Abstract
Background
The human multi-drug resistance gene (MDR1), which encodes the major trans-membrane transporter P-glycoprotein (P-gp), was found to be associated with susceptibility to cancer and response to chemotherapy. The C3435T Polymorphism of MDR1 gene was correlated with expression levels and functions of P-gp. Here, we studied the association between MDR1 C3435T polymorphism and susceptibility to Hodgkin lymphoma (HL) and patient's response to ABVD chemotherapy regimen.
Methods
a total of 130 paraffin embedded tissue samples collected from HL patients were analyzed to identify the C3435T polymorphism. As a control group, 120 healthy subjects were enrolled in the study. The C3435T Polymorphism was genotyped by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) method. Data analysis was carried out using the statistical package SPSS version 17 to compute all descriptive statistics. Chi-square and Fisher exact tests were used to evaluate the genotype distribution and allele frequencies of the studied polymorphism.
Results
these studies revealed that the frequency of T allele was significantly higher in HL patients compared to the controls (P < 0.05). In addition, the frequency of CT and TT genotypes were also significantly higher in HL patients compared to the controls (P < 0.05). No association between C3435T polymorphism and response to ABVD was detected among HL patients (P > 0.05).
Conclusions
these results suggest that MDR1 C3435T polymorphism might play a role in HL occurrence; however this polymorphism is not correlated with the clinical response to ABVD.
Keywords: Lymphoma, C3435T SNP, MDR-1
Background
Lymphomas are heterogeneous group of hematological malignancies that arise from malignant transformation of immune cells and account for 17% of all cancers in teenagers, and around 10% of childhood cancers [1]. Lymphomas are classified into two main types, Hodgkin's lymphoma (HL) and non-Hodgkin's lymphoma (NHL). The incidence of HL has risen gradually over the last few decades, representing a bimodal incidence peak, in early and late adulthood [1].
Several modalities are available to improve the overall survival in HL patients including radiotherapy, chemotherapy or combination of both [2]. However, the most commonly used regimen in the treatment of advanced stages of HL is the ABVD regimen containing doxorubicin (adriamycin), bleomycin, vinblastine and darcarbazine [3]. While more than 70% of HL patients are cured after treatment [3], about 30% of them might experience relapse after achieving initial complete remission (CR) [4]. This was attributed to the development of drug resistance, which might result from change in drug target sites or increased drug efflux by overexpression of drug transporters [5-7].
The multi-drug resistance (MDR) protein is a transporter that plays a primary role in drug resistance by affecting drug transport to cancer cells. MDR1 protein, called P-glycoprotein (P-gp), belongs to ATP-binding cassette superfamily [8]. A number of polymorphisms in the MDR1 gene were found to be of clinical importance, since they can alter drug absorption, distribution and elimination [9]. For example, the MDR1 C3435T polymorphism has been shown to affect the efficiency of chemotherapy in patients with lymphoproliferative diseases in a sample of the Europeoids of west Serbia [10].
While the association between the MDR1 C3435T polymorphism and NHL is well documented, the association between this polymorphism and HL has not been examined yet. In the present study, we investigated the association between the MDR1 C3435T polymorphism and the risk to develop HL, as well as the clinical response to ABVD chemotherapy regimen.
Methods
Studied groups
A total of 130 samples of paraffin-embedded tissue collected from HL patients were obtained from the Departments of Pathology at both Royal Medical Services and King Abdullah University Hospital. Patients included in the study are those of age more than 15-year old with HL, who received only ABVD regimen as initial chemotherapy. Patients were divided into two groups; complete response (n = 96) and relapsed disease (n = 34) according to International Workshop Criteria (IWC) [11].
Complete response (CR) was defined as 1) complete disappearance of all detectable evidence of disease on computed tomography (CT), 2) all disease-related symptoms, 3) normalization of biochemical abnormalities, 4) normal bone marrow biopsy, and 5) regression of nodes on CT of more than 1.5 cm in their axial diameter to less than 1.5 cm, and nodes of 1.1-1.5 to less than 1 cm. Relapsed disease (RD) was defined as: 1) the appearance of any new lesion 2) or increase in the size of more than 50% of previously involved sites or nodes in patients who achieved CR or Cru (uncertain). CRu corresponds to CR criteria but with a residual mass more than 1.5 cm in greatest axial diameter that has regressed by more than 75% [11].
Peripheral blood samples were collected from 120 healthy young volunteers as a control group from the same patient's geographical areas. Informed written consents were obtained from the participants in accordance with the requirements of the Institutional Review Boards of Jordan University of Science and Technology.
DNA extraction
DNA was extracted from paraffin embedded tissue samples using QIAamp DNA FFPE Tissue Kit (QIAGEN, California, USA) according to standard protocol provided by the manufacturer. Approximately, 3-5 sections of 5 μm thick were cut from each sample and used for DNA extraction. Venous blood samples were collected in EDTA tubes and obtained from young healthy control group. DNA was extracted from all blood samples using Promega wizard genomic DNA purification kit (Promega, Madison, USA). DNA samples were stored at -20°C until used.
Genotyping
The polymorphism C3435T was analyzed using polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) method. Desired DNA target sequence (197) was amplified as described by Cascorbi et al. [12] using a forward primer (5'-TGT TTT CAG CTG CTT GAT GG -3') and a reverse primer (5'-AAG GCA TGT ATG TTG GCC TC-3'). The reaction mixture of 25 μL contained 50 ng of genomic DNA, 0.5 μL of each primer, 12.5 μL of the green master mix, and 1.5-9.5 μL of deionized water. The reaction mixture was initially denatured at 94°C for 2 minutes, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 30 s. The termination elongation was performed at 72°C for 7 minutes. Successful amplification was confirmed by detection of a 197 bp band on a 2% agarose gel using a 100 bp DNA ladder. 10 μL of each PCR product was digested with 5 units of Sau3AI at 37°C overnight. The digested products were separated using 2.5% agarose gel and detected by ethidium bromide staining. Fragments obtained were 158 bp and 39 bp to the wild type genotype C/C, 197 bp to the mutant genotype T/T and 197 bp, 158 bp and 39 bp to the C/T genotype.
Statistical analysis
Data analysis was carried out using the statistical package SPSS version 17 to compute all descriptive statistics. Chi-square and Fisher exact tests were used to evaluate the genotype distribution and allele frequencies of the studied polymorphism. A P value of < 0.05 was considered statistically significant. Hardy-Weinberg equilibrium was assessed using the chi-square test. The C3435T genotypes were found to be in Hardy- Weinberg equilibrium.
Results
A hundred and thirty patients diagnosed with HL, the median age is 30 years, were included in the study. Fifty five percent are males and 47.7% have early stages of HL and complaining of B-symptoms. Most of the patients (76.2%) received 6 cycles of ABVD regimen. Other baseline characteristics of the patients are shown in Table 1. As a control, 120 healthy volunteers from the same geographical areas were enrolled (54% are males with median age of 23.5 years).
Table 1.
Demographic criteria of the patients
| Variable | Patients with Complete Remission (CR) N (%) | Patients with Relapsed Disease (RD) N (%) |
|---|---|---|
| Number | 96 | 34 |
| Age at diagnosis | ||
| Median | 31 | 27.5 |
| 15-20 | 16 (16.7) | 17 (50) |
| 21-30 | 32 (33.3) | 5 (14.7) |
| 31-40 | 18 (18.8) | 5 (14.7) |
| > 40 | 30 (31.2) | 8 (20.6) |
| Gender | ||
| Males | 50 (52.1) | 21 (61.8) |
| Females | 46 (47.9) | 13 (38.2) |
| Stage | ||
| Early stages (I &II) | 41 (42.7) | 20 (58.8) |
| Advanced stages (III & IV) | 38 (39.6) | 12 (35.3) |
| Missed data | 17 (17.7) | 2 (5.9) |
| Presence of B symptoms | ||
| Yes | 54 (56.3) | 19 (55.9) |
| No | 31 (32.3) | 13 (38.2) |
| Missed data | 11 (11.4) | 2 (5.9) |
| Bone marrow involvement | ||
| Yes | 5 (5.2) | 4 (11.8) |
| No | 91 (94.8) | 30 (88.2) |
| Histology | ||
| Nodular sclerosis | 46 (47.9) | 16 (47.1) |
| Mixed cellularity | 25 (26) | 6 (17.6) |
| Lymphocyte rich | 5 (5.2) | 3 (8.8) |
| Lymphocyte depleted | 4 (4.2) | 0 (0) |
| Nodular lymphocyte predominance | 1 (1) | 5 (14.7) |
| Classical | 7 (7.3) | 4 (11.8) |
| Missed data | 8 (8.3) | - |
| Chemotherapy regimen | ABVD: All the patients | ABVD: Initially all the patients at relapse: ICEa (8), ESHAPb (8), COPPc (3), ABVDd (8), Others: (7). |
| Number of ABVD cycles | ||
| < 6 cycles | 10 (10.4) | 6 (17.6) |
| 6 cycles | 77 (80.2) | 22 (64.7) |
| > 6 cycles | 9 (9.4) | 5 (14.7) |
aAdriamycin, Bleomycin, Vinblastine, Decarbazine; bIfosfamide, Carboplatin, Etoposide; cEtoposide, Cisplatin, Cytarabine, Methylprednisolone; dCyclophosphamide, Vincristine, Prednisolone, Procarbazine.
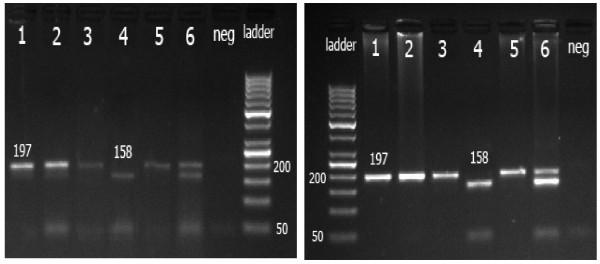
As shown in Figure 1, samples from paraffin embedded tissues and blood, were successfully genotyped using PCR-RFLP method. The mutant T allele does not carry the restriction site for Sau3AI enzyme and remains as 197 bp fragment, while the wild C allele cuts into two fragments of 158 and 39 bp.
Figure 1.
Gel electrophoresis of C3435T polymorphism from tissue samples. Left: The last lane from the right is 50 bp DNA ladder. Samples in lanes 1, 3 and 5 represent the PCR products and samples in lanes 2, 4 and 6, are the digest products of each sample, respectively. Sample in lane 2 is the mutant homozygous uncut TT genotype (197 bp). Sample in lane 4 represents the wild type cut CC genotype (158 bp and 39 bp). Sample in lane 6 represents heterozygous CT genotype (197 bp, 158 bp and 39 bp). Right: Gel electrophoresis of C3435T polymorphism from blood samples. The first lane from the left is 50 bp DNA ladder. Samples in lanes 1, 3 and 5 represent the PCR products and samples in lanes 2, 4 and 6, are the digest products of each sample, respectively. Sample in lane 2 is the mutant homozygous uncut TT genotype (197 bp). Sample in lane 4 represents the wild type cut CC genotype (158 bp and 39 bp). Sample in lane 6 represents heterozygous CT genotype (197 bp, 158 bp and 39 bp).
Results in Table 2 revealed that both C and T alleles are common in the studied population with approximately equal distribution. However, the patient group showed significantly (P value < 0.05) higher frequencies of both mutant T allele (65%) and TT homozygous mutant genotype (41%) compared to the control group. This indicates that the T allele in the C3435T polymorphism is associated with and HL occurrence.
Table 2.
Genotype and allele frequencies of C3435T polymorphism among HL patients and controls
| Genotypes & Alleles | HL patients (130) N (%) |
Controls (120) N (%) |
P-value |
|---|---|---|---|
| CC | 15 (11.5) | 37 (30.8) | |
| CT | 62 (47.7) | 48 (40.0) | 0.001 |
| TT | 53 (40.8) | 35 (29.2) | |
| Allele C | 92 (35.4) | 122 (50.8) | 0.000 |
| Allele T | 168 (64.6) | 118 (49.2) |
No significant association between the C3435T genotypes (CC, CT and TT) and alleles (C and T) with patient's baseline characteristics including patient's age, gender, specimen histology, stage of the disease and presence or absence of B-symptoms (Table 3 and 4), P value > 0.05.
Table 3.
Characteristics of patients according to C3435T genotypes
| Characteristics | CC genotype N (%) |
CT genotype N (%) |
TT genotype N (%) |
P-value |
|---|---|---|---|---|
| Age at diagnosis | ||||
| < 30 (n = 62) | 7 (46.7) | 28 (45.2) | 27 (50.9) | 0.823 |
| ≥ 30 (n = 68) | 8 (53.3) | 34 (54.8) | 26 (49.1) | |
| Gender | ||||
| Males (n = 71) | 7 (46.7) | 29 (46.8) | 35 (66) | 0.095 |
| Females (n = 59) | 8 (53.3) | 33 (53.2) | 18 (44) | |
| Histology | ||||
| NSa (n = 62) | 9 (64.3) | 32 (72.7) | 21 (60) | 0.481 |
| MCb (n = 31) | 5 (35.7) | 12 (27.3) | 14 (40) | |
| Stage | ||||
| Early stages (I &II) (n = 61) | 7 (50) | 30 (58) | 24 (53.3) | 0.842 |
| Advanced stages (III & IV) (n = 50) | 7 (50) | 22 (42) | 21 (46.7) | |
| Presence of B-symptoms | ||||
| Yes (n = 73) | 9 (60) | 36 (64.3) | 28 (60.9) | 0.920 |
| No (n = 44) | 6 (40) | 20 (35.7) | 18 (39.1) |
aNodular sclerosis; bMixed cellularity.
Table 4.
Characteristics of patients according to C3435T alleles
| Characteristics | C allele N (%) |
T allele N (%) |
Total | P-value |
|---|---|---|---|---|
| Age at diagnosis | ||||
| < 30 | 42 (45.7) | 82 (48.8) | 124 | 0.626 |
| ≥ 30 | 50 (54.3) | 86 (51.2) | 136 | |
| Gender | ||||
| Males | 43 (46.7) | 99 (58.9) | 142 | 0.059 |
| Females | 49 (53.3) | 69 (41.1) | 118 | |
| Histology | ||||
| NSa | 50 (69.4) | 74 (64.9) | 124 | 0.134 |
| MCb | 22 (30.6) | 40 (35.1) | 62 | |
| Stage | ||||
| Early stages (I &II) | 44 (55) | 78 (54.9) | 122 | 0.992 |
| Advanced stages (III & IV) | 36 (45) | 64 (45.1) | 100 | |
| Presence of B-symptoms | ||||
| Yes | 54 (62.8) | 92 (62.2) | 146 | 0.924 |
| No | 32 (37.2) | 56 (37.8) | 88 |
aNodular sclerosis; bMixed cellularity.
To verify whether different baseline characteristics of the patients might contribute to chemotherapy response, complete remission and disease relapse were studied according to the following criteria: age, gender, specimen histology, disease stage and presence or absence of B-symptoms (Table 5). None of these factors were associated with clinical response in HL patients (P value > 0.05).
Table 5.
The correlation between clinical outcome and patient's characteristics
| Baseline Factors | Complete Remission N (%) |
Relapsed Disease N (%) |
Total | P-value |
|---|---|---|---|---|
| Age at diagnosis | ||||
| < 30 | 43 (44.8) | 19 (55.9) | 62 | 0.266 |
| ≥ 30 | 53 (55.2) | 15 (44.1) | 68 | |
| Gender | ||||
| Males | 50 (52.1) | 21 (61.8) | 71 | 0.330 |
| Females | 46 (47.9) | 13 (38.2) | 59 | |
| Histology | ||||
| NSa | 46 (64.8) | 16 (72.7) | 62 | 0.490 |
| MCb | 25 (35.2) | 6 (27.3) | 31 | |
| Stage | ||||
| Early stages (I &II) | 41 (51.9) | 20 (62.5) | 61 | 0.309 |
| Advanced stages (III & IV) | 38 (48.1) | 12 (37.5) | 50 | |
| Presence of B-symptoms | ||||
| Yes | 54 (63.5) | 19 (59.4) | 73 | 0.679 |
| No | 31 (36.5) | 13 (40.6) | 44 |
aNodular sclerosis; bMixed cellularity.
Table 6 shows the genotype and allele frequencies of the C3435T polymorphism in HL patients with complete remission compared to those with relapse. No significant difference of CT and TT genotype distribution and allele frequency was found between the two groups (P value > 0.05).
Table 6.
Genotype and allele frequencies of C3435T polymorphism among patients according to the response
| Genotypes and Alleles | Complete Remission N (%) | Relapsed Disease N (%) |
P-value |
|---|---|---|---|
| CC | 12 (12.5) | 3 (8.8) | |
| CT | 44 (45.8) | 18 (52.9) | 0.729a |
| TT | 40 (41.7) | 13 (38.2) | |
| Allele C | 68 (35.4) | 24 (35.3) | 0.986 |
| Allele T | 124 (64.6) | 44 (64.7) |
aP value based on fisher exact test.
To identify possible correlation between the genotype and allele frequencies of the C3435T polymorphism and the progression free survival in relapsed group; patients were divided into two groups. The first include those having the relapse after one year of complete remission and the other group having the relapse during the first year of complete remission (Table 7). However, no significant difference in the frequencies of C3435T genotypes and the alleles was found. Thus, C3435T polymorphism seems to play no role in the progression free survival in the relapsed HL patients.
Table 7.
Genotype and allele frequencies of C3435T polymorphism among the relapsed group according to progression free survival
| Genotypes and Alleles | Progression free survival ≤ 1 year N (%) |
Progression free survival > 1 year N (%) | P-value |
|---|---|---|---|
| CC | 0 (0) | 3 (18.8) | |
| CT | 12 (66.7) | 6 (37.5) | 0.083a |
| TT | 6 (33.3) | 7 (43.7) | |
| Allele C | 12 (33.3) | 12 (37.5) | 0.720 |
| Allele T | 24 (66.7) | 20 (62.5) |
aP value based on fisher exact test.
Discussion
In this study, we investigated for the first time whether functional polymorphism C3425T in MDR1 gene could affect patient's susceptibility to HL and/or modify its response to chemotherapeutic agents. The results suggest that C3435T polymorphism plays a role in susceptibility to HL but not its response to ABVD chemotherapy. We analyzed MDR1 C3435T polymorphism in DNA isolated from paraffin embedded tissues taken from patient's lymph nodes while the same polymorphism was analyzed in the controls from peripheral blood tissues. This might raise some concern that the DNA from the two tissues is not equivalent because mutations are common during cancer progression. However, unlike most other malignant tumors, HL is characterized by low number of malignant cells that are surrounded by many non-neoplastic lymphocytes (reviewed in [13]).
The results indicate approximately equal distribution of the C and T alleles of C3425T polymorphism in the Jordanian population. This distribution is similar to that of Japanese [14], Caucasian [12], Chinese [15], Polish [16] and Malay [17] populations. However, the frequency of the T allele found in the present study is higher than that reported in Taiwanese [18], African [19], Jewish [20], Iranian [21], and Polish [22] populations, but lower than that of Czech [23] and Indian [17] populations (Table 8). Thus, the distribution of C3435T polymorphism seems to fall somewhere in the middle when compared with the Asian and European populations, which might be explained by the unique geographical location of Jordan at the crossing of Asia and Europe.
Table 8.
The frequency of 3435T allele among ethnic groups
| Ethnicity | 3435T allele Frequency (%) | Reference |
|---|---|---|
| Taiwanese (n = 110) | 37.3 | (Huang et al., 2005) |
| Japanese (n = 100) | 49.0 | (Tanabe et al., 2001) |
| Caucasians (n = 461) | 53.9 | (Cascorbi et al., 2001) |
| Africans (n = 206) | 17.0 | (Ameyaw et al., 2001) |
| Chinese in Singapore (n = 98) | 54.0 | (Balram et al., 2003) |
| Chinese in Mainland (n = 132) |
46.6 | (Ameyaw et al., 2001) |
| French (n = 227) | 46.0 | (Jeannesson et al., 2007) |
| Ashkenazi Jewish (n = 100) | 35.0 | (Ostrovsky et al., 2004) |
| Czech (n = 189) | 56.5 | (Pechandova et al., 2006) |
| Polish (n = 204) | 52.5 | (Kurzawski et al., 2006) |
| West Siberian Europeans (n = 59) |
59.0 | (Goreva et al., 2003) |
| Iranian (n = 300) | 33.5 | (Farnood et al., 2007) |
| Polish (175) | 40.0 | (Jamroziak et al., 2004) |
| Indians (n = 87) | 63.2 | (Chowbay et al., 2003) |
| Chinese (n = 96) | 53.1 | (Chowbay et al., 2003) |
| Malays (n = 92) | 51.1 | (Chowbay et al., 2003) |
| Jordanian (n = 120) | 49.2 | Present study |
Several genetic and environmental factors such as exposure to pesticides, wood dusts and chemicals were found to be associated with development of HL [24]. In here, we observed that C3435T polymorphism is significantly associated with susceptibility to HL. The homozygous mutant TT genotype and allele T frequencies were found to be higher in HL patients. Thus, our data may indicate that the C allele of C3435T polymorphism has protective role against HL. This could be explained by the low expression of T allele compared to C allele; thereby individuals with T allele are more prone to environmental toxins and carcinogens associated with HL. Previous studies suggest that the C3435T polymorphism is in linkage disequilibrium with other MDR1 polymorphisms such as C1236T and G2677T in exons 12 and 21, respectively. Thus, the contribution of those polymorphisms to susceptibility to HL observed in our study cannot be ruled out. In agreement with our results, Turgut, et al. [25] found a significant association between C3435T polymorphism and breast cancer. In the patient group, T allele frequency was significantly higher than controls. Similarly, the TT genotype of C3435T polymorphism was found to be associated with colon cancer risk [16]. The TT genotype was also associated with other malignancies such as acute lymphoblastic leukemia [22], renal cell carcinoma [26], and other diseases as ulcerative colitis [21]. In contrast, C3435T polymorphism was not associated with breast cancer in Iranian population [27]. Furthermore, C3435T variant was also not associated with acute leukemia in Turkish patients [28] and in childhood leukemia [29]. Thus, association between C3435T polymorphism and cancer development might have a population specific component. Moreover, a study by Humeny et al. [30] showed that MDR1 C3435T polymorphism is stable during carcinogenesis. Thus, it is unlikely that the observed strong association between HL and MDR1 C3435T polymorphism is due to mutations at the examined locus that are related to cancer progression.
A variety of mechanisms that may account for resistance of cancer cells to chemotherapy were described [31]. The most important one is the increase efflux of chemotherapeutic agents outside the cells by increasing the expression level of the major membrane transporter P-glycoprotein [6]. The MDR1 C3435T variant was found to alter P-gp function and expression, which might affect the disease response by modifying the pharmacokinetics of anticancer drugs. Therefore, several studies have shown the effect of C3435T MDR1 variant on disease outcome. In our study, we investigated the effect of C3435T variant on HL outcome in patients who received ABVD regimen containing common P-gp substrates adriamycin and vinblastine. According to the current results, C3435T variant was not associated with HL outcome in two groups of patients one with complete remission and the other with relapse. However, previous reports have shown that the C3435T polymorphism alters the response in different cancers. For example, the wild type genotype CC was associated with better chemotherapy response in patients with NSCLC [32,33] and in patients with SCLC [34]. On the other hand, CC genotype was linked significantly with increased risk of relapse in AML patients [35]. Furthermore, our study revealed no significant association between progression free survival and C3435T genotype and allele frequencies. However, previous studies have shown the effect of C3435T variant on survival time in cancer patients. The CC genotype was associated with a shorter overall survival in patient's with multiple myloma [36] and in patients with ALL [22] compared to both CT and TT genotypes. This difference in the results may be related to the variation in the genetic background of the studied groups, or life style or due to other unknown factors.
Results of this study show no significant association between HL response and patient's characteristics such as age, gender, HL stage, specimen histology and presence or absence of B-symptoms. In addition, the distribution of C3435T genotypes and alleles was not associated with patient's characteristics. Therefore, possibilities exist that other polymorphisms in the MDR1 gene might be involved in modulating HL response to drugs in the Jordanian population. Thus, scanning the MDR1 gene to search for common and new variants in the Jordanian population is important for future pharmacogenetic studies in this population.
In conclusion, results of this study show that C3435T polymorphism is associated with susceptibility to HL in Jordanian population. However, this variant is not correlated with the drug response or clinical parameters in HL patients.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NM, OK, OA, and AA carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. IOM participated in the sequence alignment. NM, OK, KA and OA participated in the design of the study and performed the statistical analysis. WH and IIM have participated in the study design and samples collection and preparation for perform the study. NM and KA helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Nizar M Mhaidat, Email: nizarm@just.edu.jo.
Osama Y Alshogran, Email: O_shogran@yahoo.com.
Omar F Khabour, Email: Khabour@just.edu.jo.
Karem H Alzoubi, Email: khalzoubi@just.edu.jo.
Ismail I Matalka, Email: imatalka@hotmail.com.
William J Haddadin, Email: haddadin@hotmail.com.
Ibraheem O Mahasneh, Email: ibrahimma16@yahoo.com.
Ahmad N Aldaher, Email: dnaseve@yahoo.com.
Acknowledgements
We would like to acknowledge the Jordan University of Science & Technology, Irbid, Jordan, for the financial support (Grant Number 176/2009).
References
- Morley-Jacob C, Gallop-Evans E. Update on Lymphoma. Pediatrics and child health. 2008;18:3. [Google Scholar]
- Rueda A, Olmos D, Viciana R, and Alba E. Treatment for relapse in stage I/II Hodgkin's lymphoma after initial single-modality treatment. Clin Lymphoma Myeloma. 2006;6:389–392. doi: 10.3816/CLM.2006.n.015. [DOI] [PubMed] [Google Scholar]
- Castagna L, Magagnoli M, Demarco M, and Santoro A. Lymphomas. update on cancer therapeutics. 2007. pp. 101–110.
- Quddus F, Armitage JO. Salvage therapy for Hodgkin's lymphoma. Cancer J. 2009;15:161–163. doi: 10.1097/PPO.0b013e3181a1438a. [DOI] [PubMed] [Google Scholar]
- Desoize B, Jardillier J. Multicellular resistance: a paradigm for clinical resistance? Crit Rev Oncol Hematol. 2000;36:193–207. doi: 10.1016/S1040-8428(00)00086-X. [DOI] [PubMed] [Google Scholar]
- Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, and Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- Burger H, Foekens JA, Look MP, Meijer-van Gelder ME, Klijn JG, Wiemer EA, Stoter G, Nooter K. RNA expression of breast cancer resistance protein, lung resistance-related protein, multidrug resistance-associated proteins 1 and 2, and multidrug resistance gene 1 in breast cancer: correlation with chemotherapeutic response. Clin Cancer Res. 2003;9:827–836. [PubMed] [Google Scholar]
- Ishikawa T, Hirano H, Onishi Y, Sakurai A, and Tarui S. Functional evaluation of ABCB1 (P-glycoprotein) polymorphisms: high-speed screening and structure-activity relationship analyses. Drug Metab Pharmacokinet. 2004;19:1–14. doi: 10.2133/dmpk.19.1. [DOI] [PubMed] [Google Scholar]
- Goreva OB, Grishanova AY, Mukhin OV, Domnikova NP, Lyakhovich VV. Possible prediction of the efficiency of chemotherapy in patients with lymphoproliferative diseases based on MDR1 gene G2677T and C3435T polymorphisms. Bull Exp Biol Med. 2003;136:183–185. doi: 10.1023/a:1026331326648. [DOI] [PubMed] [Google Scholar]
- Hampson FA, Shaw AS. Response assessment in lymphoma. Clin Radiol. 2008;63:125–135. doi: 10.1016/j.crad.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Cascorbi I, Gerloff T, Johne A, Meisel C, Hoffmeyer S, Schwab M, Schaeffeler E, Eichelbaum M, Brinkmann U, Roots I. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69:169–174. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- Chan WC. The Reed-Sternberg cell in classical Hodgkin's disease. Hematol Oncol. 2001;19:1–17. doi: 10.1002/hon.659. [DOI] [PubMed] [Google Scholar]
- Tanabe M, Ieiri I, Nagata N, Inoue K, Ito S, Kanamori Y, Takahashi M, Kurata Y, Kigawa J, Higuchi S, Terakawa N, Otsubo K. Expression of P-glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)-1 gene. J Pharmacol Exp Ther. 2001;297:1137–1143. [PubMed] [Google Scholar]
- Balram C, Sharma A, Sivathasan C, Lee EJ. Frequency of C3435T single nucleotide MDR1 genetic polymorphism in an Asian population: phenotypic-genotypic correlates. Br J Clin Pharmacol. 2003;56:78–83. doi: 10.1046/j.1365-2125.2003.01820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzawski M, Drozdzik M, Suchy J, Kurzawski G, Bialecka M, Gornik W, Lubinski J. Polymorphism in the P-glycoprotein drug transporter MDR1 gene in colon cancer patients. Eur J Clin Pharmacol. 2005;61:389–394. doi: 10.1007/s00228-005-0926-5. [DOI] [PubMed] [Google Scholar]
- Chowbay B, Cumaraswamy S, Cheung YB, Zhou Q, Lee EJ. Genetic polymorphisms in MDR1 and CYP3A4 genes in Asians and the influence of MDR1 haplotypes on cyclosporin disposition in heart transplant recipients. Pharmacogenetics. 2003;13:89–95. doi: 10.1097/00008571-200302000-00005. [DOI] [PubMed] [Google Scholar]
- Huang MJ, Yung LC, Chang YC, Yang YH, Ching SH. Polymorphisms of the Gene Encoding Multidrug Resistance Protein 1 in Taiwanese. Journal of Food and Drug Analysis. 2005;13:112–117. [Google Scholar]
- Ameyaw MM, Regateiro F, Li T, Liu X, Tariq M, Mobarek A, Thornton N, Folayan GO, Githang'a J, Indalo A, Ofori-Adjei D, Price-Evans DA, McLeod HL. MDR1 pharmacogenetics: frequency of the C3435T mutation in exon 26 is significantly influenced by ethnicity. Pharmacogenetics. 2001;11:217–221. doi: 10.1097/00008571-200104000-00005. [DOI] [PubMed] [Google Scholar]
- Ostrovsky O, Nagler A, Korostishevsky M, Gazit E, Galski H. Genotype and allele frequencies of C3435T polymorphism of the MDR1 gene in various Jewish populations of Israel. Ther Drug Monit. 2004;26:679–684. doi: 10.1097/00007691-200412000-00015. [DOI] [PubMed] [Google Scholar]
- Farnood A, Naderi N, Moghaddam SJ, Noorinayer B, Firouzi F, Aghazadeh R, daryani NE, Zali MR. The frequency of C3435T MDR1 gene polymorphism in Iranian patients with ulcerative colitis. Int J Colorectal Dis. 2007;22:999–1003. doi: 10.1007/s00384-007-0270-6. [DOI] [PubMed] [Google Scholar]
- Jamroziak K, Mlynarski W, Balcerczak E, Mistygacz M, Trelinska J, Mirowski M, Bodalski J, Robak T. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol. 2004;72:314–321. doi: 10.1111/j.1600-0609.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- Pechandova K, Buzkova H, Slanar O, Perlik F. Polymorphisms of the MDR1 gene in the Czech population. Folia Biol (Praha) 2006;52:184–189. doi: 10.14712/fb2006052060184. [DOI] [PubMed] [Google Scholar]
- Landgren O, Caporaso NE. New aspects in descriptive, etiologic, and molecular epidemiology of Hodgkin's lymphoma. Hematol Oncol Clin North Am. 2007;21:825–840. doi: 10.1016/j.hoc.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Turgut S, Yaren A, Kursunluoglu R, Turgut G. MDR1 C3435T polymorphism in patients with breast cancer. Arch Med Res. 2007;38:539–544. doi: 10.1016/j.arcmed.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Siegsmund M, Brinkmann U, Schaffeler E, Weirich G, Schwab M, Eichelbaum M, Fritz P, Burk O, Decker J, Alken P, Rothenpieler U, Kerb R, Hoffmeyer S, Brauch H. Association of the P-glycoprotein transporter MDR1(C3435T) polymorphism with the susceptibility to renal epithelial tumors. J Am Soc Nephrol. 2002;13:1847–1854. doi: 10.1097/01.ASN.0000019412.87412.BC. [DOI] [PubMed] [Google Scholar]
- Tatari F, Salek R, Mosaffa F, Khedri A, Behravan J. Association of C3435T single-nucleotide polymorphism of MDR1 gene with breast cancer in an Iranian population. DNA Cell Biol. 2009;28:259–263. doi: 10.1089/dna.2008.0826. [DOI] [PubMed] [Google Scholar]
- Kaya P, Gunduz U, Arpaci F, Ural AU, Guran S. Identification of polymorphisms on the MDR1 gene among Turkish population and their effects on multidrug resistance in acute leukemia patients. Am J Hematol. 2005;80:26–34. doi: 10.1002/ajh.20427. [DOI] [PubMed] [Google Scholar]
- Urayama KY, Wiencke JK, Buffler PA, Chokkalingam AP, Metayer C, Wiemels JL. MDR1 gene variants, indoor insecticide exposure, and the risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol Biomark Prev. 2007;16:1172–1177. doi: 10.1158/1055-9965.EPI-07-0007. [DOI] [PubMed] [Google Scholar]
- Humeny A, Rödel F, Rödel C, Sauer R, Füzesi L, Becker C, Efferth T. MDR1 single nucleotide polymorphism C3435T in normal colorectal tissue and colorectal carcinomas detected by MALDI-TOF mass spectrometry. Anticancer Res. 2003;23:2735–40. [PubMed] [Google Scholar]
- Larsen AK, Escargueil AE, Skladanowski A. Resistance mechanisms associated with altered intracellular distribution of anticancer agents. Pharmacol Ther. 2000;85:217–229. doi: 10.1016/S0163-7258(99)00073-X. [DOI] [PubMed] [Google Scholar]
- Pan JH, Han JX, Wu JM, Huang HN, Yu QZ, Sheng LJ. MDR1 single nucleotide polymorphism G2677T/A and haplotype are correlated with response to docetaxel-cisplatin chemotherapy in patients with non-small-cell lung cancer. Respiration. 2009;78:49–55. doi: 10.1159/000158454. [DOI] [PubMed] [Google Scholar]
- Pan JH, Han JX, Wu JM, Sheng LJ, Huang HN, Yu QZ. MDR1 single nucleotide polymorphisms predict response to vinorelbine-based chemotherapy in patients with non-small cell lung cancer. Respiration. 2008;75:380–385. doi: 10.1159/000108407. [DOI] [PubMed] [Google Scholar]
- Sohn JW, Lee SY, Lee SJ, Kim EJ, Cha SI, Kim CH, Lee JT, Jung TH, Park JY. MDR1 polymorphisms predict the response to etoposide-cisplatin combination chemotherapy in small cell lung cancer. Jpn J Clin Oncol. 2006;36:137–141. doi: 10.1093/jjco/hyi231. [DOI] [PubMed] [Google Scholar]
- Illmer T, Schuler US, Thiede C, Schwarz UI, Kim RB, Gotthard S, Freund D, Schakel U, Ehninger G, Schaich M. MDR1 gene polymorphisms affect therapy outcome in acute myeloid leukemia patients. Cancer Res. 2002;62:4955–4962. [PubMed] [Google Scholar]
- Buda G, Maggini V, Galimberti S, Martino A, Giuliani N, Morabito F, Genestreti G, Iacopino P, Rizzoli V, Barale R, Rossi AM, Petrini M. MDR1 polymorphism influences the outcome of multiple myeloma patients. Br J Haematol. 2007;137:454–456. doi: 10.1111/j.1365-2141.2007.06605.x. [DOI] [PubMed] [Google Scholar]