Abstract
Two independent lines of evidence support the localization of a schizophrenia susceptibility locus to the proximal long arm of chromosome 5. A partial trisomy of chromosome 5 (5q11.2–q13.3) cosegregates with the disorder in a Canadian family of Chinese descent, and DNA markers from proximal 5q cosegregate with schizophrenia (plus related disorders) in families of British and Icelandic descent. We constructed a human:hamster hybrid cell line (HHW 1064) whose only human complement is a chromosome 5 that is missing the trisomic region associated with schizophrenia. In combination with a “matched” cell hybrid (HHW 105) containing an intact chromosome 5, we physically mapped DNA markers relative to the trisomy. “Schizophrenia-linked” DNA markers p105–153Ra (D5S39) and p105-599Ha (D5S76) map within the trisomy and proximal to the 5q11.2 breakpoint, respectively. The hybrid cell lines HHW 105 and HHW 1064 together provide a means to identify and generate syntenic DNA markers to further investigate the location of a schizophrenia locus.
A report by Bassett et al. (1988) describes the coin-heritance of a chromosomal triplication, 5q11.2–5q13.3, with schizophrenia in a well-characterized Canadian family of Chinese descent. The two affected members of this family, a 20-year-old man and his 53-year-old uncle, share a phenotype of neuroleptic responsive schizophrenia with typical psychotic and deficit symptoms. The affected individuals also suffer mild physical anomalies which prompted clinicians to investigate and subsequently discover a chromosomal abnormality associated with the occurrence of schizophrenia in this family. High-resolution karyotyping revealed a balanced direct insertion (46, XX, inv ins) (1;5) (q32.3; q13.3–q11.2) in an unaffected relative (the mother and sister, respectively) of the two affected probands. Both affected individuals were trisomic for the translocated 5q segment, whereas other unaffected relatives had normal genomic karyotypes. This finding encouraged several laboratories to test DNA markers from the long arm of chromosome 5 for linkage to the disease phenotype in large schizophrenia pedigrees. Recently, one group has reported linkage with markers from the proximal portion of 5q to seven British and Icelandic families (Sherrington et al., 1988), while several groups report the absence of linkage in other kindreds (Kennedy et al., 1988; Kaufmann et al., 1989; St. Clair et al., 1989)
In this study we test whether DNA markers, reportedly in linkage with “schizophrenia phenotype” (Sherrington et al., 1988), map to the region of the schizophrenia-associated chromosomal triplication. Chinese hamster ovary cell line (CHO) UCW56 was fused to lymphoblastoid cells from the individual referred to above with a chromosomal rearrangement, dir ins (46, XX, inv ins) (1;5) (q32.3; q13.3–q11.2). Hybrids that contained a human chromosome 5 under selective pressure (growth at 39°C) were isolated as described previously (Dana and Wasmuth, 1982). The only human chromosome present in line HHW 1064 is the deleted chromosome 5 shown in Fig. 1. This cell line along with a “matched” control cell line (HHW 105) (Dana and Wasmuth, 1982) was used to map seven DNA markers from proximal 5q to this area.
FIG. 1.

Trypsin–Giemsa-banded metaphase chromosome preparation from hybrid HHW 1064. The Chinese hamster ovary (CHO) line UCW56 was fused to lymphoblastoid cells from an individual with the chromosomal rearrangement dir ins (46, XX, inv ins)(1;5)(q32.3; q13.3–q11.2). Hybrids that contained a human chromosome 5 under selective pressure (growth at 39°C) were isolated as described previously (4). Metaphase chromosome preparations were stained with trypsin–Giemsa (G-banded), photographed, and then destained and restained by the alkaline–Giemsa (G-11) procedure to unequivocally identify human chromosome 5 (4). The only human chromosome present in HHW 1064, the deleted chromosome 5 del (5) (5pter–5q11.2::5q13.3–5qter), is indicated by an arrow.
Several DNA markers that map to the proximal long arm of chromosomal 5 have been identified (Leppert et al., 1987; Giuffra et al., 1988). A collection of these markers, including those used in the schizophrenia linkage studies described recently, has been tested for localization to 5q11.2–q13.3. Each DNA marker was hybridized to a panel containing restriction-digested DNA from the following sources: human lymphoblast; HHW 105 (only human chromosome 5 in CHO cells) (Dana and Wasmuth, 1982); HHW 1064 (only human chromosome 5 with 5q11.2–q13.3 deletion in CHO cells); and CHO cells. Two of the resultant autoradiograms are shown in Fig. 2. DNA markers pJO110HC (D5S21), p105-599Ha (D5S76), pC11p11 (D5S71), OB7 (glucocorticoid receptor) (Hollenberg et al., 1985), and serotonin A1 receptor (G21) (Kobilka et al., 1987) were present in both HHW 105 and HHW 1064, indicating that they are located outside the deleted chromosomal region. Markers M4 (D5S6) (Dietzsch et al., 1988), p105-153Ra (D5S39), p105-798Rb (D5S78), Hex B (Korneluk et al., 1986), DHFR (dihydrofolate reductase) (Chen et al., 1984), CRI-L407 (D5S63), and CRI-L1155 (D5S51) were missing in HHW 1064 but present in HHW 105, assigning them to the deleted region, 5q11.2–q13.3. All results were confirmed with two different restriction enzyme digests.
FIG. 2.

Deletion mapping of chromosome 5 DNA markers to the trisomic region associated with schizophrenia. DNA markers were radiolabeled and hybridized to Southern blots as described previously (6). DNA samples were restriction digested with either HindIII or EcoRI, separated electrophoretically, and transferred to nylon membranes for hybridization. Each blot contained the following lanes of digested DNA: Hu, human lymphoblast DNA: Ha, Chinese hamster ovary DNA: 5, hybrid cell line HHW 105 containing a normal human chromosome 5 (4); 5−, hybrid cell line HHW 1064, containing a human chromosome 5 missing 5q11.2–q13.3. Radiolabeled bacteriophage λ DNA digested with EcoRI is the size marker. (A) D5S21 (pJO110HC), HindIII digest; (B) D5S39 (p105-153Ra), HindIII digest.
Figure 3 shows a physical map of the DNA markers tested for inclusion or exclusion to the region 5q11.2–q13.3. Seven of the eleven markers tested are within the deleted region (indicated in bold type). The genetic order and distances of several of the markers have been previously reported and are indicated to the right of the figure. On the basis of the combined physical and genetic data, we predict the following sequence of marker and gene loci: D5S21 and D5S76 lie outside and proximal to the deletion; D5S6, D5S39, HexB, D5S78, D5S63, D5S51, and DHFR lie within the deletion; and pC11p11 and OB7 map outside and distal to the deletion. The serotonin A1 receptor probe G21 also maps outside the deletion region (Kobilka et al., 1987). HexB has previously been mapped within the trisomy region on the basis of dosage estimation (Wood et al., 1988), which is consistent with our result.
FIG. 3.
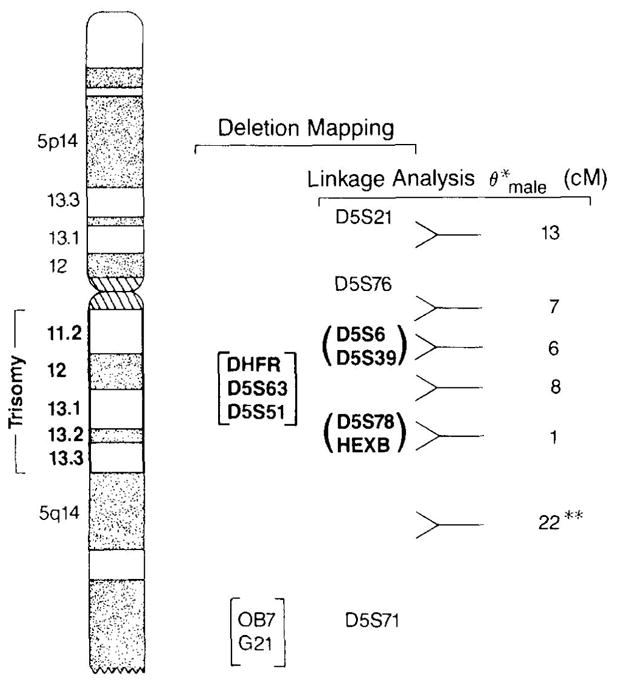
Map of the trisomic region. The trisomic region is indicated to the left of chromosome 5. Seven markers mapped within the trisomic region (loci indicated in bold type) and five markers mapped outside the region of chromosome 5. The male recombination fractions for some of the markers are listed to the right. Markers listed within parentheses may be reversed in order, and markers listed in brackets are of unknown genetic order. *Male recombinant fractions as reported in Ref. (8); **male recombinant fraction between D5S78 and D5S71 as reported in Ref. (15).
The estimated genetic distance from D5S76 to D5S6 is 7–8 cM (Guiffra, Kennedy, and Kidd, personal communication). Although it is also possible that D5S6 maps distal to D5S39, physical and genetic mapping indicates that D5S76 and D5S6 border the proximal translocation breakpoint at 5q11.2. We estimate that the genetic distance between D5S6 and HexB/D5S78 is 13–20 cM (male recombination fraction), which provides a minimum size estimate of the trisomic region.
To directly address the concordance of the genetic linkage (Sherrington et al., 1988) and karyotypic (Bassett et al., 1988) evidence for a schizophrenia locus on chromosome 5, we have hybridized DNA markers used in the schizophrenia linkage studies to hybrid cell lines containing either a normal human chromosome 5 or a chromosome 5 that is missing the region of trisomy. One of the schizophrenia-linked markers lies within the trisomy region, while the other maps proximal to the 5q11.2 translocation breakpoint, which is consistent with, though not proof of, a schizophrenia susceptibility locus at 5q11.2–5q13.3. A human:hamster hybrid cell line (HHW 1064) which harbors a chromosome 5 deleted for the trisomic region will provide a useful resource for cloning and mapping DNA markers in the vicinity of the proposed disease locus. We are currently using HHW 1064 along with the “matched” hybrid cell HHW 105 to enrich by subtraction hybridization (Kunkel et al., 1985; Casna et al., 1986) for DNA sequences from the deletion region. We are also screening chromosome 5 cell-sorted libraries (American Type Culture Collection) for DNA sequences that map to 5q11.2–q13.3 based on hybridization to the deletion panel illustrated in Fig. 2 (Gilliam et al., 1987b).
The isolation of additional informative DNA markers from proximal 5q should allow verification of the schizophrenia map position and (if verified) better definition of the disease gene region. Although the independent reports of a schizophrenia locus at 5q are very encouraging, the complexity of data analysis, which necessarily involves several assumptions (for example, mode of inheritance, penetrance of the susceptibility locus, and determination and “boundaries” of disease status), still leaves some ambiguity regarding the weight of evidence in favor of a disease locus at 5q. If a chromosome 5 mutation accounts for only a small proportion of schizophrenia cases, it may prove difficult to verify the linkage studies with additional family analysis. Support for the initial findings can be obtained through development of additional polymorphic DNA markers in the 5q11.2–13.3 region, since markers that lie between D5S76 and D5S39 must support linkage in the British and Icelandic families if the assumptions regarding disease characterization are correct.
If the trisomy at proximal 5q and the DNA markers D5S39 and D5S76 identify an identical susceptibility locus, then the cell hybrids will be useful for mapping the locus. The imposition of physical breakpoint boundaries for the disease locus at 5q11.2 and 5q13.3 should, in concert with statistically determined genetic confidence intervals, facilitate the definition of an “obligate chromosomal region” that contains a susceptibility locus and that is sufficiently limited to allow physical mapping of the entire region (Smith and Cantor, 1986). If the genetic confidence interval spans a translocation breakpoint, the physical boundary will serve to delimit the obligate disease locus region. At present, both the genetic and the karyotypic data define chromosomal regions much too large for physical mapping studies, which are a necessary prelude to a systematic search for the susceptibility locus. Identification of additional highly polymorphic DNA markers from the trisomic region will allow multipoint linkage analysis with the disease locus and it is hoped, will further refine its location.
This study verifies that the two independently derived map positions for a schizophrenia susceptibility locus do actually overlap. Construction of a hybrid cell line lacking just the relevant region of 5q (q11.2–q13.3) provides a resource to facilitate the physical and genetic characterization of the locus.
Acknowledgments
We thank Kris Kumaraswamy and Dr. Kamna Das for their assistance in the growth and maintenance of hybrid cell lines. The following individuals generously provided DNA clones: Drs. M. Caron, S. Hollenberg, A. E. Retief, R. A. Gravel, and A. W. Nienhuis. This work was supported by the W. M. Keck Foundation (TCG) and NIH Grant CM25339 (J.J.W.). C.A.K. is supported by PSA Award 5K11MH00682. P.P.P. and A.S.B. are W. M. Keck Foundation Scholars.
References
- 1.Bassett AS, Jones BD, McGillivray BC, Pantzer JT. Partial trisomy chromosome 5 cosegregating with schizophrenia. Lancet. 1988;1:799–801. doi: 10.1016/s0140-6736(88)91660-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casna NJ, Novak DF, Hsu MT, Ford FP. Genomic analysis. II. Isolation of high molecular weight DNA following differential methylase protection and Formamide–PERT hybridization. Nucleic Acids Res. 1986;14:7285–7303. doi: 10.1093/nar/14.18.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen MJ, Shimada T, Noulton AD, Cline A, Humphries RK, Maizel J, Nienhuis AW. The functional human dihydrofolate reductase gene. J Biol Chem. 1984;259:3933–3943. [PubMed] [Google Scholar]
- 4.Dana S, Wasmuth JJ. Selective linkage disruption in human–Chinese hamster cell hybrids: Deletion mapping of the leuS, emtB, and chr genes on human chromosome 5. Mol Cell Biol. 1982;2:1220–1228. doi: 10.1128/mcb.2.10.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietzsch E, Retief AE, Lotze MJ, Warnich L, Nicholson DL, Fox MF, Fricke J, du Plessis L, Oosthuizen CJJ. An anonymous human single copy genomic clone, D5S6 (M4), on chromosome 5 identifies a three allele RFLP. Nucleic Acids Res. 1988;14:1923. doi: 10.1093/nar/14.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilliam TC, Tanzi RE, Haines JL, Bonner TI, Faryniarz AG, Hobbs WJ, MacDonald ME, Cheng SV, Folstein SE, Conneally PM, Wexler NS, Gusella JF. Localization of the Huntington’s disease gene to a small segment of chromosome 4 flanked by D4S10 and the telomere. Cell. 1987a;50:565–571. doi: 10.1016/0092-8674(87)90029-8. [DOI] [PubMed] [Google Scholar]
- 7.Gilliam TC, Healey ST, MacDonald ME, Stewart GD, Wasmuth JJ, Tanzi RE, Roy JC, Gusella JF. Isolation of polymorphic DNA fragments from human chromosome 4. Nucleic Acids Res. 1987b;15:1445–1458. doi: 10.1093/nar/15.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiffra LA, Kennedy JL, Castiglione CM, Evans RM, Wasmuth JJ, Kidd KK. Glucocorticoid receptor maps to the distal long arm of chromosome 5. Cytogenet Cell Genet. 1988;49:313–314. doi: 10.1159/000132686. [DOI] [PubMed] [Google Scholar]
- 9.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature (London) 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufmann CA, Delisi LE, Lehner T, Gilliam TC. Physical mapping and linkage analysis of a susceptibility locus for schizophrenia on chromosome 5q. Schizophrenia Bull. 1989 doi: 10.1093/schbul/15.3.441. in press. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy JL, Guiffra LA, Noises HW, Cavilli-Sforza LL, Pakstis AJ, Kidd JR, Castiglione CM, Sjogren B, Wetterberg L, Kidd KK. Evidence against linkage of schizophrenia to markers on chromosome 5 in a northern Swedish pedigree. Nature (London) 1988;336:167–170. doi: 10.1038/336167a0. [DOI] [PubMed] [Google Scholar]
- 12.Kobilka BK, Frielle T, Collins S, Xang-Feng T, Kobilka TS, Franke U, Lefkowitz RJ, Caron MG. An intronless gene encoding a potential member of the family of receptors coupled to guanine nucleotide regulatory proteins. Nature (London) 1987;329:75–79. doi: 10.1038/329075a0. [DOI] [PubMed] [Google Scholar]
- 13.Korneluk RG, Mahuran DJ, Neote K, Klavins MH, O’Dowd BF, Tropak M, Willard HF, Anderson M, Lowden JA, Gravel RA. Isolation of cDNA clones coding for the alpha-subunit of human beta-hexosaminidase. J Biol Chem. 1986;261:8407–8413. [PubMed] [Google Scholar]
- 14.Kunkel LM, Monaco AP, Middleworth W, Ochs HD, Latt SA. Specific cloning of DNA fragments absent from the DNA of a male patient with an X chromosome deletion. Proc Natl Acad Sci USA. 1985;82:4778–4782. doi: 10.1073/pnas.82.14.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leppert M, Dobbs M, Scambler P, O’Connell P, Nakamura Y, Stauffer D, Woodward S, Burt R, Hughes J, Gardner E, Lathrop M, Wasmuth J, Lalouel J-M, White R. The gene for familial polyposis coli maps to the long arm of chromosome 5. Science. 1987;238:1411–1413. doi: 10.1126/science.3479843. [DOI] [PubMed] [Google Scholar]
- 16.Sherrington R, Brynjolfsson J, Petursson H, Potter M, Dudleston K, Barraclough B, Wasmuth J, Dobbs M, Gurling H. Localization of a susceptibility locus for schizophrenia on chromosome 5. Nature (London) 1988;336:164–167. doi: 10.1038/336164a0. [DOI] [PubMed] [Google Scholar]
- 17.Smith CL, Cantor CR. Approaches to physical mapping of the human genome. Cold Spring Harbor Symp Quant Biol. 1986;51:115–122. doi: 10.1101/sqb.1986.051.01.014. [DOI] [PubMed] [Google Scholar]
- 18.St Clair D, Blackwood D, Muir W, Baillie D, Hubbard A, Wright A, Evans JH. No linkage of chromosome 5q11–q13 markers to schizophrenia in Scottish families. Nature (London) 1989;339:305–309. doi: 10.1038/339305a0. [DOI] [PubMed] [Google Scholar]
- 19.Wood S, Bassett AS, Jones BD, Langlois S, Pantzar TP, McGillivray BC. Partial chromosome 5 trisomy and schizophrenia in two families members. Proceedings, XVIth International Congress of Genetics; Toronto, Canada. 1988. [Google Scholar]


