragment common to all tyra-3 isoforms was amplified. The following primers were used, which include the T7 sequence (underlined):
taatacgactcactatagggagagaaaatggcagcaggacttt
taatacgactcactatagggagaatcctcgcagtctgtggagt
in vitro transcription was performed with RiboMAX kit (Promega). dsRNA was injected at 1.2 μg/μL into the gonads of adult hermaphrodites. Eggs laid 24 and 48 hours after injection were used for the behavioural assays.
Octanol Avoidance Assay
Avoidance assays were conducted essentially as described 53. In brief, ~20 three-day old animals were picked off of their growth plates food into a transfer plate without bacteria where they were allowed to crawl and rid themselves of bacteria. Animals were then transferred onto an NGM plate without food. After 40 minutes, a microcapillary with 30% octanol (v/v diluted fresh every day in ethanol) was presented in front of the animal’s nose. The time to reverse was recorded. If animals did not reverse within 20 seconds, the assay was stopped. Animals were presented with odor 1–3 times per experiment, with at least 3 minutes of rest interval. We replicated published results demonstrating that tyra-3 null mutants had more rapid responses than N2 in the presence of exogenous serotonin and tyramine 29 but also observed more rapid responses in the absence of exogenous neuromodulators, as shown in Supplementary Fig. 6.
Cell Ablations
For leaving behaviour assays ASK was ablated with a laser microbeam as described 54. BAG was killed using split human caspase 3 fragments 55 expressed from flp-17 and glb-5 promoters that overlapped only in BAG. For lysine chemotaxis assays, ASK was killed using a mouse caspase 1 gene expressed from the sra-9 promoter 56. The ASK strain was a generous gift from Ryuzo Shingai.
Supplementary Material
Acknowledgments
We thank Ryuzo Shingai for strains and Patrick McGrath and members of the Bargmann lab for discussions. A.B. was supported by the Secretaría de Educación Pública of Mexico and by The Rockefeller University. C.I.B. and L.K. are Investigators of the Howard Hughes Medical Institute. This work was supported by HHMI and by NIH grant GM089972.
Footnotes
Author Contributions
A.B. and C.I.B. designed experiments, A.B. conducted experiments, M.V.R. constructed strains for QTL mapping, M.T. developed tracking methods, A.B., M.V.R., L.K., and C.I.B. analyzed and interpreted results, and A.B. and C.I.B. wrote the paper.
Author Information
The authors declare no competing financial interests.
References
- 1.Flint J, Mackay TF. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens DW, Brown JS, Ydenberg RC. Foraging: behavior and ecology. University of Chicago Press; 2007. [Google Scholar]
- 3.Charnov EL. Optimal foraging, the marginal value theorem. Theor Popul Biol. 1976;9:129–136. doi: 10.1016/0040-5809(76)90040-x. [DOI] [PubMed] [Google Scholar]
- 4.March JG. Exploration and exploitation in organizational learning. Organization science. 1991;2:71–87. [Google Scholar]
- 5.Barrett HC, Fiddick L. Evolution and risky decisions. Trends in Cognitive Sciences. 2000;4:251–252. doi: 10.1016/s1364-6613(00)01495-9. [DOI] [PubMed] [Google Scholar]
- 6.Aston-Jones G, Cohen JD. An integrative theory of locus coeruleus-norepinephrine function: adaptive gain and optimal performance. Annu Rev Neurosci. 2005;28:403–450. doi: 10.1146/annurev.neuro.28.061604.135709. [DOI] [PubMed] [Google Scholar]
- 7.Harvey SC. Non-dauer larval dispersal in Caenorhabditis elegans. J Exp Zool B Mol Dev Evol. 2009;312B:224–230. doi: 10.1002/jez.b.21287. [DOI] [PubMed] [Google Scholar]
- 8.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 10.Pradel E, et al. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipton J, Kleemann G, Ghosh R, Lints R, Emmons SW. Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J Neurosci. 2004;24:7427–7434. doi: 10.1523/JNEUROSCI.1746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gloria-Soria A, Azevedo RB. npr-1 Regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 2008;18:1694–1699. doi: 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 13.Styer KL, et al. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322:460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 16.Gray JM, et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- 17.Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol. 2006;16:649–659. doi: 10.1016/j.cub.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 18.Bretscher AJ, Busch KE, de Bono M. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:8044–8049. doi: 10.1073/pnas.0707607105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGrath PT, et al. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macosko EZ, et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osborne KA, et al. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. doi: 10.1126/science.277.5327.834. [DOI] [PubMed] [Google Scholar]
- 22.Reaume CJ, Sokolowski MB. cGMP-dependent protein kinase as a modifier of behaviour. Handb Exp Pharmacol. 2009:423–443. doi: 10.1007/978-3-540-68964-5_18. [DOI] [PubMed] [Google Scholar]
- 23.Hong RL, Witte H, Sommer RJ. Natural variation in Pristionchus pacificus insect pheromone attraction involves the protein kinase EGL-4. Proc Natl Acad Sci U S A. 2008;105:7779–7784. doi: 10.1073/pnas.0708406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinmetz LM, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 26.Edwards AC, Mackay TF. Quantitative trait loci for aggressive behavior in Drosophila melanogaster. Genetics. 2009;182:889–897. doi: 10.1534/genetics.109.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legare ME, Bartlett FS, 2nd, Frankel WN. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson MJ, Edwards JD, Septiningsih EM, Harrington SE, McCouch SR. Substitution mapping of dth1.1, a flowering-time quantitative trait locus (QTL) associated with transgressive variation in rice, reveals multiple sub-QTL. Genetics. 2006;172:2501–2514. doi: 10.1534/genetics.105.050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wragg RT, et al. Tyramine and octopamine independently inhibit serotonin-stimulated aversive behaviors in Caenorhabditis elegans through two novel amine receptors. JNeurosci. 2007;27:13402–13412. doi: 10.1523/JNEUROSCI.3495-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackay TF. Quantitative trait loci in Drosophila. Nat Rev Genet. 2001;2:11–20. doi: 10.1038/35047544. [DOI] [PubMed] [Google Scholar]
- 31.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 32.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakabayashi T, Kitagawa I, Shingai R. Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci Res. 2004;50:103–111. doi: 10.1016/j.neures.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Hallem EA, Sternberg PW. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:8038–8043. doi: 10.1073/pnas.0707469105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer M, et al. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron. 2009;61:865–879. doi: 10.1016/j.neuron.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frokjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suo S, Kimura Y, Van Tol HH. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J Neurosci. 2006;26:10082–10090. doi: 10.1523/JNEUROSCI.0819-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alkema MJ, Hunter-Ensor M, Ringstad N, Horvitz HR. Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron. 2005;46:247–260. doi: 10.1016/j.neuron.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 39.Greer ER, Perez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 2008;8:118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirri JK, McPherson AD, Donnelly JL, Francis MM, Alkema MJ. A tyramine-gated chloride channel coordinates distinct motor programs of a Caenorhabditis elegans escape response. Neuron. 2009;62:526–538. doi: 10.1016/j.neuron.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crocker A, Sehgal A. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28:9377–9385. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roeder T. Tyramine and octopamine: ruling behavior and metabolism. Annu Rev Entomol. 2005;50:447–477. doi: 10.1146/annurev.ento.50.071803.130404. [DOI] [PubMed] [Google Scholar]
- 43.Hoyer SC, et al. Octopamine in male aggression of Drosophila. Current Biology. 2008;18:159–167. doi: 10.1016/j.cub.2007.12.052. [DOI] [PubMed] [Google Scholar]
- 44.Yalcin B, et al. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- 45.Young LJ, Nilsen R, Waymire KG, MacGregor GR, Insel TR. Increased affiliative response to vasopressin in mice expressing the V1a receptor from a monogamous vole. Nature. 1999;400:766–768. doi: 10.1038/23475. [DOI] [PubMed] [Google Scholar]
- 46.Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 47.Gerke J, Lorenz K, Ramnarine S, Cohen B. Gene-environment interactions at nucleotide resolution. PLoS Genet. 6:e1001144. doi: 10.1371/journal.pgen.1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS One. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 50.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 51.Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechnique. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 52.Ahringer J WormBook. The C. elegans Research Community, WormBook. Apr 6, 2006. Reverse genetics. [Google Scholar]
- 53.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 54.Bargmann CI, Avery L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 1995;48:225–250. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chelur DS, Chalfie M. Targeted cell killing by reconstituted caspases. Proc Natl Acad Sci U S A. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim K, et al. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science. 2009;326:994–998. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.