Abstract
Polyvalent oligonucleotide-nanoparticle conjugates possess several unique emergent properties including enhanced cellular uptake, high antisense bioactivity, and nuclease resistance, which hypothetically originate from the dense packing and orientation of oligonucleotides on the surface of the nanoparticle. In this communication, we describe a new class of polyvalent nucleic acid nanostructures (PNANs), which comprise only crosslinked and oriented nucleic acids. We demonstrate that these particles are capable of effecting high cellular uptake and gene regulation without the need of a cationic polymer co-carrier. The PNANs also exhibit cooperative binding behavior and nuclease resistance properties.
The natural defenses of biological systems for exogenous oligonucleotides, such as synthetic antisense DNA and siRNA, present many challenges for the delivery of nucleic acids in an efficient, non-toxic and non-immunogenic fashion.1–3 Indeed, because nucleic acids are negatively charged and prone to enzymatic degradation, researchers have historically relied on transfection agents such as cationic polymers,4–6 liposomes,7 and modified viruses8 to facilitate cellular entry and protect DNA from degradation. However, each of these materials is subject to several drawbacks, which include toxicity at high concentrations, inability to be degraded biologically, and severe immunogenicity.9 The recently introduced polyvalent nucleic acid-nanoparticle (NP) conjugates (inorganic nanoparticles densely coated with highly oriented oligonucleotides) pose one possible solution of circumventing these problems in the context of both antisense and RNAi pathways.10–15 Remarkably, these highly negatively charged structures (zeta potential <-30 mV) do not require cationic transfection materials or additional particle surface modifications and naturally enter all cell lines tested to date (over 50, including primary cells). Further work has shown the cellular uptake of these particles to be dependent upon DNA surface density; higher densities lead to higher levels of particle uptake.16 The high-density polyvalent nucleic acid surface layer is believed to recruit scavenger receptors from the cells that facilitate endocytosis.16,17 Moreover, the ion cloud associated with the high-density oligonucleotide shell combined with steric inhibition at the surface of the particles inhibits enzymatic nucleic acid degradation and activation of the enzymes that trigger the innate immune response of certain cells.18,19 These surprising observations pose the question, will polyvalent nucleic acids alone (lacking a particle core) readily enter cells without the need for a cationic polymer co-carrier? If so, one could create new gene regulation, drug delivery and imaging systems based solely on high density polyvalent nucleic acid nanostructures (PNANs) and bypass some of the issues that surround inorganic particle-based systems,20–22 especially those pertaining to long term toxicity.23–25 Herein, we report a strategy for synthesizing monodisperse, spherical high-density polyvalent DNA and DNA-RNA hybrid nanostructures with no inorganic cores. These new nanostructures are composed entirely of oligonucleotides, maintain the unique binding properties of nucleic acid inorganic nanoparticle conjugates (vide infra), exhibit the ability to enter cells without co-carriers, and engage the RNAi pathway in human cells.
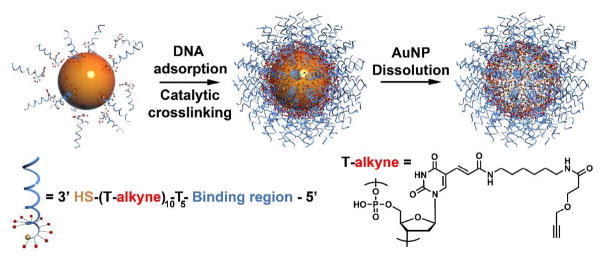
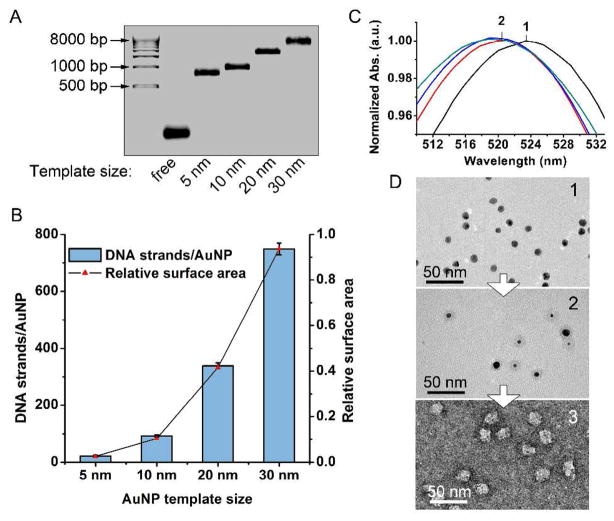
Recently, we discovered that propargyl-ether-modified polymers could be adsorbed and crosslinked on AuNP surfaces in water.26 Subsequent oxidative dissolution of the gold core yields hollow nanostructures that closely match the size and shape of the AuNP template. This novel approach takes advantage of the AuNP’s ability to act both as a template and a catalyst. To synthesize the PNANs, we prepared a novel class of oligonucleotides with a sequence of thymine (T) bases modified with propargyl ether groups linked through an amidohexylacrylamido linker to the 5 position of the T base, and a subsequent spacer of five unmodified T bases for flexibility and accessibility of the remaining sequence of ca. 20 base pairs (Scheme 1). The final nucleotides can be composed of either DNA or RNA designed for recognition purposes in the context of antisense or siRNA gene knockdown, respectively. These structures readily adsorb onto the surfaces of citrate stabilized gold nanoparticles (5 nm, 10 nm, 20 nm, and 30 nm evaluated in this study) and can be loaded to approximately 90 strands per 10 nm particle (at a density of ca. 0.28 strands/nm2). Crosslinking takes place between the propargyl groups on the surface of the particle and along the modified bases, creating a densely packed, crosslinked shell of DNA. Indeed, the loading is approximately twice the loading of oligonucleotides with only alkylthiol groups under optimized conditions.27,28 Upon removal of the gold nanoparticle template with 1 μM aqueous KCN, PNANs with the shape of the original AuNP template are formed. The size of the PNANs can also be easily controlled by choosing AuNP templates of appropriate sizes (5 – 30 nm); larger PNANs made of more DNA strands show lower mobility in agarose gel electrophoresis (Figure 1A, 1B and S1).
Scheme 1.
Synthesis of the PNAN from alkyne-modified 3′ thiolated DNA.
Figure 1.
(A) Agarose gel electrophoresis (1%) of PNANs synthesized from a range of template sizes (5, 10, 20, 30 nm). (B) Number of DNA strands per AuNP determined using 32P-radiolabeled DNA for AuNPs of different sizes (5–30 nm). Line shows the relative surface area of respective AuNPs. (C) UV-Vis spectra corresponding to the TEM images in (D) during the dissolution process, showing a blue shift of the plasmon resonance from 524 nm to 518 nm as the AuNP core is oxidatively dissolved. (D) TEM of alkyne-DNA modified AuNPs (1), PNANs encasing partially dissolved AuNPs (2), and fully formed PNANs negatively stained with uranyl acetate (3).
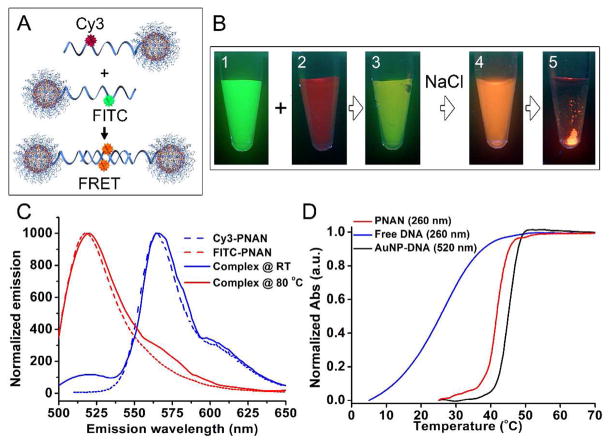
In contrast with citrate-capped gold nanoparticles, which rapidly aggregate and turn blue due to plasmon resonance red shifting upon the addition of KCN,29 the gold particles encased in a shell of alkyne-modified DNA do not aggregate during oxidative dissolution but rather slowly lose color with a concomitant blue shift in the plasmon resonance from 524 to 515 nm suggesting a decrease in size of the particles (original size = 10 nm, Figure 1C).30 Indeed, transmission electron microscopy (TEM) images of samples of the alkyne-DNA coated AuNPs during this dissolution process are consistent with the conclusion that the gold core is slowly dissolved and eventually yields hollow structures with a cross-linked shell of DNA (visualized by uranyl acetate staining, Figure 1D and Figure S1). Highly polyvalent DNA nanostructures often exhibit a high degree of cooperative binding with complementary nucleic acids, as evidenced by their association and thermal dissociation behavior.31 In principle, the PNAN structures should exhibit high cooperativity as manifested in a sharp melting transition when complexed with particles with complementary sequences. We prepared two sets of PNANs with strands that are complementary to each other, and labeled them internally with fluorescein and Cy3, respectively; these two dyes can form a Fluorescence Resonance Energy Transfer (FRET) pair when they encounter each other through hybridization of the particles (Figure 2A). When these particles are combined in Nanopure™ water, one only observes the green fluorescence of fluorescein under UV illumination. However, when the NaCl concentration is raised to 150 mM, the orange fluorescence from the Cy3 due to FRET is immediately observed, and within minutes macroscopic DNA interconnected assemblies of PNANs form and begin to precipitate (Figure 2B). The green to orange color change is reversible with increasing temperature, and is analogous to the red to purple plasmonic shift observed for aggregated DNA-AuNPs.13 These visual observations are further confirmed with fluorescence emission measurements. Pre-hybridized PNANs show a dominant FRET signal at 565 nm with minimal donor fluorescence following excitation at 450 nm (Figure 2C). Upon melting, the FRET signal disappears and donor fluorescence is observed at 520 nm.
Figure 2.
(A) Schematic of two PNANs designed to exhibit FRET upon hybridization, each having either a FITC or a Cy3 dye incorporated in the alkyne-DNA strand. (B) Photographs of PNANs under UV illumination: (1) FITC labeled PNANs, (2) Cy3 labeled PNANs, (3) mixture of FITC and Cy3 labeled PNANs in Nanopure™ water, (4) solution from 3 at 150 mM NaCl, (5) solution from 4 after 30 min. (C) FRET signal (solid blue) is observed when FRET PNAN pair is hybridized at room temperature. Upon melting, the FRET signal (565 nm) disappears and donor fluorescence (520 nm) becomes dominant (solid red). Dashed lines show respective Cy3- or FITC-PNAN fluorescence. (D) Melting analysis of free DNA, DNA-AuNP conjugate, and PNAN, each hybridized to complimentary DNA/particle. PNANs exhibit a sharper melting transition similar to DNA-AuNPs, compared with free DNA duplexes
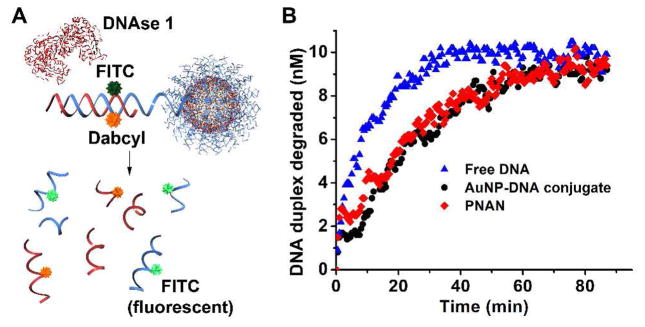
We then analyzed the melting properties of these aggregates by UV-vis spectroscopy, as the position and breadth of the melting transition temperature (Tm) of the assembled nanoparticles are important parameters for evaluating the quality of the PNANs. The free strand duplexes of the same sequences exhibit a Tm of ~ 24 °C and a typical broad FWHM (full width at half maximum, >10 °C) of the derivative of the melting curve. Remarkably, the aggregates exhibit an increased (Tm of ca. 42 °C) and much more narrow melting transition (FWHM of the derivative ca. 2 °C), indicative of polyvalent DNA interactions (Figure 2D). The melting behavior is identical to the undissolved AuNP system, with a slight decrease in Tm (ca. 3 °C), which is likely due to the swelling of the particles (with a concomitant decrease in DNA density) as observed by DLS (Figure S2). These data demonstrate that the recognition of the DNA binding region of the propargyl-ether modified strand is unaffected by the crosslinking chemistry occurring near the 3′ region of the strand. Another measure of PNAN DNA density is their ability to resist nuclease degradation due to their high local sodium ion concentration and steric inaccessibility of the nucleases to the DNA.18 We synthesized PNANs that were modified with an internal Dabcyl quencher and hybridized them to complementary fluorescein-labeled reporter strands (~ 10 per particle). The fluorescence of the reporter strand is quenched when hybridized to the Dabcyl-functionalized PNANs; however, when introduced to an enzyme specific for DNA duplex degradation (DNase I), the fluorophores are released and an increase in fluorescence can be observed (Figure 3A). Upon incubation with DNase I, the duplexes of the PNANs are more slowly degraded (rate ca. 4 times slower) as compared to the free duplexes, and nearly identical to PNANs that have not had their Au core dissolved (Figure 3B).
Figure 3.
(A) Schematic of nuclease degradation assay. DNAse I is used to degrade FITC-labeled DNA strand hybridized to a Dabcyl-labeled PNAN, wherein FITC fluorescence is quenched. Upon enzyme action, the fluorescence signal increases due to release of FITC-containing fragments, and a degradation rate can be determined. (B) DNA degradation following addition of DNAse I to free DNA duplexes, DNA-AuNPs and PNANs monitored by fluorescence intensity.
With confirmation that PNANs retain cooperative binding and nuclease resistance properties similar to those of the gold nanoparticle system, we evaluated their ability to enter human cells and facilitate gene knockdown via the RNAi pathway. Human squamous carcinoma cells (SCC12) were incubated with PNANs synthesized from 5′ Cy5-labeled DNA for 24 hours in the absence of any cationic transfection agents. Strikingly, PNANs readily enter these cells despite being composed only of negatively charged oligonucleotides. Entry of PNANs into SCC12 cells was qualitatively analyzed by confocal fluorescence microscopy (Figure 4A); however fluorescence labeling in this case may suffer from self-quenching due to the closely packed DNA strands that comprise the PNAN, and therefore is not ideal for quantitative measurements. Radiolabeling provides a quantitative way of measuring cellular uptake and internalization of the PNANs, and to this end, we synthesized PNANs from 5′ 32P-radiolabeled DNA, as the radioactivity of the free strand could be correlated to both strands per particle and particle concentration. Remarkably, PNANs enter cells in very high numbers (5.27 × 106 particles per cell) after 24 h incubation at 10 nM concentration, and at nearly identical levels to the DNA-AuNP system (5.09 × 106 particles per cell) (Figure 4B). It was previously revealed that DNA-AuNP conjugates recruit extracellular proteins as a first step in their cellular internalization, a phenomenon that results in an increase in the hydrodynamic diameters of the particles upon incubation in cell culture media 16. We monitored the size of the PNANs in media by DLS. Comparing with the DNA-AuNP system, which showed an increase of size from 29.1 ± 0.5 nm to 36.7 ± 2.4 nm after 30 min incubation with cell growth media containing 10% FBS, the PNANs also showed a size increase from 40.2 ± 2.9 nm to 48.3 ± 7.8 nm. These data demonstrate that high density polyvalent DNA by itself can facilitate cellular entry without the need for a cationic co-carrier or physical membrane disruption, such as electroporation or sonication, which usually lead to reduction in cell survival rates.32,33 Indeed, no cytotoxicity is observed for PNANs even at high concentrations (Figure 4C). Conversely, Lipofectamine2000 reduces cell survival rate to 0% when one attempts to transfer 20% of the DNA delivered by PNAN into SCC12 cells.
Figure 4.
(A) Confocal micrograph of SCC12 cells incubated with 1 nM Cy5-labeled PNANs for 24 h. Cell nuclei are stained with Hoechst 33342. (B) Cell uptake of DNA-AuNP conjugates and PNANs quantified by using radiolabeled alkyne-DNA and expressed as the number of particles per cell. Both constructs show similar high cell uptake that is dependent on concentration. (C) Cytotoxicity measurement of PNANs and Lipofectamine™ 2000 as measured by MTT assay. (D) EGFR gene knockdown using DharmaFECT™, siRNA-AuNP conjugates, and PNANs, determined by qRT-PCR. Inset shows western blot of EGFR when PNAN is used compared to non-treated cells. GAPDH is used as an internal reference.
After establishing that PNANs can enter cells through polyvalent-DNA mediated uptake, we synthesized siRNA-based PNANs targeting the epidermal growth factor receptor (EGFR) expressed in SCC12 cells. To compare the effectiveness of the siRNA functionalized PNANs to conventional transfection systems, SCC12 cells were incubated with DharmaFECT™-complexed EGFR siRNA, EGFR siRNA-AuNPs, scrambled siRNA-AuNPs, EGFR siRNA-functionalized PNANs, and scrambled siRNA functionalized PNANs at final concentrations of 5 nM for 72 hours. The cells were then harvested and analyzed for their protein content by Western blotting and mRNA levels by quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR). Protein levels of EGFR were significantly reduced by EGFR siRNA-AuNPs and EGFR siRNA PNANs, relative to internal control Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Furthermore, mRNA levels of EGFR were reduced by 81% when AuNP-siRNA conjugates were used, and by 82% when siRNA-based PNANs were used (Figure 4D).
The results reported herein show that it is possible to use nanostructures composed entirely of nucleic acids to effect cellular transfection and gene regulation, provided the nucleic acids are presented in a high density and oriented form. The work challenges the notion that one must complex nucleic acids with positively charged synthetic polymers, lipids, or metal ions to create effective platforms for transfecting the nucleic acids across cell membranes. Indeed, these PNANs are the first core-free structures with high density DNA shells that are capable of cellular internalization and gene knockdown. As such, they have the potential to bypass some of the disadvantages of conventional co-carrier routes, including issues pertaining to immunogenicity and nucleic acid life time. Previous efforts in developing agents targeted for gene therapy have largely focused on the properties of the transfection co-carrier.34,35 The results here imply that future studies should look beyond the chemical and biological traits of the co-carrier, to include the morphology, density and arrangement of the nucleic acid construct itself.
Supplementary Material
Acknowledgments
C.A.M. acknowledges the NCI-CCNE and NSF for support of this research. E.A. acknowledges the NDSEG for a Graduate Research Fellowship. A.E.P. acknowledges Northwestern University for a Ryan Fellowship. The transmission electron microscopy was carried out in the EPIC facility of NUANCE Center at Northwestern University. NUANCE Center is supported by NSF-NSEC, NSF-MRSEC, Keck Foundation, the State of Illinois and Northwestern University. Infrared Spectroscopy and mass spectrometry was conducted at the IMSERC facility at Northwestern University.
Footnotes
Supporting Information Available: Experimental procedures, DNA/RNA sequences and additional characterization data. This materials is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nishikawa M, Huang L. Hum Gene Ther. 2001;12:861. doi: 10.1089/104303401750195836. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead KA, Langer R, Anderson DG. Nature Rev Drug Discov. 2009;8:129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim WJ, Bonoiu AC, Hayakawa T, Xia C, Kakimoto MA, Pudavar HE, Lee KS, Prasad PN. Int J Pharm. 2009;376:141. doi: 10.1016/j.ijpharm.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Mintzer MA, Simanek EE. Chem Rev. 2009;109:259. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 5.McAllister K, Sazani P, Adam M, Cho MJ, Rubinstein M, Samulski RJ, DeSimone JM. J Am Chem Soc. 2002;124:15198. doi: 10.1021/ja027759q. [DOI] [PubMed] [Google Scholar]
- 6.Zhang K, Fang HF, Wang ZH, Li Z, Taylor JSA, Wooley KL. Biomaterials. 2010;31:1805. doi: 10.1016/j.biomaterials.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang L, Li S. Nature Biotechnol. 1997;15:620. doi: 10.1038/nbt0797-620. [DOI] [PubMed] [Google Scholar]
- 8.Young LS, Searle PF, Onion D, Mautner V. J Pathol. 2006;208:299. doi: 10.1002/path.1896. [DOI] [PubMed] [Google Scholar]
- 9.Amiji MM. Polymeric Gene Delivery: Principles and Applications. CRC Press; 2004. [Google Scholar]
- 10.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 11.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. J Am Chem Soc. 2009;131:2072. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryou SM, Kim S, Jang HH, Kim JH, Yeom JH, Eom MS, Bae J, Han MS, Lee K. Biochem Biophys Res Commun. 2010;398:542. doi: 10.1016/j.bbrc.2010.06.115. [DOI] [PubMed] [Google Scholar]
- 13.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 14.Agbasi-Porter C, Ryman-Rasmussen J, Franzen S, Feldheim D. Bioconjugate Chem. 2006;17:1178. doi: 10.1021/bc060100f. [DOI] [PubMed] [Google Scholar]
- 15.Cobley CM, Chen JY, Cho EC, Wang LV, Xia YN. Chem Soc Rev. 2011;40:44. doi: 10.1039/b821763g. [DOI] [PubMed] [Google Scholar]
- 16.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Nano Lett. 2007;7:3818. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Bioconjugate Chem. 2010;21:2250. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2008 doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. Mol Pharmaceutics. 2009;6:1934. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokolova V, Epple M. Angew Chem Int Ed. 2008;47:1382. doi: 10.1002/anie.200703039. [DOI] [PubMed] [Google Scholar]
- 21.Chompoosor A, Saha K, Ghosh PS, Macarthy DJ, Miranda OR, Zhu ZJ, Arcaro KF, Rotello VM. Small. 2010;6:2246. doi: 10.1002/smll.201000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohs A, Duan H, Kairdolf B, Smith A, Nie S. Nano Res. 2009;2:500. doi: 10.1007/s12274-009-9046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhabra G, et al. Nat Nanotechnol. 2009;4:876. doi: 10.1038/nnano.2009.313. [DOI] [PubMed] [Google Scholar]
- 24.Nel A, Xia T, Madler L, Li N. Science. 2006;311:622. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y, Neuss S, Leifert A, Fischler M, Wen F, Simon U, Schmid G, Brandau W, Jahnen-Dechent W. Small. 2007;3:1941. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- 26.Zhang K, Cutler JI, Zhang JA, Zheng D, Auyeung E, Mirkin CA. J Am Chem Soc. 2010;132:15151. doi: 10.1021/ja107224s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill HD, Millstone JE, Banholzer MJ, Mirkin CA. ACS Nano. 2009;3:418. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Anal Chem. 2006;78:8313. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniel MC, Astruc D. Chem Rev. 2004;104:293. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 30.Link S, Wang ZL, El-Sayed MA. J Phys Chem B. 1999;103:3529. [Google Scholar]
- 31.Jin RC, Wu GS, Li Z, Mirkin CA, Schatz GC. J Am Chem Soc. 2003;125:1643. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 32.Lefesvre P, Attema J, van Bekkum D. Bmc Mol Biol. 2002:3. doi: 10.1186/1471-2199-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyber JA, Andrews J, DEmanuele A. Pharm Res. 1997;14:750. doi: 10.1023/a:1012198321879. [DOI] [PubMed] [Google Scholar]
- 34.Itaka K, Ishii T, Hasegawa Y, Kataoka K. Biomaterials. 2010;31:3707. doi: 10.1016/j.biomaterials.2009.11.072. [DOI] [PubMed] [Google Scholar]
- 35.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.