Abstract
Background
E. coli belonging to the phylogenetic group B2 are linked to Inflammatory Bowel Disease (IBD). Studies have shown that antimicrobials have some effect in the treatment of IBD, and it has been demonstrated that E. coli Nissle has prophylactic abilities comparable to 5-aminosalicylic acid (5-ASA) therapy in ulcerative colitis. The objective of this study was to test if ciprofloxacin and/or E. coli Nissle could eradicate IBD associated E. coli in the streptomycin-treated mouse intestine.
Results
After successful colonization with the IBD associated E. coli strains in mice the introduction of E. coli Nissle did not result in eradication of either IBD associated strains or an E. coli from a healthy control, instead, co-colonization at high levels were obtained. Treatment of mice, precolonized with IBD associated E. coli, with ciprofloxacin for three days alone apparently resulted in effective eradication of tested E. coli. However, treatment of precolonized mice with a combination of ciprofloxacin for 3 days followed by E. coli Nissle surprisingly allowed one IBD associated E. coli to re-colonize the mouse intestine, but at a level 3 logs under E. coli Nissle. A prolonged treatment with ciprofloxacin for 7 days did not change this outcome.
Conclusions
In the mouse model E. coli Nissle can not be used alone to eradicate IBD associated E. coli; rather, 3 days of ciprofloxacin are apparently efficient in eradicating these strains, but surprisingly, after ciprofloxacin treatment (3 or 7 days), the introduction of E. coli Nissle may support re-colonization with IBD associated E. coli.
Introduction
The etiology of Inflammatory Bowel Disease (IBD) is so far unknown, it is, however, believed that both genetic and environmental factors are involved in the pathogenesis of IBD. Some characteristic pathological elements in IBD, such as mucosal inflammation, mucosal ulcers, and in the case of Crohn's disease, epithelioid cell granulomas also occur in infectious diseases. Therefore, many different bacteria, viruses and other microorganisms have been suspected to cause IBD. It is now well established that luminal factors in the intestine are involved in the inflammatory process of Crohn's disease (CD) and ulcerative colitis (UC). For example, diversion of the continuity of the intestines results in healing of the resting gut, whereas the inflammation will return when continuity is reestablished [1]. Furthermore, several animal models have documented the participation of intestinal bacteria in the inflammatory process [2]. More importantly, the recent finding of a defect in the caspase recruitment domain family, member 15 (NOD2/CARD15), gene among CD patients, has reawakened the search for specific pathogenic microorganisms in IBD [3]. NOD2/CARD15 is believed to be involved in the innate immune system including the production of defensins, and defects in this gene could indicate that the host is more susceptible to microorganisms [4]. It has accordingly been shown that the number of viable internalized S. typhimurium in Caco2 cells was higher when the Caco2 cells were transfected with a variant NOD2 expression plasmid associated with Crohn's disease [5].
Escherichia coli are among the most interesting bacteria in the human gut. Certain E. coli types with specific virulence factors are involved in childhood diarrhea, (enteropathogenic E. coli), tourist diarrhea (enterotoxigenic E. coli), and bloody diarrhea associated with hemolytic uremic syndrome (verotoxin-producing E. coli). Already in the 1970's, it was found that hemolytic E. coli were linked to active UC [6]. Furthermore, E. coli was linked to CD since an abundance of specific adherent-invasive E. coli (AIEC) was found in resected ileum from patients with CD [7], [8]. Interestingly, AIEC isolated from patients with CD have the ability to survive and replicate within the phagocytes without inducing cell death and AIEC-infected macrophages secrete large amounts of Tumor Necrosis Factor-α [9] It was shown that E. coli are very predominant in inflamed mucosa of patients with UC. Furthermore, these strains are “active”, based on 16 S rRNA PCR, and overrepresented in comparison with the microbiota of healthy controls, who generally had a higher biodiversity of the active microbiota [10]. Moreover, it has been demonstrated by ribosomal intergenic spacer analysis that Enterobacteriaceae are more abundant in tissue samples from patients with IBD compared to controls, and after culture, specific phylogenetic groups, B2 and D, of E. coli were found to be more frequent among patients with IBD [11]. Recently we showed that significantly more patients with active IBD were found to be infected with B2 E. coli strains with at least one positive extraintestinal pathogenic E. coli (ExPEC) gene compared to IBD patients with inactive disease [12]. In addition, an exuberant inflammatory response to E. coli has been demonstrated among patients with UC [13]. In a meta-analysis of placebo-controlled trials it was concluded that antimicrobials have some effect in the treatment of both CD [14], and ulcerative colitis [15]. Furthermore, the probiotic E. coli strain (E. coli Nissle), originally isolated during World War I from a soldier who withstood a severe outbreak of diarrhea affecting his detachment, prevents relapse of UC just as well as 5-aminosalicylic acid (5-ASA) and E. coli Nissle has also been proposed for maintenance therapy of Crohn's disease [16], [17]. Part of E. coli Nissle's probiotic abilities are presumably linked to its ability to prevent colonization of the gut with pathogenic microorganisms by producing two microcins and being able to produce a strong biofilm [18], [19]. These facts make it plausible that a combination of an antimicrobial and E. coli Nissle could be efficient in eradicating IBD associated E. coli and possibly in treating patients with IBD. The streptomycin treated mouse model was developed based on the observation that streptomycin treatment eliminated the gram-negative flora from the gut, leaving almost intact the gram-positive flora and the strict anaerobes making this model ideal for testing the ability of gram negatives to colonize the gut [20].
Our objective was, in order to understand the possible role of antimicrobials and E. coli Nissle in the treatment of patients with IBD, to test the ability of ciprofloxacin and/or E. coli Nissle to eradicate precolonized IBD associated E. coli strains (B2 strains isolated from patients with active IBD) in the streptomycin treated mouse intestine.
Methods
Ethics Statement
All animal experiments were approved by The Animal Experiments Inspectorate, The Danish Ministry of Justice (Permission no. 2007/561-1430). Animal experiments were performed by skilled personnel. Permission for collection of human specimens and recruitment of participants was obtained from the Regional Ethics Committee for Copenhagen County Hospitals (Permission no. KA03019) and all participants gave their informed written consent.
Clinical data
The IBD E. coli strains were collected from patients with active ulcerative colitis (IBD1 and IBD2) and from a healthy control person, disease activity was confirmed by sigmoidoscopy, both IBD strains were of the phylogenetic group B2 and the control strain was of the phylogenetic group A. E. coli strains IBD1, IBD2 and the control strain were previously molecularly characterised [12] and designated as p7, p25 and c17 respectively. Both strains isolated from patients with IBD were found to be positive in five of six genes associated with extraintestinal pathogenic E. coli (ExPEC), while the strain from the control patient was negative for all tested ExPEC genes.
Bacterial strains and media
All strains were tested for antimicrobial susceptibility against a panel of 18 different antimicrobials in a variety of concentrations by using Sensititre® DKMVN2. 20 ml of Muller Hilton broth (SSI nr. 1054, Copenhagen, DK) were inoculated with 10 µl of a 0.5 MacFarland (McF) solution. The inoculated Muller Hilton broth was distributed using a Sensititre AutoInoculator (TREK Diagnostic Systems, LTD, England) in the 96×microtitreplate (50 µl/well) containing the antibiotics. The microtitterplate were incubated for 18–22 h at 37°C where after the growth were visualised in Sensititre SeniTouch™ (TREK Diagnostic Systems, LTD, England) manually reading (growth/no growth) but analysed using Sensititre® Windows Software SWIN® (TREK Diagnostic Systems, LTD, England). The tested antimicrobials were amoxicillin+clavulanic acid (2∶1) 2/1–32/16 mg/L, ampicillin 1–32 mg/L, apramycin 4–32 mg/L µg/ml, cefpodoxime 0.125–4 mg/L, ceftiofur 0.5–8 mg/L, cephalothin 4–32 mg/L, chloramphenicol 2–64 mg/L, ciprofloxacin 0.03–4 mg/L, colistin 4–16 mg/L, florfenicol 2–64 mg/L, gentamicin 1–32 mg/L, nalidixic acid 4–64 mg/L, neomycin 2–32 mg/L, spectinomycin 16–256 mg/L, streptomycin 4–64 mg/L, sulphamethoxazole 64–1024 mg/L, tetracycline 2–32 mg/L and trimethoprim 4–32 mg/L. E. coli IBD1 and the E. coli control were susceptible to all antimicrobials tested. E. coli IBD2 strain was found resistant to ampicillin, sulphamethoxazole, streptomycin, trimethoprim and cephalothin. First a spontaneous streptomycin mutant of the E. coli IBD1 strain and the E. coli strain from a healthy control were constructed then a genetic marker transposon Tn7 kanamycin resistant gene cassette was inserted downstream of the coding region of the gene [21]. A spontaneous streptomycin and rifampicin resistant mutant of Nissle 1917, DSM 6601 was used as the probiotic strain (hereafter called Nissle). All resistant mutants were analyzed for any inhibiting/enhancing metabolic and/or growth changes and none were found. All strains were grown in Luria-Bertani (Sigma-aldrich, St. Louis, USA) media at 37°C overnight. Unless otherwise stated, the antimicrobials and chemicals used in this study were of analytical grade and obtained from Sigma-aldrich, St. Louis, USA.
Mouse colonization experiments
Six-to-eight-week-old, outbreed albino female CFW1 mice (Harlan Laboratories, Netherlands) were used for the studies. The mice were caged in groups of three mice, and cages were changed weekly. The mice had unlimited access to food and continuously received water containing streptomycin sulphate (5 g/L) prior to inoculation with the strains and throughout the experiment. Each experiment included three mice colonized individually. Faecal samples from each mouse were tested prior to inoculation for the presence of indigenous E. coli with similar resistance. Inoculum suspension was prepared by overnight cultures (∼109 CFU/mouse) resuspended in 20% (w/v) sucrose. Each mouse was given 100 µL bacterial suspensions orally. Faecal samples were collected individually with a 2 to 3 days interval and the numbers of CFU were determined by serially dilution and spread on selective agar plates. Selection between strain were conducted using LB-plates containing: 100 mg/L streptomycin and 25 mg/L kanamycin (for IBD1 and control strain), 100 mg/L streptomycin and 50 mg/L ampicillin (for IBD2) and 100 mg/L streptomycin and 100 mg/L rifampicin were used as selective plates for Nissle. The mice were euthanized and experimental duration time was between 3 to 6 weeks.
Treatment of colonized mice with E. coli Nissle
The E. coli strains (IBD1, IBD2 or control) were given orally at aprox 5×108 CFU/mouse once. After 6 days mice were inoculated with the probiotic strain E. coli Nissle given orally, aprox 8×109 CFU/mouse. Colonization levels were tested daily through out the experiment by faecal cultures.
Treatment with ciprofloxacin for 3 days
The E. coli strains (IBD1, IBD2 or control) were given orally at aprox 8×108 CFU/mouse once. The strains were allowed to colonize the mouse intestine for 6 days before initiation of subcutaneous injections of ciprofloxacin (Ciproxin, 2 mg/ml Bayer) treatment every 6th hour (0.2 mg/mouse) for 3 days. Colonization levels were tested daily through out the experiment by faecal cultures.
Treatment of colonized mice with combinations of ciprofloxacin and E. coli Nissle
E. coli strains (IBD1, IBD2 or control) were given orally at aprox 8×108 CFU/mouse. The strains were allowed to colonize the mouse intestine for 6 days before initiation of subcutaneous injections of ciprofloxacin (Ciproxin, 2 mg/ml Bayer) every 6th hour (0.2 mg/mouse) for 3 day or 7 days. After antibiotic treatment mice were inoculated with the probiotic strain E. coli Nissle orally with aprox 9×109 CFU/mouse daily. Colonization levels were tested daily throughout the experiment by faecal cultures
Verifications of inoculated strains
Colonies on the selective plates were continuously verified as the inoculated strains by plasmid profile, PCR or Biochemical assays (MiniBactE, Statens Serum Institut, Diagnostica, Hillerød, Denmark) throughout the experiment.
Plasmid purification was described in detail by Schjørring et al [22]. PCR was used when verifying strains with the kanamycin resistance gene cassette insert. The primers that were used to detect the kanamycin gene cassette were: F-5′GAT GCT GGT GGC GAA GCT GT -3′, R 5′-GAT GAC GGT TTG TCA CAT GGA-3′; Original wild type (kans) and the inoculated strain were used as negative/positive controls. Expand High Fidelity PCR system (2.6 U/reaction) (ROCHE Diagnostics GmbH, Mannheim, Germany) and boiling lysate were used. All PCR amplifications were performed in Peltier Thermal Cycler DNA engine DAYD™ (VWR™ international, Albertslund, Denmark) using the following PCR program: 2 min at 94°C; 30 cycles of 15 s at 94°C, 30 s at 55°C and 3 min at 72°C; and 7 min at 72°C. The PCR products were run at 50V on a 0.8% SeaKem® LE agarose gel (Lonza Rockland, ME, USA).
Results
Treatment of colonized mice with E. coli Nissle
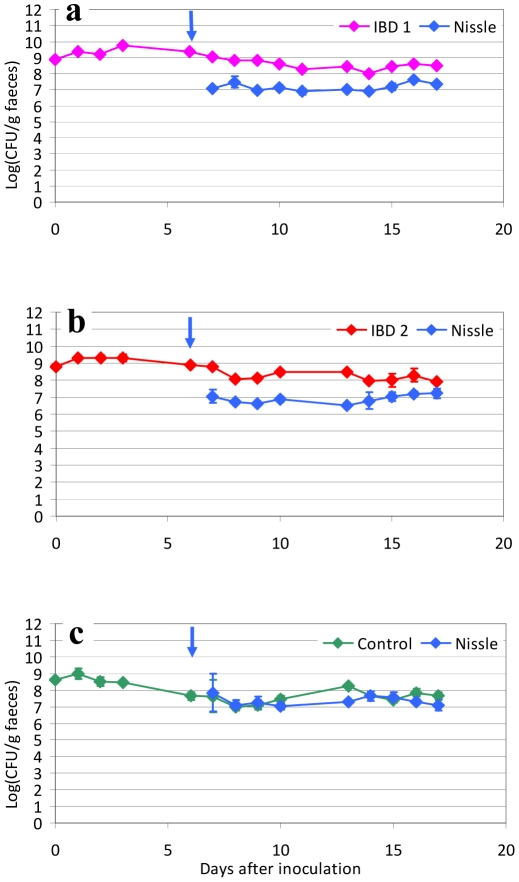
To test if E. coli Nissle alone was efficient in eradication of IBD associated E. coli, mice were precolonized by two E. coli strains isolated from patients with active ulcerative colitis (IBD1 and IBD2) and one E. coli strain isolated from a healthy person. Groups of three mice were inoculated at day 0 with one of the strains IBD1, IBD2 or the control strain, and colonization was obtained at a stable level which was demonstrated by subsequent culture of faecal samples from the inoculated mice. On the sixth day after inoculation, E. coli Nissle was introduced by an inoculum of 109 CFU given for three days. The introduction of E. coli Nissle did not result in eradication or any notable changes in number of CFU in samples taken from the mice until day 17 (11 days after the introduction of E. coli Nissle) of either IBD1, IBD2 or the E. coli from a healthy control; however, co-colonization was obtained (fig. 1 a–c). The two pathogenic strains IBD1 and IBD2 did show some colonization advantage compared to E. coli Nissle, since E. coli Nissle did colonize 1 log under the precolonized strain, however at a stable level (fig. 1, a and b). Mice inoculated with the commensal E. coli strain isolated from a healthy control co-colonized with E. coli Nissle and resulted in colonization at equal levels (fig. 1 c).
Figure 1. Inoculation of E. coli Nissle in mice pre-colonized with IBD associated E. coli or a control strain.
Sets of three mice were used in each experiment. CFU of the inoculation suspension of IBD/control strain are shown at day 0 (∼109 CFU/mouse). Blue arrow indicates the inoculation of E. coli Nissle strain at day 6 (∼109 CFU/mouse). Each graph represents the CFU counts of three mice and bars represent Standard error of the means (SEM). Detection Limit (DL) at 20 CFU/g faeces.
Treatment of colonized mice with ciprofloxacin for 3 days
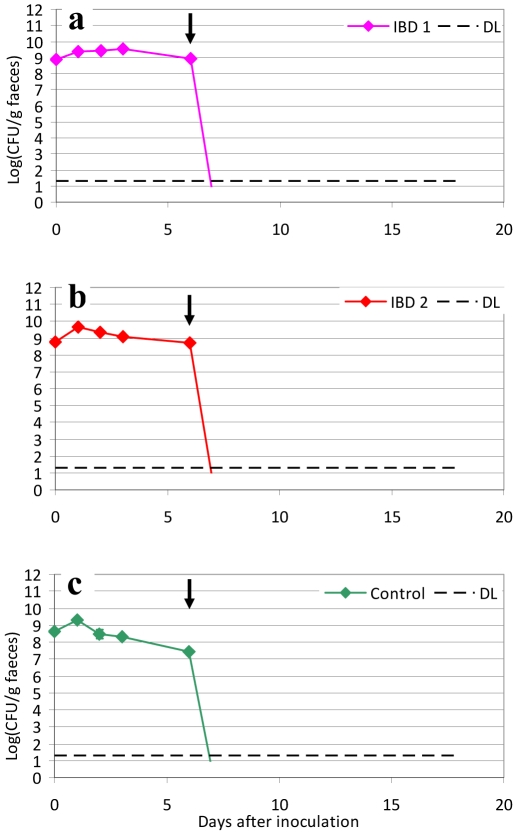
Mice were precolonized with the three test strains by inoculation of mice with 109 CFU and achieving a stable colonization level. On the 6th day mice were treated with subcutaneous injections of ciprofloxacin 0.2 mg/mouse every 6th hour (7 am, 1 pm, 7 pm and 1 am) from day 6 to 8 after inoculation. Culture of faecal samples from the mice revealed an apparent efficient eradication of both IBD1 and IBD2, and of the strain isolated from a healthy person (control), with no E. coli detected for the following nine days (fig. 2 a–c), with a detection limit of 20 CFU/g faeces.
Figure 2. Three days treatment with ciprofloxacin in mice pre-colonized with IBD associated E. coli or a control strain.
Sets of three mice were used in each experiment. CFU counts of the inoculated strains from faecal samples of mice. CFU of the inoculation suspension is shown at day 0 (∼109 CFU/mouse). Black arrow indicates the initiation of ciprofloxacin treatment from day 6–9 (0.2 mg/mouse) every 6 h. Detection limit 20 CFU/g faeces (dotted line). Each graph represents the CFU counts of three mice and bars represent SEM. Detection Limit (DL) at 20 CFU/g faeces.
Treatment of colonized mice with combinations of ciprofloxacin and E. coli Nissle
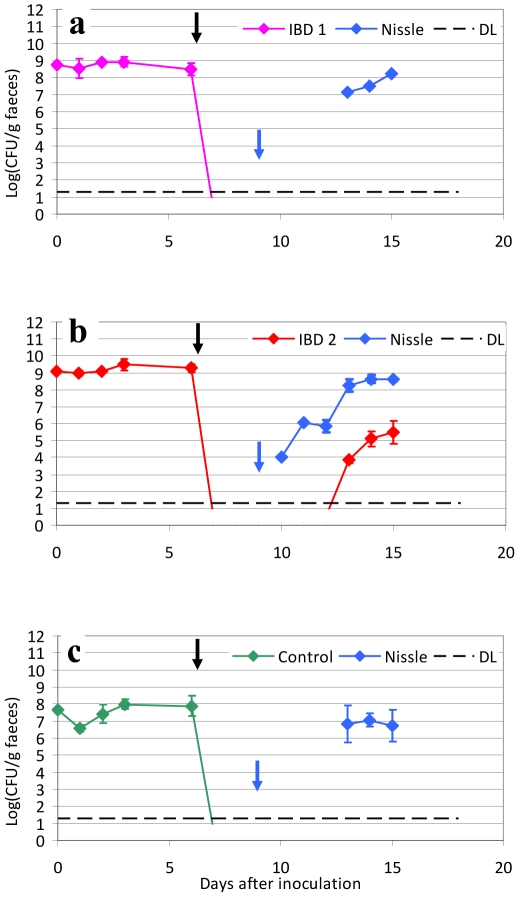
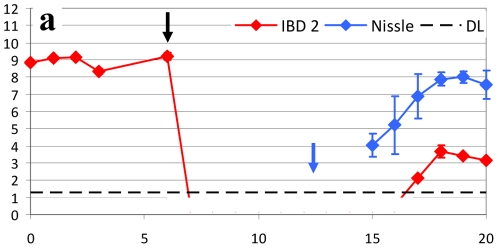
Since E. coli Nissle has previously been shown to be efficient as a prophylactic treatment in IBD, we aimed at testing the possibility of colonizing the streptomycin treated mouse intestine with E. coli Nissle after clearing the precolonized IBD associated E. coli strains from the mouse gut with 3 days of ciprofloxacin treatment. However, combination of ciprofloxacin for 3 days followed by E. coli Nissle for 3 days resulted in lack of colonization with E. coli Nissle (data not shown). Instead we chose to inoculate mice with E. coli Nissle 109 CFU daily for the rest of the study. This is in accordance with the clinical approach in IBD patients treated with E. coli Nissle. With daily inoculation, E. coli Nissle was found in stable numbers in faecal samples from the tested mice (fig. 3 a–c). One of the tested IBD strains (IBD2) was surprisingly able to reappear after 4 days but only reaching a level 3 logs below E. coli Nissle (fig. 3 b). In a new set of experiments subcutaneously ciprofloxacin treatment was prolonged from three to seven days in all, however, this did not prohibit the IBD2 strain from reappearing at day 3 after the initiation of subsequent E. coli Nissle treatment (fig. 4). Within the study time the IBD2 strain reached a level of 4 to 5 log below E. coli Nissle. The reappeared IBD 2 strain was tested to see if sensitivity to ciprofloxacin was changed, and at day 19 one colony in each mouse were found to be ciprofloxacin resistant.
Figure 3. Three days treatment with ciprofloxacin and a subsequent treatment with E. coli Nissle daily in mice pre-colonized with IBD associated E. coli or a control strain.
Sets of three mice were used in each experiment. CFU counts of the inoculated strains from faecal samples of mice. CFU of the inoculation suspension is shown at day 0 (∼109 CFU/mouse). Black arrow indicates the initiation of ciprofloxacin treatment from day 6–9 (0.2 mg/mouse) every 6 h. Blue arrow indicates initiation of inoculation with E. coli Nissle strain at high levels (∼109 CFU/mouse) every day throughout the experiment. Each graph represents the CFU counts of three mice and bars represent SEM. Detection Limit (DL) at 20 CFU/g faeces.
Figure 4. Treatment with ciprofloxacin for 7 days followed by inoculation with E. coli Nissle.
Sets of three mice were used in each experiment. CFU counts of the inoculated strains from faecal samples of mice. CFU of the inoculation suspension is shown at day 0 (∼109 CFU/mouse). Black arrow indicates the initiation of ciprofloxacin treatment from day 6–13 (0.2 mg/mouse) every 6 h. Blue arrow indicates initiation of inoculation with E. coli Nissle strain at high levels (∼109 CFU/mouse) every day throughout the experiment. IBD associated E. coli strain 2 was apparently eradicated by 7 days of treatment with ciprofloxacin; however, strain 2 once again reappeared under treatment with E. coli Nissle. Each graph represents the CFU counts of three mice and bars represent SEM. Detection Limit (DL) at 20 CFU/g faeces (dotted line).
Discussion
The eradication of bacteria involved in the pathogenesis of IBD and their permanent replacement with non-pathogenic, probiotic bacteria may provide a new option for the treatment of IBD and for the maintenance of remission.
The probiotic E. coli strain Nissle 1917 was reported to maintain remission of ulcerative colitis and pouchitis and to prevent colitis in different murine models of colitis [16], [23]–[25]. Although E. coli Nissle was originally isolated in 1917, the underlying mechanism of its beneficial effect in various intestinal diseases, including ulcerative colitis still remains elusive. Recently, it was shown in the streptomycin treated mouse model that E. coli Nissle can limit the growth of pathogenic E. coli O157 when administrated as treatment in precolonized mice [26]. It has been suggested that the different nutrient utilization of the different strains colonizing the intestine play an important role in the colonization ability of the strains [27]. Theoretically it is possible that E. coli Nissle ousts other more harmful bacteria involved in the pathogenesis of ulcerative colitis. Furthermore, E. coli of the phylogenetic group B2, having several virulence factors in common with ExPEC, were found in patients with active IBD. E. coli Nissle is also of the phylogenetic group B2 thus sharing many traits with members of this group (including uropathogenic E. coli) [19]. An intriguing mechanism in the prophylactic effect of Nissle in IBD could be that Nissle is able to inactivate IBD associated E. coli or that Nissle is able to hinder re-infection with IBD associated E. coli. In the present study the introduction of E. coli Nissle for 3 days, after precolonization of streptomycin treated mice with IBD associated E. coli, did not eradicate the infection with the IBD associated E. coli. Although E. coli Nissle 1917 is known to produce microcins M and H47 [18], it did not seem to be efficient in eradicating the IBD associated E. coli, but instead co-colonization occurred. This does not rule out that E. coli Nissle in the human intestine interact with the possible harmful IBD associated E. coli by blocking their attachment to epithelial cells. This theory could be supported by a study showing E. coli Nissle's ability to block the adherence of AIEC in vitro experiments with an intestinal cell line [28]. Moreover in the mouse model E. coli do not seem to adhere to the epithelial cells, instead E. coli are found situated in the mucus layer [29]. As previously mentioned, it has been demonstrated that different E. coli strains, including experiments with E. coli Nissle, can co-exist based on the utilization of different nutrients [26]. This indicates that Nissle and IBD associated E. coli are not competing for the same nutrients in the streptomycin treated mouse intestine. However, it was also shown that infection with three different non-pathogenic E. coli including E. coli Nissle were able to prevent recolonization with a pathogenic (enterohemorhagic) E. coli [26]. Therefore it is possible that E. coli Nissle in the human intestine, in the presence of other gram-negative bacteria, would be successful in preventing recolonization with IBD associated E. coli.
Ciprofloxacin for three days was efficient in eradicating the colonization of the IBD associated E. coli, and subsequent colonization with E. coli Nissle was also possible, however, only with repeated inoculation. Surprisingly, it was found that one of the IBD associated E. coli strains reappeared after ciprofloxacin treatment, when mice were inoculated with E. coli Nissle. A longer duration of ciprofloxacin treatment for 7 days did not solve this problem. Apparently in the presence of E. coli Nissle this IBD associated E. coli strain reemerged, however at lower levels than before. Several mechanisms could be speculated, first of all a subset of IBD2 E. coli became ciprofloxacin resistant and they could therefore survive at a dormant level. Their reawakening is perhaps effectuated by nutrients made available by E. coli Nissle, by crosstalk or by E. coli Nissle promoting the adherence of the IBD associated E. coli in the mucus layer, since E. coli Nissle has a marked ability to form biofilm [19]. It is obvious, that a combined treatment with ciprofloxacin and E. coli Nissle does encompass several challenges; ciprofloxacin treatment should be followed by continuous E. coli Nissle treatment, and the reintroduction of some IBD associated E. coli could be facilitated by E. coli Nissle. It must however be remembered that colonization is only the first step in infection and that the streptomycin-treated mouse intestine is only a model for IBD associated E. coli colonization and not pathogenesis. Future studies will have to elucidate the mechanisms of co-colonization, and the mechanisms by which Nissle stimulates the growth of dormant possibly ciprofloxacin resistant IBD associated E. coli in the streptomycin treated mouse intestine. In conclusion, the role of E. coli Nissle in eradication of IBD associated E. coli is questionable. E. coli Nissle alone did not eradicate any of the tested E. coli strains in the mouse model; instead, co-colonization occurred, and although ciprofloxacin apparently eradicated the IBD associated E. coli, inoculation with daily doses of E. coli Nissle resulted in reappearance of some IBD associated E. coli strains in the streptomycin treated mouse intestine.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: These authors have no support or funding to report.
References
- 1.Janowitz HD, Croen EC, Sachar DB. The role of the faecal stream in Crohn's disease: an historical and analytic review. Inflamm Bowel Dis. 1998;4(1):29–39. doi: 10.1097/00054725-199802000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Madsen KL. Inflammatory bowel disease: lessons from the IL-10 gene-deficient mouse. Clin Invest Med. 2001;24(5):250–7. [PubMed] [Google Scholar]
- 3.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 4.Fellermann K, Wehkamp J, Herrlinger KR, Stange EF. Crohn's disease: a defensin deficiency syndrome? Eur J Gastroenterol Hepatol. 2003;15(6):627–34. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Hisamatsu T, Suzuki M, Reinecker HC, Nadeau WJ, McCormick BA, et al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124(4):993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 6.Cooke EM, Ewins SP, Hywel-Jones J, Lennard-Jones JE. Properties of strains of Escherichia coli carried in different phases of ulcerative colitis. Gut. 1974;15(2):143–6. doi: 10.1136/gut.15.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115(6):1405–13. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 8.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127(2):412–21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 9.Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, et al. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69(9):5529–37. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokol H, Lepage P, Seksik P, Doré J, Marteau P. Temperature gradient gel electrophoresis of Faecal 16S rRNA reveals active Escherichia coli in the Microbiota of patients with ulcerative colitis. J Clin Microbiol. 2006;44(9):3172–7. doi: 10.1128/JCM.02600-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogentic group in inflammatory bowel disease. Gut. 2007;56:669–75. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, et al. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol. 2009;9:171. doi: 10.1186/1471-2180-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marks DJ, Rahman FZ, Novelli M, Yu RC, McCartney S, et al. An exuberant inflammatory response to E. coli: Implications for the pathogenesis of ulcerative colitis and pyoderma gangrenosum. Gut. 2006;55(11):1662–3. doi: 10.1136/gut.2006.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feller M, Huwiler K, Schoepfer A, Shang A, Furrer H, et al. Long-term antibiotic treatment for Crohn's disease: systematic review and meta-analysis of placebo-controlled trials. Clin Infect Dis. 2010;50(4):473–80. doi: 10.1086/649923. [DOI] [PubMed] [Google Scholar]
- 15.Rahimi R, Nikfar S, Rezaie A, Abdollahi M. A meta-analysis of antibiotic therapy for active ulcerative colitis. Dig Dis Sci. 2007;52(11):2920–5. doi: 10.1007/s10620-007-9760-1. [DOI] [PubMed] [Google Scholar]
- 16.Kruis W, Schutz E, Fric P, Fixa B, Judmaier G, et al. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther. 1997;11(5):853–8. doi: 10.1046/j.1365-2036.1997.00225.x. [DOI] [PubMed] [Google Scholar]
- 17.Malchow HA. Crohn's disease and Eshcerichia coli. A new approach in therapy to maintain remission of colonic Crohn's disease? J Clin Gastroenterol. 1997;25(4):653–8. doi: 10.1097/00004836-199712000-00021. [DOI] [PubMed] [Google Scholar]
- 18.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors RepA, Cir, Fiu and IroN. Microbiology. 2003;149:2257–70. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 19.Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics. 2010;283(5):469–84. doi: 10.1007/s00438-010-0532-9. [DOI] [PubMed] [Google Scholar]
- 20.Myhal ML, Laux DC, Cohen PS. Relative colonizing abilities of human fecal and K12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur J Clin Microbiol. 1982;1:186–192. doi: 10.1007/BF02019621. [DOI] [PubMed] [Google Scholar]
- 21.McKenzie GJ, Craig NL. Fast, easy and efficient: site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 2006;6:39. doi: 10.1186/1471-2180-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schjørring S, Struve C, Krogfelt KA. Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J Antimicrob Chemother. 2008;62(5):1086–93. doi: 10.1093/jac/dkn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzela L, Kascak M, Vavrecka A. Induction and maintenance of remission with nonpathogenic Escherichia coli in patients with pouchitis. Am J Gastroenterol. 2001;96:3218–3219. doi: 10.1111/j.1572-0241.2001.05294.x. [DOI] [PubMed] [Google Scholar]
- 24.Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon AT. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomized trial. Lancet. 1999;354:635–639. doi: 10.1016/s0140-6736(98)06343-0. [DOI] [PubMed] [Google Scholar]
- 25.Schultz M, Strauch UG, Linde HJ, Watzl S, Obermeier F, et al. Preventive effects of Escherichia coli strain Nissle 1917 on acute and chronic intestinal inflammation in two different murine models of colitis. Clin Diagn Lab. 2004;11:372–378. doi: 10.1128/CDLI.11.2.372-378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, et al. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun. 2009;77(7):2876–86. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76(3):1143–52. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boudeau J, Glasser A-L, Julien S, Colombel JF, Darfeuille-Michaud A. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn's disease.. Aliment Pharmacol Ther. 2003;18:45–56. doi: 10.1046/j.1365-2036.2003.01638.x. [DOI] [PubMed] [Google Scholar]
- 29.Poulsen LK, Lan F, Kristensen CS, Hobolth P, Molin S, et al. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun. 1994;62:5191–4. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]