Abstract
In the plant genus Silene, separate sexes and sex chromosomes are believed to have evolved twice. Silene species that are wholly or largely hermaphroditic are assumed to represent the ancestral state from which dioecy evolved. This assumption is important for choice of outgroup species for inferring the genetic and chromosomal changes involved in the evolution of dioecy, but is mainly based on data from a single locus (ITS). To establish the order of events more clearly, and inform outgroup choice, we therefore carried out (i) multi-nuclear-gene phylogenetic analyses of 14 Silene species (including 7 hermaphrodite or gynodioecious species), representing species from both Silene clades with dioecious members, plus a more distantly related outgroup, and (ii) a BayesTraits character analysis of the evolution of dioecy. We confirm two origins of dioecy within this genus in agreement with recent work on comparing sex chromosomes from both clades with dioecious species. We conclude that sex chromosomes evolved after the origin of Silene and within a clade that includes only S. latifolia and its closest relatives. We estimate that sex chromosomes emerged soon after the split with the ancestor of S. viscosa, the probable closest non-dioecious S. latifolia relative among the species included in our study.
Introduction
Many plants with separate sexes (dioecy) evolved this sex system much more recently than the main animal model systems (mammals, Drosophila and birds). Plants such as Silene latifolia, Carica papaya, Bryonia dioica and Fragaria may thus be suitable for studying the first steps in sex chromosome evolution [1], [2], [3].
Silene latifolia and its dioecious close relatives, S. dioica, S. marizii, S. heufellii and S. diclinis [4], have an XY chromosomal sex determination system. In a distinct group of dioecious species with no chromosome heteromorphism, S. otites, S. colpophylla and S. acaulis which are closely related (see [5] and our results below), separate sexes probably evolved independently [4], [6], [7]. Many other Silene species are non-dioecious. These are either hermaphroditic, or else gynodioecious (with some individuals hermaphroditic and others female), or gynodioecious with gynomonoecious individuals (having both hermaphroditic and female flowers). Dioecy in Silene therefore probably evolved from hermaphroditism via gynodioecy (rather than from monoecy, a common ancestral state for dioecious plants, in which individuals have separate male and female flowers [8], [9], [10], [11]).
Evolution of dioecy from hermaphroditism via gynodioecy requires at least two mutations [12]: first a male-sterility mutation creates females and establishes a polymorphism for females and hermaphrodites (gynodioecy), and the hermaphrodite individuals (with male function) may then evolve increased maleness through partial or complete female-sterility mutations. Because the second mutation, suppressing female functions, lowers the reproductive fitness of the females carrying the first mutation, linkage is required between the genes that undergo the mutations [12]. This predicts that a single autosome in an ancestral species evolves into a proto-sex chromosome pair (as opposed to chromosome rearrangements bringing the sex-determining genes together from different ancestral chromosomes to form a non-recombining region carrying the male- and female-sterility factors inferred genetically, see [12], [13]). The initial linkage may be followed by selection for reduced genetic recombination in the sex-determining region. Cessation of recombination can occur in a stepwise manner, beginning in the region flanking the sex determining locus and then spreading to other parts of the sex chromosomes, leading to the large non-recombining genome regions now found on many sex chromosomes, including those of S. latifolia [13], [14].
Once recombination stops between a gene or region on the sex chromosome pair, sequences in the region start diverging, and sequence divergence can indicate the age of the sex chromosome system. In S. latifolia, divergence has been estimated between multiple loci spread across the X and Y chromosomes. The maximum nucleotide sequence divergence between X and Y chromosomes is ∼20%, suggesting that recombination stopped in the oldest such region between 5 and 10 MYA [15], [16].
This X-Y sequence divergence is similar to the estimated divergence for the same loci between S. latifolia and some non-dioecious Silene species, including the gynodioecious S. vulgaris [17], [18] and the hermaphrodite S. conica [19]. The estimated date of the split of Lychnis and Silene from outgroups is ∼12.4 MY ago, and that between the different Silene subgroups 7.9–9.5 MY [20], similar to the date estimated above for initiation of X-Y divergence. The possibility must thus be considered that sex chromosomes in Silene evolved early in the evolutionary history of the genus, and could have been secondarily lost in ancestors of the non-dioecious Silene species. Under the evolutionary model for dioecy outlined above, a single mutation can cause reversion to gynodioecy or to full hermaphroditism (through loss of a major female-sterility mutation). It is therefore important to test whether dioecy is ancestral (and hermaphroditism a reversion from this) in plant taxa used to study early stages of sex chromosome evolution.
In Silene, it is important to consider the possibility of reversion to hermaphroditism, because reversion is probably common in plants [21], and reverted species are unsuitable as outgroups for inferring directions of changes. For example, the inference that a single pair of autosomal chromosomes in an ancestral Silene species evolved into a sex chromosome pair (as the population genetics outlined above predicts) is based on the mapping of homologues of genes in the non-recombining regions of the Y chromosomes of S. latifolia to a single S. vulgaris autosome [22]. However, if S. vulgaris had a dioecious ancestor, it could be incorrect to infer that the sex chromosomes evolved without translocations from a state like that in S. vulgaris. Similarly, one could not infer the ancestral states at orthologues of sex-determining loci by using the hermaphrodite S. viscosa [23].
The only sequence-based study available that is relevant for selecting outgroups for studying the origin of dioecy in Silene used the internal transcribed spacer of the ribosomal DNA (ITS regions) in the nuclear genome, and included 25 Silene species [4]. This has remained the accepted phylogeny relevant for the evolution of dioecy in Silene, and is consistent with evolution of different sex-determining genes in S. colpophylla (a relative of S. otites and S. acaulis), based on the finding that its orthologues of 4 sex-linked S. latifolia genes are in one S. colpophylla linkage group, but are not sex-linked [24].
However, such phylogenies based on a single genomic region represent only that region's evolutionary history, which may differ substantially from that in other genome regions, and may not represent the species' ancestry. Other phylogenetic work on Silene has added chloroplast sequences and some nuclear sequences, including likely single-copy genes [25], but mainly examined deeper relationships in the family Caryophyllaceae, often including few members of one subgenus when studying the phylogenetic relationships of another subgenus [25], [26], [27], [28], [29], [30], or have focused on a particular group [7], [31], rather than including all species relevant for the study of the evolution of sex-determination within Silene. New analyses of DNA sequences are therefore needed, ideally using multiple unlinked loci. Here we report the first multi-gene study in Silene, including both sections with dioecious species, Melandrium and Otites [20], [32]. Our taxonomic sampling also includes other Silene species from subgenera Behen and Silene (in the terminology of [25]), Lychnis and, as an outgroup for rooting trees, we used a Petrocoptis species [20], [25]. Our inclusion of suitably selected species to help infer the relationships, enabled us to use BayesTraits analysis to infer the evolution of dioecy in a phylogenetic context that allows for reversals.
We used nuclear genes, because sequences from the cytoplasmic genome lack frequent recombination, and do not yield independent phylogenies. In Silene as in other plants, cytoplasmic genomes introgress more frequently than nuclear genes [20], which blur true phylogenetic relationships. Also, transpositions of large regions of chloroplast sequences to the S. latifolia Y chromosome [33], and perhaps to other chromosomes, can confuse phylogenetic inferences. Furthermore, in Silene these genomes may have had major changes in mitochondrial mutation rates [34], [35], experienced complex mutational processes, and/or have been under positive selection [36], [37], [38]. To deal with the difficulties of data from multiple, independently evolving genome regions, we employed several recently developed approaches for multi-gene phylogenetic analysis.
Materials and Methods
Plant material
Following [4] we chose fourteen species representing the diversity of breeding systems within Silene (see Table S1, which also gives voucher details and GenBank accession numbers of the genes sequenced). The 14 species include Lychnis coronaria and L. flos-jovis as Lychnis appears as a close sister group to Silene in the chloroplast+ITS phylogeny [20]. We also sequenced the genes from either Petrocoptis hispanica, a close outgroup species of Silene+Lychnis [25], [29], [30] or Dianthus (for the PGK gene, which did not amplify from P. hispanica). Plants were grown from seeds collected in the wild, or obtained from the Royal Botanic Garden Edinburgh's wild collections.
Molecular methods
Genomic DNA was extracted from young leaves using the FastDNA kit (Q-BioGene). We sequenced 10 autosomal genes and 3 genes that are sex-linked in S. latifolia, S. dioica and S. diclinis (Tables S2 and S3). The autosomal genes were chosen from S. latifolia ESTs [39]. The putative functions of the loci newly sequenced for this study (ABCtr, ATUB, ADPGph, ClpP3, ELF, LIP21, OxRZn, PSIcentII, PGK and 2A10) are listed in Table S2, together with the primers used for sequencing, and the PCR conditions. The sex-linked loci sequenced were SlXY4 [40], SlXY7 [16], and SlXYCyp [16]. Intron-exon boundaries were determined by alignment with cDNA sequences of A. thaliana homologues, to avoid primer sites spanning introns; most sequences include coding regions and introns.
Sequencing and alignment
Sequences were obtained by direct sequencing of PCR products after removal of excess primers and unincorporated dNTPs using shrimp alkaline phosphatase and ExoI respectively (ExoSAP-IT; Amersham Biosciences). Both strands were sequenced using BigDye Cycle Terminator protocols (Amersham Biosciences) on an ABI377 automated capillary sequencer. Whenever we found more than one sequence in autosomal genes, or when the primers amplified both X and Y copies of sex-linked genes, PCR products were cloned into TOPO TA Cloning kits (Invitrogen) and sequenced as described above.
To detect and exclude paralogues (which can confuse estimates of species relationships), direct sequences were obtained from several plants for as many species as possible; for dioecious species, DNA was sampled from both male and female plants (except for S. heuffelii, for which only one male was available). The sequences often included heterozygous sites, and, from such individuals, both sequences were determined after cloning. Sequences for each gene were inspected using Sequencher version 4.0.5 (Gene Codes Corporation) and aligned using Clustal X version 1.81 [41] using the default parameter values. When each species had <1% divergence between the sequences amplified with a set of primers, the sequences were assumed to be alleles of a single-copy locus. Loci not satisfying this criterion were discarded (in total, about the same number of loci as the number analysed, data not shown). The alignments were modified manually using BioEdit version 7.0.4.1 [42]. Poorly aligned positions were removed using Gblocks [43] with the “relaxed block selection” option [44]. The resulting alignments total 5582 bp (Table S3 shows the numbers of sites per gene). ITS1 and ITS2 sequences were retrieved from GenBank (1503 bp, or 1195 bp after Gblocking).
Concatenation of genes
When several sequences were available for a given gene in a species, we used the software ScaFos to select the sequence from the alignments after GBlocking (see above), the one with the smallest average divergence from the other conspecific sequences [45]. Fast evolving species were not excluded, and we used minimal evolutionary distances with a gamma distribution, with maximal 50% missing sites (the other options used were: minimal length to select a sequence = 0%, making chimera = YES). We then used Concaterpillar, a hierarchical clustering method based on likelihood-ratio testing that identifies genes with congruent phylogenies, and very efficiently identifies loci that can be concatenated for estimating phylogenies [46].
Phylogenetic tree reconstruction
Individual locus trees were constructed using PhyML [47], [48] with the General Time-Reversible (GTR) model, the estimated base frequencies, percentage of variants and alpha parameter for the gamma distribution, and with 4 or 8 rate categories. We used the GTR+Γ as it is the most sophisticated model available for nucleotide sequences but other simpler models gave the same conclusions (data not shown). Support for nodes was evaluated with 100 replicates of non-parametric bootstrapping, again using the maximum likelihood optimality criterion. For the sex-linked loci, only X sequences were included; Y sequences were not used for the phylogeny, because of their unusually fast sequence evolution [49], [50], [51].
We used three different approaches to combine the results from different genes. First, we estimated PhyML trees using concatenated sequences (with the parameters above). By default, we used GTR+Γ. For the dataset used to build the tree shown in Figure 1A, we tested the effect of the models for sequence evolution. jModeltest [52] recommends the “transitional model”+invariant+gamma model (TIM+I+Γ) and the Tamura-Nei+invariant+gamma model (TrN+I+Γ) as the best of the 88 models tested for the sequences using respectively the Akaike and Bayesian information criteria (AIC and BIC). We thus built the tree shown Figure 1A using TrN+I+Γ (best model using BIC criterion, see [53]), which is implemented in the PhyML. Note however that the topology of this tree is not affected by model selection since TrN+I+Γ, GTR+Γ, or the model-averaged option available in jModelTest give identical topologies.
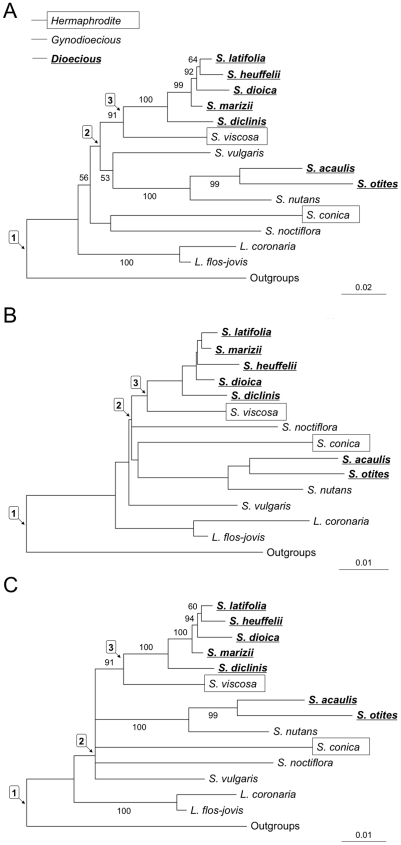
Figure 1. Phylogeny of the Silene species studied.
Only bootstrap values >50% are shown. The breeding systems of the species are indicated in the figure. The outgroup for rooting the tree for the genus Silene was usually P. hispanica (see Tables S2 and S3 for details). For the sex-linked genes, only X sequences were included. A) Maximum likelihood tree obtained with PhyML using the concatenated alignment with 12 genes (LIP21 and the outgroup sequences for 2A10, clpP3, ELF were excluded by Concaterpillar). B) Dataset is as in A, but tree obtained by combining all 12 gene trees using the SDM method. C) Consensus tree built from the trees in A and B. Labels 1, 2 and 3 in the trees indicate the nodes used in the BayesTraits analysis (see main text).
Second, we built supertrees from the individual gene trees using the Super Distance Matrix (SDM) method, which normalises tree lengths before combining the trees; this is less sensitive to rate heterogeneity than the first method [54]. We then imported the supermatrix of squared distances into BIONJ [55]; this method does not yield bootstrap values.
Genes with trees that differ from the species tree can produce misleading species trees when the latter is estimated from concatenated genes [56]. Methods incorporating an explicit model of coalescence [57] have been developed specifically to deal with deep coalescence. To allow for lineage sorting and other population genetic events that can result in different genes having incongruent trees (e.g. [58], [59]), we therefore also analyzed the multi-locus data using a third approach, available through the R package phybase [60]. As explained in Text S2, this includes two methods. STAR is based on the average ranks of coalescence events, and STEAC employs average coalescence times [61].
Tree topologies were compared by the Approximately Unbiased (AU) test, using Treefinder [62].
Analysis of character evolution
We analysed the evolution of breeding systems in Silene, following [63] with the BayesMultiState function in BayesTraits [63]. Breeding system data followed [4]. For the two Lychnis species, S. noctiflora and S. acaulis, we allowed two possible character states, because for these species both gynodioecy and hermaphroditism are recorded (Lychnis and S. noctiflora) or gynodioecy and dioecy (S. acaulis) [64], [65], [66]. The results were not qualitatively affected by allowing two character states, or only one (data not shown). We used maximum likelihood to estimate the probabilities for three different states, dioecy, gynodioecy and hermaphroditism, at several relevant nodes, and the model allowed transitions and reversals between each of these states.
Distribution of pairwise divergence values
To compute the d S values for S. latifolia from other species, or X-Y d S values for S. latifolia sex-linked genes, we extracted the coding regions of each gene and used codeml in PAML [67]. The mean d S values for each comparison of S. latifolia with other groups of species were compared using a non-parametric sign-test with setting either 0.06 (d S value for XY2) or 0.16 (d S value for XY1) as theoretical values. The test is significant when the null hypothesis (the number of observed d S values>the theoretical value = the number of observed d S values<the theoretical value) is rejected.
Results and Discussion
The phylogenetic relations of the Silene species studied
Several trees from individual genes show the expected grouping together of S. latifolia, S. dioica, S. marizii, S. heufellii and S. diclinis (6/13 have significant bootstrap support, and 2 have some bootstrap support, see Figure S1). S. acaulis and S. otites (and the gynodioecous species S. nutans) also often form a clade (6/13 trees), as also do the two Lychnis species (7/13 trees). However, the topologies and relationships among these three clades differ among the individual loci. We therefore used several approaches to estimate the species tree using combined data.
We first used concatenated sequences. To test whether concatenating different gene sequences is appropriate, we used Concaterpillar. This requires the same set of species for all the loci, limiting our analysis to only 11 of the 14 ingroup species, and only 9 genes, including the ITS sequences (Table S3). However, excluded species (S. nutans, S. heuffelii, or S. marizii), are all very close relatives of included ones, and their exclusion had little effect on the results. With this reduced dataset, the LIP21 gene falls into a distinct block (top part of Table S4, block 3). Additionally excluding outgroup sequences for 2A10, clpP3, ELF, Concaterpillar recovered a single block with the 7 newly sequenced autosomal genes together with ITS (lower part of Table S4, block 1). This concatenation confirms the early divergence of Lychnis relative to the species included in the present study [25], [29], [30], and also the distinctness of the two dioecious clades inferred using ITS (Figure S2 and Text S1). Also S. latifolia and its close dioecious relatives (S. dioica, S. diclinis, S. heuffelii, S. marizii) are a monophyletic group, as in the individual gene trees. However, sequence information from these species was probably lost in removing poorly aligned regions, in order to include all the species (see Methods). Because this Gblock pre-processing is unnecessary for such close relatives, we also analysed the raw alignments of just these species. Using maximum likelihood (Figure S3), we get slightly different relationships among the dioecious species, which are, however, fully consistent with those in the trees presented below in Figure 1.
A PhyML tree using a concatenation of the Concaterpillar block of 7 non-ITS sequences described above, plus the 5 other genes (Figure 1A) yields the same statistically supported clades as just the 7 genes, and the bootstrap values are as good. S. viscosa is now closest to the S. latifolia group species (although statistical support is weak), and S. vulgaris more distant. This is consistent with genomic in situ hybridization results [68], and is biologically plausible, because S. viscosa (unlike S. vulgaris) can hybridize with S. latifolia, suggesting a close relationship [23]. However, a chloroplast tree [20] suggests a closer relationship for S. vulgaris than S. viscosa. In that tree, as in ours, S. conica no longer appears to be a close relative of S. latifolia, a major difference from previous conclusions based on few genome regions. Identical results were found with 4 or 8 site categories, and also using RaxML [69] with 25 site categories and Treefinder with various optimization methods (data not shown), and also with other models of sequence evolution (see Methods). The SDM tree combining the 12 gene trees (Figure 1B) is similar to the PhyML tree.
Species trees using STAR or STEAC (Figure S4 and Text S2) are very similar to Figure 1B and are fully consistent with the consensus tree shown in Figure 1C. These methods are based on a coalescent model, and can thus handle incongruencies due to incomplete lineage sorting, which can adversely affect concatenation-based methods [56]. Moreover, as they use either average coalescence times (STEAC) or ranked coalescence times (STAR), the results are not biased by missing data. Finally, the rank-based STAR method is robust against differences in evolutionary rates or branch length [61].
The evolution of dioecy
We ran BayesTraits using topologies in Figures 1A and 1B, which were based on the alignment of the 12 genes, and do not differ significantly (AU test; p value: 0.995), and on a consensus topology built from these trees using Treefinder (Figure 1C). In all analyses, dioecy is unlikely at deep nodes (nodes 1 and 2 in Table 1). Gynodioecy or hermaphroditism are the most probable ancestral states for Silene, and the results for node 2 support the belief that dioecy evolved at least twice in the genus. Our results for node 3 suggest, with less certainty, that dioecy evolved after the split of S. latifolia, and its close dioecious relatives from the common ancestor of S. latifolia and S. viscosa (see Table 1).
Table 1. Probabilities of dioecy and other sexual systems at three critical nodes in the Silene phylogeny, estimated using BayesTraits.
| Node and description of the node | Phylogeny used in the analysis | Probability dioecious | Probability gynodioecious | Probability hermaphrodite | Probability non-dioecious |
| Node 1 | Figure 1A | <10−4 | 0.457 | 0.543 | >0.999 |
| Root of the tree | Figure 1B | 0.240 | 0.388 | 0.372 | 0.760 |
| Figure 1C | <10−4 | 0.488 | 0.512 | >0.999 | |
| Node 2 | Figure 1A | <10−4 | 0.687 | 0.313 | >0.999 |
| Common ancestor of all dioecious species, | Figure 1B | 0.488 | 0.198 | 0.313 | 0.511 |
| including otites and latifolia groups | Figure 1C | <10−4 | 0.540 | 0.460 | >0.999 |
| Node 3 | Figure 1A | <10−4 | 0.478 | 0.522 | >0.999 |
| Common ancestor of the latifolia group and | Figure 1B | 0.623 | 0.115 | 0.262 | 0.377 |
| S. viscosa | Figure 1C | 0.113 | 0.401 | 0.486 | 0.887 |
Two possible character states were allowed for Lychnis, S. noctiflora and S. acaulis (see text).
It seems unlikely that our sparse sampling of Silene species could lead to erroneous inference of the ancestral character state. Fundamental relationships within Silene are well enough established [25], [30], [70] to be confident that Lychnis is basal to the species studied here, and that the two major subgenera (Behen and Silene) within our set of Silene species each includes one of the two clades of dioecious species discussed above [4], [20]. We included in our analysis species from both these major Silene clades, as well as suitable outgroups. Many other species that were not included in our analysis are however, known to be hermaphrodites [4]. Thus, our sample over-represents dioecious species, which is conservative, because any bias would be towards inferring a dioecious ancestor.
Furthermore, simulations of taxon sampling [71] show that it is more important to include basal species (as in our sample) than many late-diverging species; when a molecular clock is not applicable, adding more taxa can even decrease the quality of inference (in our trees, some differences in branch lengths are large, see Figures 1, S2 and Text S1). When a molecular clock is assumed, adding more taxa increases the probability of inferring the correct state for the ancestor, but not greatly, and improvements occur mainly when the character changes rapidly [72]. If there are fewer character changes over the ancestry than assumed in these models, as is probably true in Silene, given the fairly short time-scale of evolution within the genus, and the low likelihood that dioecy will evolve, due to the need for at least 2 genetic changes (see Introduction), the effect is minor, even when as few as 16 out of more than 500 taxa are sampled [72]. Therefore, although we included only 14 Silene species out of 700 estimated in the genus [26], [27], [32], [73], [74], this should not be a serious problem, particularly if the species omitted are mostly hermaphrodites or gynodioecious, although including more taxa would, of course, change the probabilities in Table 1.
The phylogeny of sex-linked genes and the evolution of sex chromosomes
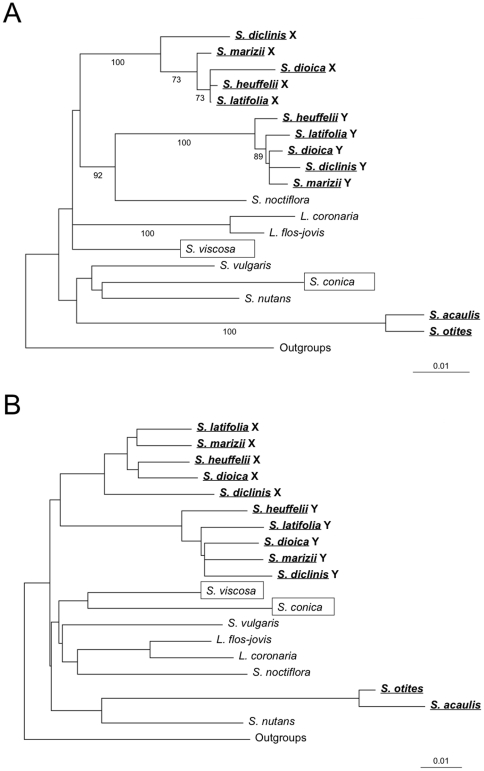
To independently test for an early emergence of dioecy in Silene, followed by loss of dioecy in some taxa, we also analysed X and Y sequences of the three genes that are sex-linked in S. latifolia and the four closely related dioecious species. If the sequences of a gene include a monophyletic X and Y group of sequences, we can infer that sex chromosomes evolved in the lineage ancestral to the S. latifolia group of dioecious species. Genes estimated to have stopped recombining soon after the sex chromosomes first evolved [15], [16] are the most relevant; of these, the SlXY7 tree is fully consistent with the combined data, whereas the S. otites sequences of the SlXY4 homologue branch with the Y-linked sequences, but non-significantly (AU test p = 0.563, see also Figure S1). The concatenation and supertree approaches using the whole X and Y sequence dataset also support this conclusion. The SDM tree (Figure 2B) also supports evolution of the sex chromosomes after the origin of the genus. In the PhyML tree (Figure 2A), S. noctiflora branches with the Y sequences, but this topology is not significantly better than one with X and Y sequences forming a monophyletic group (AU test p = 0.136).
Figure 2. Concatenate tree and supertree for sex-linked genes.
A) PhyML tree (GTR with gamma distribution) for the concatenated alignments of the three sex-linked genes. Bootstrap values >50% are shown. B) SDM tree from the three individual sex-linked gene trees.
Comparing the age of S. latifolia sex chromosomes with species divergence times within Silene
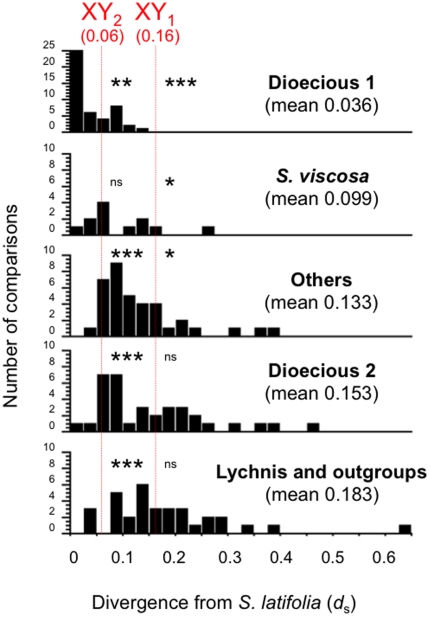
Finally, if the sex chromosomes in S. latifolia (plus its closest relatives) evolved within the genus, divergence between S. latifolia and most of the species in the genus should exceed the maximum X-Y divergence. As another test, we therefore computed pairwise synonymous divergence values (d S) between S. latifolia and the other species sampled, using only X-linked sequences for the S. latifolia sex-linked genes, and compared them with X-Y d S values for three S. latifolia sex-linked genes, SlXY4, SlXY7 and SlCyp-XY (Figure 3). Using a non-parametric sign-test (see Methods), we show in Figure 3 that the average X-Y d S value for the most highly diverged sex-linked genes, SlXY4 and SlXY7 [15], [16], [75] (i) is, as expected, significantly higher than the values between the close dioecious relatives of S. latifolia, (ii) is marginally significantly higher than the value from S. viscosa and also S. vulgaris, S. conica or S. noctiflora, and (iii) does not significantly exceed the higher divergence between S. latifolia and the other group of dioecious species (S. otites and S. acaulis), or the Lychnis species (or the outgroups). Overall, these results are consistent with an origin of sex chromosomes within an ancestor of S. latifiolia and its close dioecious relatives, rather than the alternative of an earlier origin. As expected, the divergence of SlCyp-X and -Y started much more recently [15], [16], [76], and our analysis places this event long after the split from the S. otites group of dioecious species, and probably after the split from S. viscosa (although the test in this case is non-significant). We find no clear evidence for reversions from dioecy to hermaphroditism or gynodioecy among dioecious species related to S. latifolia. The date when dioecy evolved in the ancestor of S. otites remains unknown, but it should be possible to estimate this date once the sex-determining chromosome of these species is identified.
Figure 3. Divergence between sex chromosomes and between Silene species.
Distribution of synonymous divergence values (mean d S values, estimated using PAML) from pairwise comparisons between S. latifolia and the other species for the 13 genes. One such comparison is for S. viscosa, and the others are for the following groups of species: Dioecious 1 = S. dioica, S. diclinis, S. marizii, S. heuffelii, Dioecious 2 = S. otites, S. acaulis, S. nutans (although, as noted in the text, most S. nutans populations are gynodioecious), Others = S. vulgaris, S. conica, S. noctiflora, Lychnis and outgroups = Lychnis flos-jovis, Lychnis coronaria and outgroups. In addition, vertical arrows indicate the average d S between the S. latifolia X and Y sequences for SlXY4 and SlXY7, which belong to the oldest stratum of X-Y divergence (symbolised by XY1), and for SlCyp-XY (labelled XY2), a gene which belongs to the “intermediate stratum” that stopped recombining considerably later than the first two genes [15], [16], [75]. For each panel in Figure 3, we performed a sign-test (see Methods) of whether either the XY1 or the XY2 d S value is significantly different from the mean in the panel (e.g. for the top panel, we tested S. latifolia vs. Dioecious 1). Significant differences are indicated as follows: * p<10−1, ** p<10−2, *** p<10−3; ns = non-significant differences.
Supporting Information
Individual gene trees for our 13 genes. Trees were obtained with PhyML (see Material and Methods) with 100 non-parametric bootstrap replicates (only values >50% are shown). Note that some of the sequences were excluded from the further analyses, as follows: cDNA sequences from S. latifolia for ABCtr, PGK, SlXY4 and SlXYCyp, whose sequence lengths were too different, and the Petrocoptis sequence of OxRZn because of doubts of the orthology of the sequence. For each gene, there is one outgroup sequence. As noted in Table S2 and footnote 4 of Table S3, the outgroup species used to root the tree is usually Petrocoptis hispanica; this sequence was the outgroup for 8 of the 13 trees shown, including all the X-linked genes, but, for PGK, a Dianthus sequence was used, and Lychnis for ATUB-A and OxRZn.
(TIFF)
Other inferred phylogenies of the Silene species studied. For details about sequences and species, see Table S3. Only bootstrap values >50% are shown. A) Maximum parsimony tree of the ITS sequences from 12 species from [4] that were also studied by ourselves. The different mating systems are indicated, and stars indicate bootstrap values larger than 50%. B) Maximum likelihood tree (PhyML) of the ITS sequences from 13 of our species. C) Maximum likelihood tree of 7 autosomal genes concatenated by Concaterpillar (see Table S4) with all the species (except outgroups for 3 genes: 2A10, ClpP3 and ELF, see main text and Table S4) obtained with PhyML. Identical results were found with 4 or 8 categories of sites. In trees C and D, and also those in Figure 1 of the main text, only X sequences were included for the sex-linked genes. D) Dataset as in C, but the tree was obtained by combining all the 7 gene trees using the SDM method. In C, S. conica and S. noctiflora diverge early, but in D they group with S. vulgaris and S. viscosa.
(TIFF)
Tree for dioecious species, rooted using S. diclinis (as this species is early diverging in all trees in Figure 1 and Figure S2). The tree was obtained by PhyML on the concatenate of all genes without Gblocking. Values indicate the results of non-parametric bootstrapping with 100 replicates (only values >50% are shown).
(TIFF)
Estimated species trees, using all genes except for LIP21 (12 loci). A) Tree using STAR. B) Tree using STEAC. In both cases, the branch lengths are proportional to the bootstrap support, which is given as a percentage at each branch. As for the individual locus trees, the outgroup species was Petrocoptis hispanica for all genes except PGK, where a Dianthus sequence was used, and ATUB-A and OxRZn where no outgroups were available.
(TIFF)
Provenance and voucher details of plants used to assess autosomal and sex-linked gene phylogenies in Silene .
(DOC)
Primers used to amplify genomic DNA of the genes studied, lengths of the sequences studied, and the outgroup used.
(DOC)
List of species and sequences.
(DOC)
Results of tests for combining alignments using Concaterpillar.
(DOC)
Discussion on the differences in branch length in our trees.
(RTF)
Methods and results for the STAR and STEAC analysis.
(RTF)
(RTF)
Acknowledgments
We thank the following colleagues for plant samples: H. Prentice (Lund University) for S. noctiflora, Royal Botanic Garden Edinburgh for S. acaulis, Lychnis spp. and Petrocoptis, D.A. Filatov (Oxford University) for S. heufelii, M. Looseley and T. Meagher (University of St. Andrews) for plants of S. marizii, and B. Oxelman for S. viscosa. We thank Dr. T. Vanhala for help with analysing cDNA sequences. We thank Sophie Abby for help with Concaterpillar, and Ben Evans and Liang Liu for their help with BEST, STEAC and STAR. We thank B. Oxelman for his helpful comments on a previous version of the ms.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: DC, AF and EK acknowledge support from Natural Environment Research Council (United Kingdom), and GM and JK from Agence National de la Recherche (grant number ANR-08-JCJC-0109). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Charlesworth D, Guttman DS. The evolution of dioecy and plant sex chromosome systems. In: Ainsworth CCe., editor. Sex Determination in Plants. Oxford: 1999. pp. 25–49. [Google Scholar]
- 2.Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, et al. A primitive Y chromosome in Papaya marks the beginning of sex chromosome evolution. Nature. 2004;427:348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- 3.Spigler R, Lewers K, Main D, Ashman T-L. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity. 2008;101:507–517. doi: 10.1038/hdy.2008.100. [DOI] [PubMed] [Google Scholar]
- 4.Desfeux C, Maurice S, Henry JP, Lejeune B, Gouyon PH. Evolution of reproductive systems in the genus Silene. Proc Biol Sci. 1996;263:409–414. doi: 10.1098/rspb.1996.0062. [DOI] [PubMed] [Google Scholar]
- 5.Eggens F, Popp M, Nepokroeff M, Wagner W, Oxelman B. The origin and number of introductions of the Hawaiian endemic Silene species (Caryophyllaceae). American Journal of Botany. 2007;94:210–218. doi: 10.3732/ajb.94.2.210. [DOI] [PubMed] [Google Scholar]
- 6.Mrackova M, Nicolas M, Hobza R, Negrutiu I, Monéger F, et al. Independent origin of sex chromosomes in two species of the genus Silene. Genetics. 2008;179:1129–1133. doi: 10.1534/genetics.107.085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frajman B, Eggens F, Oxelman B. Hybrid origins and homoploid reticulate evolution within Heliosperma (Sileneae, Caryophyllaceae) — a multigene phylogenetic approach with relative dating. Systematic Biology. 2009;58:328–345. doi: 10.1093/sysbio/syp030. [DOI] [PubMed] [Google Scholar]
- 8.Darwin CR. The Different Forms of Flowers on Plants of the Same Species. London: John Murray; 1877. 352 [Google Scholar]
- 9.Renner SS, Ricklefs RE. Dioecy and its correlates in the flowering plants. Amer J Bot. 1995;82:596–606. [Google Scholar]
- 10.Renner SS, Won H. Repeated evolution of dioecy from monoecy in Siparunaceae (Laurales). Systematic Biology. 2001;50:700–712. doi: 10.1080/106351501753328820. [DOI] [PubMed] [Google Scholar]
- 11.Dorken ME, Barrett SCG. Sex determination and the evolution of dioecy from monoecy in Sagittaria latifolia (Alismataceae). Proceedings of the Royal Society of London B. 2004;271:213–219. doi: 10.1098/rspb.2003.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlesworth B, Charlesworth D. A model for the evolution of dioecy and gynodioecy. Amer Nat. 1978;112:975–997. [Google Scholar]
- 13.Westergaard M. The mechanism of sex determination in dioecious plants. Adv Genet. 1958;9:217–281. doi: 10.1016/s0065-2660(08)60163-7. [DOI] [PubMed] [Google Scholar]
- 14.Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- 15.Nicolas M, Marais G, Hykelova V, Janousek B, Laporte V, et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 2005;3:e4. doi: 10.1371/journal.pbio.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filatov DA, Moneger F, Negrutiu I, Charlesworth D. Low variability in a Y-linked plant gene and its implications for Y-chromosome evolution. Nature. 2000;404:388–390. doi: 10.1038/35006057. [DOI] [PubMed] [Google Scholar]
- 18.Laporte V, Filatov DA, Kamau E, Charlesworth D. Indirect evidence from DNA sequence diversity for genetic degeneration of Y-chromosome in dioecious species of the plant Silene: the SlY4/SlX4 and DD44-X/DD44-Y gene pairs. J Evol Biol. 2005;18:337–347. doi: 10.1111/j.1420-9101.2004.00833.x. [DOI] [PubMed] [Google Scholar]
- 19.Matsunaga S, Isono E, Kejnovsky E, Vyskot B, Dolezel J, et al. Duplicative transfer of a MADS box gene to a plant Y chromosome. Mol Biol Evol. 2003;20:1062–1069. doi: 10.1093/molbev/msg114. [DOI] [PubMed] [Google Scholar]
- 20.Rautenberg A, Hathaway L, Oxelman B, Prentice HC. Geographic and phylogenetic patterns in Silene section Melandrium (Caryophyllaceae) as inferred from chloroplast and nuclear DNA sequences. Molecular Phylogenetics and Evolution. 2010;57:978–971. doi: 10.1016/j.ympev.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Heilbuth JC. Lower species richness in dioecious clades. American Naturalist. 2000;156:221–241. doi: 10.1086/303389. [DOI] [PubMed] [Google Scholar]
- 22.Filatov DA. Evolutionary history of Silene latifolia sex chromosomes revealed by genetic mapping of four genes. Genetics. 2005;170:975–979. doi: 10.1534/genetics.104.037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zluvova J, Lengerova M, Markova M, Hobza R, Nicolas M, et al. The inter-specific hybrid Silene latifolia×S. viscosa reveals early events of sex chromosome evolution. Evol Dev. 2005;7:327–336. doi: 10.1111/j.1525-142X.2005.05038.x. [DOI] [PubMed] [Google Scholar]
- 24.Mrackova M, Nicolas M, Hobza R, Negrutiu I, Moneger F, et al. Independent origin of sex chromosomes in two species of the genus Silene. Genetics. 2008;179:1129–1133. doi: 10.1534/genetics.107.085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popp M, Oxelman B. Evolution of a RNA polymerase gene family in Silene (Caryophyllaceae) — incomplete concerted evolution and topological congruence among paralogues. Systematic Biology. 2004;53:914–932. doi: 10.1080/10635150490888840. [DOI] [PubMed] [Google Scholar]
- 26.Oxelman B, Lidén M. Generic boundaries in the tribe Sileneae (Caryophyllaceae) as iInferred from nuclear rDNA sequences. Taxon. 1995;44:525–542. [Google Scholar]
- 27.Oxelman B, Lidén M, Berglund D. Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Systematics and Evolution. 1997;206:393–410. [Google Scholar]
- 28.Oxelman B, Lidén M, Rabeler R, Popp M, Oxelman B. A revised generic classification of the tribe Sileneae (Caryophyllaceae). Nordic Journal of Botany. 2001;20:743–748. [Google Scholar]
- 29.Fior S, Karis PO, Casazza G, Minuto L, Sala F. Molecular phylogeny of the Caryophyllaceae (Caryophyllales) inferred from chloroplast matK and nuclear rDNA ITS sequences. American Journal of Botany. 2006;93:399–411. doi: 10.3732/ajb.93.3.399. [DOI] [PubMed] [Google Scholar]
- 30.Erixon P, Oxelman B. Reticulate or tree-like chloroplast DNA evolution in Sileneae (Caryophyllaceae)? Molecular Phylogenetics and Evolution. 2008;48:313–325. doi: 10.1016/j.ympev.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Frajman B, Oxelman B. Reticulate phylogenetics and phytogeographical structure of Heliosperma (Sileneae, Caryophyllaceae) inferred from chloroplast and nuclear DNA sequences. Molecular Phylogenetics and Evolution. 2007;43:140–155. doi: 10.1016/j.ympev.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Desfeux C, Lejeune B. Systematics of euromediterranean Silene (Caryophyllaceae) - evidence from a phylogenetic analysis using ITS sequences. Comptes Rendus de l'Academie des Sciences Serie III-Sciences de la Vie-Life Sciences. 1996;319:351–358. [PubMed] [Google Scholar]
- 33.Kejnovsky E, Kubat Z, Hobza R, Lengerova M, Sato S, et al. Accumulation of chloroplast DNA sequences on the Y chromosome of Silene latifolia. Genetica. 2006;128:167–175. doi: 10.1007/s10709-005-5701-0. [DOI] [PubMed] [Google Scholar]
- 34.Mower J, Touzet P, Gummow J, Delph L, Palmer J. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sloan D, Oxelman B, Rautenberg A, Taylor D. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol. 2009;9:260. doi: 10.1186/1471-2148-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapralov MV, Filatov DA. Molecular adaptation during adaptive radiation in the Hawaiian endemic genus Schiedea. PLoS ONE. 2007;1:e8. doi: 10.1371/journal.pone.0000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muir G, Filatov D. A selective sweep in the Chloroplast DNA of dioecious Silene (Section Elisanthe). Genetics. 2007;177:1239–1247. doi: 10.1534/genetics.107.071969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erixon P, Oxelman B. Whole-gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS ONE. 2008;3:e1386. doi: 10.1371/journal.pone.0001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergero R, Qiu S, Forrest A, Borthwick H, Charlesworth D. Ms in preparation. [DOI] [PMC free article] [PubMed]
- 40.Atanassov I, Delichere C, Filatov DA, Charlesworth D, Negrutiu I, et al. Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol Biol Evol. 2001;18:2162–2168. doi: 10.1093/oxfordjournals.molbev.a003762. [DOI] [PubMed] [Google Scholar]
- 41.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 42.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 43.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 44.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 45.Roure B, Rodriguez-Ezpeleta N, Philippe H. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evolutionary Biology. 2007;7:S2. doi: 10.1186/1471-2148-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leigh J, Susko E, Baumgartner M, Roger A. Testing congruence in phylogenomic analysis. Systematic Biology. 2008;57:104–115. doi: 10.1080/10635150801910436. [DOI] [PubMed] [Google Scholar]
- 47.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 48.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online — a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research. 2005;33:W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filatov DA, Charlesworth D. Substitution rates in the X- and Y-linked genes of the plants, Silene latifolia and S. dioica. Mol Biol Evol. 2002;19:898–907. doi: 10.1093/oxfordjournals.molbev.a004147. [DOI] [PubMed] [Google Scholar]
- 50.Marais GA, Nicolas M, Bergero R, Chambrier P, Kejnovsky E, et al. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr Biol. 2008;18:545–549. doi: 10.1016/j.cub.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 51.Qiu S, Bergero R, Forrest A, Kaiser VB, Charlesworth D. Nucleotide diversity in Silene latifolia autosomal and sex-linked genes. Proc Biol Sci. 2010;277:3283–3290. doi: 10.1098/rspb.2010.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 53.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 54.Criscuolo A, Berry V, Douzery E, Gascuel O. SDM: a fast distance-based approach for (super) tree building in phylogenomics. Systematic Biology. 2006;55:740–755. doi: 10.1080/10635150600969872. [DOI] [PubMed] [Google Scholar]
- 55.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 56.Edwards SV, Liu L, Pearl DK. High-resolution species trees without concatenation. Proceedings of the National Academy of Sciences of the USA. 2007;104:5936–5941. doi: 10.1073/pnas.0607004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rannala B, Yang Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linder C, Rieseberg L. Reconstructing patterns of reticulate evolution in plants. American Journal of Botany. 2004;91:1700–1708. [PMC free article] [PubMed] [Google Scholar]
- 59.Cummings M, Neel M, Shaw K. A genealogical approach to quantifying lineage divergence. Evolution. 2008;62:2411–2422. doi: 10.1111/j.1558-5646.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 60.Liu L, Yu L. Phybase: an R package for species tree analysis. Bioinformatics. 2010;26:962–963. doi: 10.1093/bioinformatics/btq062. [DOI] [PubMed] [Google Scholar]
- 61.Liu L, Yu L, Pearl D, Edwards S. Estimating species phylogenies using coalescence times among sequences. Syst Biol. 2009;58:468–477. doi: 10.1093/sysbio/syp031. [DOI] [PubMed] [Google Scholar]
- 62.Jobb G, Haeseler Av, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evolutionary Biology. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Systematic Biology. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 64.Jürgens A, Witt T, Gottsberger G. Pollen grain numbers, ovule numbers and pollen-ovule ratios in Caryophylloideae: correlation with breeding system, pollination, life form, style number, and sexual system. Sex Plant Reprod. 2002;14:279–289. [Google Scholar]
- 65.Matsunaga S, Yagisawa F, Yamamoto M, Uchida W, Nakao S, et al. LTR retrotransposons in the dioecious plant Silene latifolia. Genome. 2002;45:745–751. doi: 10.1139/g02-026. [DOI] [PubMed] [Google Scholar]
- 66.Touzet P, Delph L. The effect of breeding system on polymorphism in mitochondrial genes of Silene. Genetics. 2009;181:631–644. doi: 10.1534/genetics.108.092411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 68.Markova M, Michu E, Vyskot B, Janousek B, Zluvova J. An interspecific hybrid as a tool to study phylogenetic relationshipsin plants using the GISH technique. Chromosome Research. 2007;15:1051–1059. doi: 10.1007/s10577-007-1180-8. [DOI] [PubMed] [Google Scholar]
- 69.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 70.Popp M, Oxelman B. Inferring the history of the polyploid Silene aegaea (Caryophyllaceae) using plastid and homoeologous nuclear DNA sequences. Molecular Phylogenetics and Evolution. 2001;20:474–481. doi: 10.1006/mpev.2001.0977. [DOI] [PubMed] [Google Scholar]
- 71.Li G, Steel M, Zhang L. More taxa are not necessarily better for the reconstruction of ancestral character states. Systematic Biology. 2008;57:647–653. doi: 10.1080/10635150802203898. [DOI] [PubMed] [Google Scholar]
- 72.Salisbury B, Kim J. Ancestral state estimation and taxon sampling density. Syst Biol. 2001;50:557–564. [PubMed] [Google Scholar]
- 73.Mabberley D. The Plant Book. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- 74.Lidén M, Popp M, Oxelman B. A revised generic classification of the tribe Sileneae (Caryophyllaceae). Nordic Journal of Botany. 2001;20:513–518. [Google Scholar]
- 75.Bergero R, Charlesworth D, Filatov DA, Moore RC. Defining regions and rearrangements of the Silene latifolia Y chromosome. Genetics. 2008;178:2045–2053. doi: 10.1534/genetics.107.084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rautenberg A, Filatov D, Svennblad B, Heidari N, Oxelman B. Conflicting phylogenetic signals in the SlX1/Y1 gene in Silene. BMC Evol Biol. 2008;8:299. doi: 10.1186/1471-2148-8-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual gene trees for our 13 genes. Trees were obtained with PhyML (see Material and Methods) with 100 non-parametric bootstrap replicates (only values >50% are shown). Note that some of the sequences were excluded from the further analyses, as follows: cDNA sequences from S. latifolia for ABCtr, PGK, SlXY4 and SlXYCyp, whose sequence lengths were too different, and the Petrocoptis sequence of OxRZn because of doubts of the orthology of the sequence. For each gene, there is one outgroup sequence. As noted in Table S2 and footnote 4 of Table S3, the outgroup species used to root the tree is usually Petrocoptis hispanica; this sequence was the outgroup for 8 of the 13 trees shown, including all the X-linked genes, but, for PGK, a Dianthus sequence was used, and Lychnis for ATUB-A and OxRZn.
(TIFF)
Other inferred phylogenies of the Silene species studied. For details about sequences and species, see Table S3. Only bootstrap values >50% are shown. A) Maximum parsimony tree of the ITS sequences from 12 species from [4] that were also studied by ourselves. The different mating systems are indicated, and stars indicate bootstrap values larger than 50%. B) Maximum likelihood tree (PhyML) of the ITS sequences from 13 of our species. C) Maximum likelihood tree of 7 autosomal genes concatenated by Concaterpillar (see Table S4) with all the species (except outgroups for 3 genes: 2A10, ClpP3 and ELF, see main text and Table S4) obtained with PhyML. Identical results were found with 4 or 8 categories of sites. In trees C and D, and also those in Figure 1 of the main text, only X sequences were included for the sex-linked genes. D) Dataset as in C, but the tree was obtained by combining all the 7 gene trees using the SDM method. In C, S. conica and S. noctiflora diverge early, but in D they group with S. vulgaris and S. viscosa.
(TIFF)
Tree for dioecious species, rooted using S. diclinis (as this species is early diverging in all trees in Figure 1 and Figure S2). The tree was obtained by PhyML on the concatenate of all genes without Gblocking. Values indicate the results of non-parametric bootstrapping with 100 replicates (only values >50% are shown).
(TIFF)
Estimated species trees, using all genes except for LIP21 (12 loci). A) Tree using STAR. B) Tree using STEAC. In both cases, the branch lengths are proportional to the bootstrap support, which is given as a percentage at each branch. As for the individual locus trees, the outgroup species was Petrocoptis hispanica for all genes except PGK, where a Dianthus sequence was used, and ATUB-A and OxRZn where no outgroups were available.
(TIFF)
Provenance and voucher details of plants used to assess autosomal and sex-linked gene phylogenies in Silene .
(DOC)
Primers used to amplify genomic DNA of the genes studied, lengths of the sequences studied, and the outgroup used.
(DOC)
List of species and sequences.
(DOC)
Results of tests for combining alignments using Concaterpillar.
(DOC)
Discussion on the differences in branch length in our trees.
(RTF)
Methods and results for the STAR and STEAC analysis.
(RTF)
(RTF)