Abstract
Huntington’s disease (HD) is an inherited disorder characterized by neuronal dysfunction and degeneration in striatum and cerebral cortex. Although the signaling pathways involved in HD are not yet clearly elucidated, mutant huntingtin protein is a key factor in the induction of neurodegeneration. The mutant huntingtin protein alters intracellular Ca2+ homeostasis, disrupts intracellular trafficking and impairs gene transcription. In this review, I emphasize the effects of mutant huntingtin protein in Ca2+ handling and transcriptional factors. Transcriptional alterations are key factors in the deficits of several proteins involved in the cellular machinery. These proteins include neurotrophic factors such as brain-derived neurotrophic factor, fibroblast growth factor, glial-cell-line-derived neurotrophic factor, ciliary neurotrophic factor and neurturin that have been suggested to restore neuronal dysfunction, improve behavioral deficits and prolong the survival in animal models of HD. An understanding of the molecular pathways involved in neurodegeneration will shed light on the choice of neurotrophic factors targeting a specific neuronal population in HD and will consequently overcome behavioral deficits.
Keywords: neurotrophic factors, neurturin, mutant huntingtin protein, neuroprotection
INTRODUCTION
Huntington’s disease (HD) is an inherited disorder caused by expansion of a polyglutamine repeat within exon 1 of the huntingtin gene on chromosome 4 (Huntington’s Disease Collaborative Research Group) (1). The mutation of expanded CAG encodes the polyglutamine is the product for the mutant huntingtin protein, a key player in HD. Findings have shown that individuals carry from 6 to 35 CAG repeats are unaffected. The full penetrance occurs when the number of repeats exceeds 35; the gene encodes a version of huntingtin protein that leads to HD (2).
Alteration of the huntingtin protein is a key factor in the induction of dysfunction or neurodegeneration, both of which lead to HD. It is not clear how the mutated huntingtin protein induces neuronal dysfunction and neuronal degeneration. There is a possibility that HD is caused by accumulation of the polyglutamine fragments in the cytoplasm and nucleus. Consequently, the neuropathology includes neuronal atrophy in the cerebral cortex and the striatum, forebrain regions that process a wide range of information for behavioral output (3, 4). GABAergic medium spiny neurons are the major degenerated neurons in HD (3, 5, 6). It is noteworthy that the examination of postmortem brains from advanced HD patients shows degeneration of other brain regions, including the hippocampus, the angular gyrus, and the lateral tuberal nuclei of the hypothalamus (5, 7-9).
Currently, there is no treatment for HD; however, target compounds including neurotrophic factors have been considered to play a potentially significant role in neuroprotection. Studies have shown that there is a link between the mutant huntingtin protein and cellular proteins including neurotrophic factors (10-14). The regulation of the expression of neurotrophic factors and their receptors may play an important role in neuroprotection. Studies have used exogenous neurotrophic factors in HD animal models in order to establish trophic requirements of neurons. Among these neurotrophic factors are brain-derived neurotrophic factor (BDNF), fibroblast growth factor (FGF), glial cell line-derived neurotrophic factor (GDNF), ciliary neurotrophic factor (CNTF) and neurturin. These neurotrophic factors have been demonstrated to play a key role against the progression of HD and behavioral deficits found in animal models and patients. In this review paper, I discuss mutant huntingtin protein in HD with emphasis on its role in calcium homeostasis and apoptosis. The roles of mutant huntingtin protein in transcriptional factors and endocytosis-vesicluar transport are also discussed. Finally, I discuss the role of neurotrohpic factors in HD.
Mutant huntingtin protein in HD
Mutant huntingtin protein can interact with other cellular proteins, leading to the progression of HD (15). The formation of neuronal intranuclear inclusions that contain mutant huntingtin protein causes neuronal degeneration in transgenic HD mouse models (16). The huntingtin protein itself is a cytoplasmic protein that interacts with vesicular and cytoskeletal proteins [for review, see ref. (10)]. Furthermore, studies demonstrated that huntingtin protein plays an important role in intracellular trafficking, including membrane recycling, clathrin-mediated endocytosis, neuronal transport and postsynaptic signaling (17-22). Thus, mutant huntingtin protein is likely to have an impact on a wide range of cellular functions. In addition, mutant huntingtin protein interacts with transcriptional regulatory proteins (23-25). Moreover, the expanded polyglutamine repeats facilitate the interactions of mutant huntingtin protein with huntingtin protein-associated proteins selectively expressed in the striatum and cortex. Among these proteins are calmodulin, huntingtin protein-associated protein (HAP-1), huntingtin protein-interacting proteins (HIP-1 and 2) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The interactions of the mutant huntingtin protein with these proteins induce protein dysfunction and lead to toxicity characteristic of HD (16, 26-30). Thus, the mutant huntingtin protein may trigger a cascade of several intracellular pathways that cause death of some neurons, including medium spiny neurons, although huntingtin protein is expressed in all types of cells.
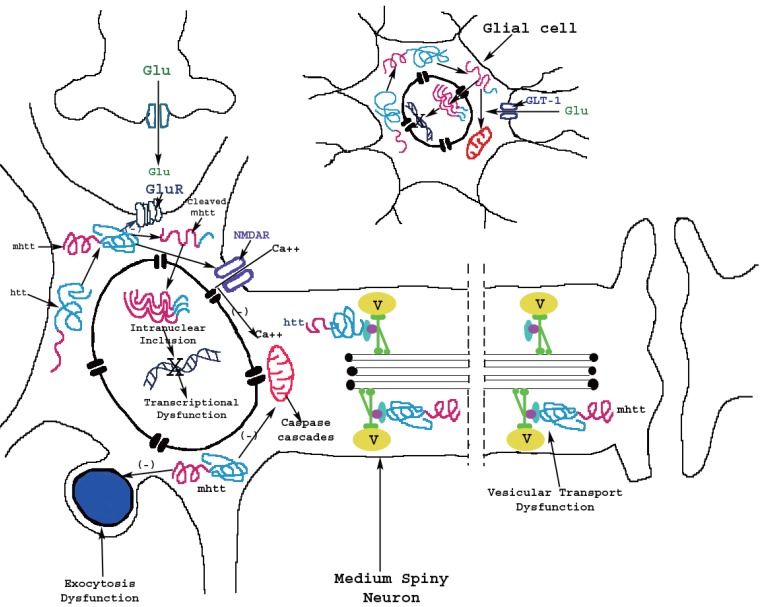
There is little known about the role of glia in HD neuropathology. A previous study has shown that mutant huntingtin protein accumulates in nuclei of glial cells in the brain of HD mouse models (31). Interestingly, intranuclear mutant huntingtin protein in glial cells increases with age (Fig. 1) and was found to be correlated with disease progression in the R6/2 transgenic HD mouse model, which shows neuropathological symptoms around 6-8 weeks and often dies after 12 weeks of age (16).
Figure 1.
Model for mechanism of actions of mutant huntingtin protein (mhtt) in medium spiny neuron and glial cell in Huntington’s disease. The huntingtin protein is transformed to mhtt through unknown mechanism, genetically or environmentally, in both neuron and glial cell. There are several theories suggesting different actions of mhtt: 1) mhtt protein may interact with metabotropic (GluR) or ionotropic glutamatergic (NMDA) receptors and alters their function, 2) the mhtt protein may bind to cellular transport components and induce vesicular transport (V) or exocytosis dysfunction. Moreover, mhtt protein might be proteolytically cleaved in amino-terminal fragments which form β-sheet structures. In cytoplasm, the cleaved mhtt may interact with mitochondria and alters their function. In addition, the cleaved mhtt can enter the nucleus and forms intranuclear aggregates or intranuclear inclusions, which induce transcriptional dysfunction. Both mutant full-length and cleaved forms of htt may form soluble monomers, oligomers or large insoluble aggregates. Similar mechanism of the actions of cleaved mhtt is suggested to occur in glial cells. The cleaved mhtt may alter glutamatergic system such as glutamate transporter 1 (GLT-1) and consequently alters the uptake of glutamate.
Effects of mutant huntingtin protein in calcium homeostasis. Although the mechanism of action of mutant huntingtin protein is not clear, evidence has shown that it may induce mitochondrial dysfunction [for review, see ref. (32, 33)]. There is an interrelationship between mitochondrial dysfunction and dysregulation of transcriptional factors in HD [for review, see (34)]. Defects of mitochondrial respiratory chain activity have been shown in striatum of postmortem brains of patients suffering from HD and also in R6/2 HD mouse models (35, 36). Isolated mitochondria from lymphoblasts of HD patients and from brains of the YAC72 transgenic HD mouse (yeast artificial chromosome, length of polyglutamine is 72) have shown deficit in intracellular Ca2+ (37). Moreover, mutant huntingtin protein induces impairment of Ca2+ homeostasis in cloned striatal cells (38). Cells expressing mutant huntingtin protein show a reduction in mitochondrial Ca2+ uptake compared to wild type cells. The mutant huntingtin protein-induced lower Ca2+ loads were attenuated in the presence of ADP; the decreases in the uptake of Ca2+ were abolished in the presence of permeability transition pore inhibitors (38). Moreover, a fragment of mutant huntingtin protein may be directly bound with mitochondria (39) (Fig. 1); this has been shown at the ultrastructural level in the brain of YAC72 HD mouse model (37). Although the mechanism of action of mutant huntingtin protein in mitochondrial Ca2+ handling is still unknown, one possibility is that mutant huntingtin protein acts directly on the ion permeability of the mitochondrial membrane (37). Interestingly, several genes related to calcium signaling, including copine V, striatin, SCNβ4 and α-actinin2, are altered in the R6/1 transgenic HD mouse model (40). The highest level of gene expression is found in the subunit of the sodium channel, SCNβ4, with a decrease of its expression level in striatum of R6/1 HD mouse model compared to wild type. It has been suggested that sodium levels are directly dependent on intracellular Ca2+ levels through sodium-calcium exchanger (41). The reduction in SCNβ4 expression in R6/1 HD mice may have a dramatic effect on intracellular calcium accumulation (41). Decreases in Ca2+ signaling genes are found in HD mouse models (42). Interestingly, mutant huntingtin protein-induced alteration of calcium signaling was found to lead to apoptosis of medium spiny neurons in the YAC128 HD mouse model (43). Alteration of intracellular Ca2+ homeostasis may be a factor in the induction of apoptosis and consequent neurodegeneration in HD.
Several lines of evidence suggest that stimulation of glutamatergic receptors such as ionotropic [N-methyl-D-aspartate receptors, NMDA receptors (subunits NR1/NR2R)] and metabotropic (mGluR5) glutamate receptors alter Ca2+ homeostasis in striatal medium spiny neurons in HD models (43-45). The overstimulation of these receptors, through application of excess glutamate, results in mitochondrial Ca2+ overload leading to apoptosis of medium spiny neurons. Excess glutamate might be associated with impaired glutamate transport, as it was demonstrated in HD animal models from studies performed by us and others (46-49). Alteration in glutamate uptake might be linked to a deficit in one of the major glutamate transporters, a glial glutamate transporter 1 (GLT1), as it was demonstrated in our recent study (50). GLT1 is one of the proteins that might be altered by mutant huntingtin protein (Fig. 1).
Evidence indicates that perturbations of Ca2+ homeostasis may lead to excitotoxicity and, consequently, apoptosis (43, 51). Activation of NR1/NR2B NMDA receptors induces a Ca2+ influx; activation of mGluR5 leads to production of InsP3 and Ca2+ release via InsP3R1 (43, 52). The mutant huntingtin protein alters the Ca2+ handling in medium spiny neurons of HD mouse model through NMDA and mGlutamate receptors. This results in an overload of cytosolic Ca2+ along with an excess of mitochondrial Ca2+ storage, which lead to cytochrome c release into the cytosol, inducing apoptosis through activation of the caspase cascade (53-55). The caspases convey the apoptotic signal in a proteolytic cascade, with caspases cleaving and activating other caspases that subsequently degrade cellular targets, leading to cell death. Upon mitochondrial stress through disruption of Ca2+ homeostasis, the release of cytochrome c may interact with Apaf-1, causing self-cleavage and activation of caspase-9. The effector of caspase cascade, such as caspase-3, -6 and -7, is downstream of the activator caspases and acts to cleave various cellular targets. Recent studies demonstrated that caspase-6 is a key factor in the cleavage of mutant huntingtin protein in HD (56). Thus, cleavage of mutant huntingtin protein by caspase-6 is an important event in mediating neuronal dysfunction and possibly neurodegeneration. Interestingly, the cleavage of mutant huntingtin protein is dependent on the brain region. The cleavage at two N-terminal sites (A and B) was predominant in the cortex, whereas cleavage occurred at one N-terminal (A) and a C-terminal site in the striatum (57). In addition, inhibition of digestion of mutant huntingtin protein by both caspase-3 and 6 inhibitors was found to reduce apoptosis in vitro, which suggests that caspase inhibitors may be a key factor in the prevention of HD (58-60). Inhibitors of the apoptotic cascade may be used as a tool for prevention of cell death in HD (61). Moreover, the depletion of huntingtin protein has been found to induce activation of caspase-3, and the overexpression of this protein caused a reverse action of caspase-3, its inhibition (62). Huntingtin protein was found to interact with active caspase-3 at high affinity, but mutant huntingtin protein binds to caspase-3 at lower affinity. These findings suggest a mechanism whereby caspase-mediated huntingtin protein depletion results in an amplification cascade leading to further caspase-3 activation, resulting in neuronal dysfunction and neuronal death.
Effects of mutant huntingtin protein in nucleus. As shown in Fig. 1, mutant huntingtin protein impairs gene transcription through either intranuclear aggregate formation or sequestration to transcription factors that play a key role in HD [for review, see ref. (70, 71)]. Important transcriptional factors including p53, cAMP response-element binding protein (CREB)-binding protein (CBP), co-activator CA150, specificity protein 1 (SP1), co-activators TAFII130 and TFIID, and TATA-binding protein (TBP) can be recruited to intranuclear aggregates (72-75). There are interactions between these transcriptional factors and other associated proteins that may interact with SP1 in the regulation of gene expression. It has been demonstrated that huntingtin protein may strengthen the bridge between DNA-bound transcription factor SP1 and TFIID-associated proteins and consequently stimulate gene expression (72, 76, 77). The cAMP-responsive element (CRE) and CBP play a critical role in HD [for review see ref. (70)]. Alteration of CRE-regulated genes has been found in HD mouse models (78) and HD patients (79). The CBP and CRE-mediated transcription have been suggested to be affected by the coactivator TAFII130, which is also found in aggregates of CREB-dependent transcription (80). Additionally, mutant huntingtin protein may disrupt the interaction of SP1 and TAFII130 by formation of aggregates. Increased association of mutant huntingtin protein with SP1 has been found in brain extracts from HD patients. Consequently, the association of SP1 and TAFII130 was found to be reduced in brains of HD patients (72). Moreover, SP1 interacts with N-terminal huntingtin protein fragments in the nucleus of both transfected cells and in brains of HD mice (81). These findings suggest that shorter N-terminal huntingtin protein fragments, responsible for misfolding and aggregation, are more likely to bind SP1 and may inhibit its activity. Interestingly, this effect of huntingtin protein can be reversed by a molecular chaperone (Hsp40), which reduces the misfolding of mutant huntingtin protein.
There are other transcriptional factors that may interact with normal or mutant huntingtin protein in the nucleus. Among them, CA150 transcriptional factor has been found to interact with normal and mutant huntingtin protein (82). CA150 protein levels have been found to be increased in HD brain samples. There are also nuclear repressors that have been shown to interact with huntingtin protein, including N-CoR and C-terminal binding protein (CtBP) (73, 83). The mechanism of the repression appears to occur through the formation of a complex of repressor proteins including the N-CoR, mSin3, histone deacetylases and CtBP. The relocalization of repressor proteins in HD brains may alter transcription, which plays a role in HD neuropathology (83).
Effects of mutant huntingtin protein in endocytosis and axonal vesicular transport. Mutant huntingtin protein has been found to be involved in clathrin-mediated endocytosis. Dysregulation of endocytosis occurs with the interactions of mutant huntingtin protein with proteins that play a role in clathrin-mediated endocytosis (Fig. 1). Moreover, dysregulation of endocytosis is mediated through interactions of mutant huntingtin protein with its associated proteins, HIP1, HIP12, HIP14, PACSIN1 and SH3GL3, known as accessory factors in clathrin-dependent synaptic vesicle endocytosis (25, 30, 63-68). The interactions of mutant huntingtin protein with multiple accessory factors involve several steps that lead to dysregulation of clathrin-mediated endocytosis [for review, see ref. (12)]. Mutant huntingtin protein also is involved in vesicular transport processes in axons. In normal physiological situations, huntingtin protein and HAP1 are transported anterogradely and retrogradely along microtubules in axons (69). There is interaction of the complex huntingtin protein and HAP1 with dynactin, which influences the mobility of dynein in vesicular transport. Huntingtin protein and HAP1 stabilize dynein-dynactin complex of vesicles and consequently enable transport along microtubules in endocytosis processes (12). However, if huntingtin protein is mutated, dysfunctional interaction occurs, which leads to impairment of the anterograde and retrograde transport. Neurotrophic factors may be involved in this transport. Alterations of the transport of neurotrophic may be critical in cell survival.
Role of neurotrophic factors in neuroprotection in HD
Neurotrophic factors play an important role in the prevention of apoptosis and cell differentiation. Neurotrophic factors are released by glial cells, neurons and other types of cells including endothelial and fibroblast cells. Deficiency of neurotrophic factors affects the neuroplasticity of the central nervous system (CNS) and consequently leads to neural death. A chronic deficit of neurotrophic factors involves several target tissues that may play a key role in degeneration of distinct neuronal populations in the adult (84). In addition, the deficiency of endogenous neurotrophic factors is considered critical for the progression of degeneration in neurodegenerative diseases, including HD (85-87).
Neurotrophic or growth factors. Neurotrophic factors are critical for cell differentiation, neuronal growth, and neuronal survival (88, 89). Among these neurotrophic factors, BDNF belongs to the neurotrophin family, which includes nerve growth factor (NGF), neurotrophin 3 (NT-3), and neurotrophin 4/5 (NT-4/5), neurotrophin-6 (NT-6), and neurotrophin-7 (NT-7) (90-92). The biological effects of neurotrophins (93-95) are mediated by high-affinity tyrosine kinase (Trk) receptors (96), although all neurotrophins also bind to a low-affinity receptor, p75LNTR (97). NGF binds TrkA receptors, NT-4 and BDNF preferentially activate TrkB receptors, and NT-3 interacts with TrkC receptors. Neurotrophins are expressed by glial cells and neurons. Neuronal survival and/or neuronal differentiation also involve proteins other than neurotrophins, most notably the members of GDNF, which is a cloned member of the transforming growth factor (TGF) β-superfamily (98). Neurturin is another member of the TGFβ family that presents a GDNF-structurally-related neurotrophic action. Although GDNF and neurturin act through the same receptor complex (c-ret/GFRα), GDNF has binding preference for GFRα-1 and neurturin for GFRα-2 (99-101). CNTF is one of the neurotrophic factors that is distinct from neurotrophins in both structural and biological actions (102, 103). CNTF acts through CNTF receptor alpha and leukemia inhibitory factor receptor (104, 105). The roles of BDNF, FGF-2, GDNF, Neurturin, and CNTF are the factors that are involved in neuroprotection in HD.
Brain Derived Neurotrophic Factor in HD. BDNF is considered a particularly important trophic factor in HD. BDNF is produced by cortical neurons and transported to projection sites in the striatum, and it acts on striatal neuronal survival (106-109). In addition, the nigrostriatal pathway is considered another source of BDNF production and may play a key role in HD (110). The level of BDNF is decreased in the cortex and striatum of HD patients, which is possibly due to decrease in BDNF transcription (11, 13, 111). In animals, a reduction in the level of BDNF mRNA and its protein is found in cortex, striatum, hippocampus and/or cerebellum of transgenic HD mouse models (13, 14, 112-118).
Down-regulation of BDNF in HD mice is related to the length of CAG repeats and the levels of expression (119). This suggests that the decrease of the level of BDNF depends on both the number of CAG repeats and the level of expression of mutant huntingtin protein (119). However, expression of mutant huntingtin protein appears sufficient to alter the level of BDNF (13, 14). A recent study reported that HAP1 interacts with the prodomain of BDNF, however, this interaction is diminished with the presence of mutant huntingtin protein (120). Huntingtin protein is associated with vesicular structures and microtubules, which play an important role in intracellular trafficking (12). Mutant huntingtin protein has been found to impair the transport of BDNF (11, 121). These findings indicate that huntingtin protein promotes BDNF transport, and loss or mutation of huntingtin protein may contribute to deficit of BDNF, which leads to pathogenesis.
The cellular effects of BDNF are mediated through its receptor, TrkB. Reduction of TrkB receptors has been found in transgenic exon-1 and full-length knock-in HD mouse models as well as postmortem HD human brain (122, 123). Interestingly, the overexpression of mutant huntingtin protein may be required for the down-regulation of TrkB levels. Although, the precise mechanism through which mutant huntingtin protein decreases TrkB is still unclear, the expression of TrkB is regulated by CREB, which binds to the second cAMP-responsive element site of one of the TrkB gene promoters and then stimulates TrkB expression (124). Additionally, down-regulation of a regulatory BDNF gene, c-AMP-responsive element, was found in HD mice (13, 42, 113, 124-126). Moreover, mutant huntingtin protein may impair CREB-mediated transcription, which contributes to the reduction of TrkB expression found in HD. Thus, TrkB may contribute to the alteration of the neurotrophic effect in HD models. Together, these findings suggest that loss of trophic maintenance in animal HD models and in HD patients may be related to deficits in BDNF and to a decrease in TrkB expression.
Increasing the levels of BDNF in the cortico-striatal pathway might promote cell survival and, in turn, regulate genes that are transcriptionally disrupted in HD. Therapeutic approaches targeting an increase in BDNF might be a strategy to slow or prevent HD (10, 127). The effect of adenovirus-mediated transfer of the BDNF gene was studied in quinolinic-acid lesioned rat striatum. Intrastriatal injections of adenovirus encoding BDNF demonstrated neuroprotection of striatal neurons (128). Moreover, adenoassociated viral (AAV) vector-mediated gene delivery of BDNF induced a neuroprotective effect in quinolinic-acid treated rats. AAV-BDNF vector provides neuroprotection of striatal neurons in quinolinic-acid HD rats (129). These suggest that local BDNF gene delivery has therapeutic value for the treatment of neurodegeneration in HD. Moreover, another approach was developed to engineer cells that express BDNF and release it continuously (130-133). One of these studies demonstrated slight neuroprotection by the BDNF-secreting cells (130). However, the other studies using BDNF-secreting cells have shown significant improvement in motor performance and reduction in damaged striatal neurons (131-133). The authors of these studies suggested that the dose of BDNF plays a critical role in neuroprotection. The overexpression of BDNF may have a secondary side effect such as increase in neuronal excitability.
Fibroblast Growth Factor in HD. Among different types of FGF, FGF-2 type has been found to protect neurons exposed to toxins or excitatory amino acids (134). Other studies have shown that FGF-2 protects and exerts trophic effects on striatal neurons and stimulates proliferation of striatal neural stem cells (135-137). In addition, FGF-2 promotes neurogenesis, leads to neuroprotection and consequently prolongs survival of R6/2 transgenic mice (138). The increase of neurogenesis through the application of FGF-2 may be associated with the migration of nascent neurons in the subventricular zone toward the striatum, where these neurons become medium spiny neurons as the principal component to replace the lost in HD models. The use of FGF-2 may be considered a potential treatment strategy for the replacement of the lost of neurons in HD models.
Glial cell line-Derived Neurotrophic Factor in HD. GDNF is an important factor for the treatment of neurodegenerative diseases, including HD (139-142). GDNF prevents neurodegeneration of striatal calbindin- and parvalbumin-immunoreactive neurons in a lesion model of HD. The neuroprotection is specific to striatal, medium spiny neurons (141). GDNF is protective for striatal neurons of the indirect pathway, GABA/substance P neurons, which project to the internal segment of globus pallidus and/or substantia nigra pars reticulata (143).
Moreover, transplantation of mouse striatum infected with lentivirus expressing GDNF into the striatum of pre-symptomatic N171-82Q mice maintained motor function and prevented neuronal loss (144). Studies using AAV encoding GDNF have demonstrated a deficit of this trophic factor in N171-82Q transgenic HD mouse model (145). Viral delivery of GDNF induces structural and functional neuroprotection in this HD mouse model. In contrast, a study using R6/2 transgenic HD mouse model did not show any neuroprotective effects with GDNF viral delivery (146). This difference exists because R6/2 transgenic mice have human cDNA that encodes mutant huntingtin protein with a larger number of repeats than those of N171-82Q transgenic mice. Importantly, GDNF treatment fails its neuroprotective effect when it is applied after the time when the inclusions of mutant huntingtin protein have been formed in the striatum (146). Thus, GDNF might be efficient when delivered at the time of the expression of mutant huntingtin protein. The criteria of the application of this trophic factor in a specific age of the progression of HD may apply to all neurotrophic factors.
Neurturin in HD. Neurturin is a neurotrophic factor of the TGFβ family structurally related to GDNF. High doses of neurturin have been found to protect striatal neurons from death in both kainic-acid and quinolinic-acid HD models (147). The neuroprotective effect of neurturin requires repeated injections to maintain high levels within the striatum for a long period of time (148). A recent study has used AAV vector encoding for the trophic factor neurturin in 3-nitropropionic acid (3NP)-treated rats. AAV-neurturin-3NP application induces neuroprotection as compared to saline-3NP-treated rats (149). The advantage of AAV vectors is that they can have long-term gene expression. Thus, the neuroprotective effect of neurturin may persist for several years without repeated manipulations or surgeries (149-151). Another approach was used for neuroprotection of striatal neurons in quinolinic-acid treated rats (HD model). A fibroblast cell line engineered to over-express neurturin was injected into adult rat striatum one day before quinolinic acid injection; grafting of the neurturin-secreting cell line showed a more specific and efficient trophic effect on striatal neurons of the indirect pathway (152).
The neurotrophic action of neurturin is more specific to certain striatal neuronal populations. In quinolinic-acid HD model, neurturin selectively protects striatal neuron projections of the indirect circuit, which is the first to enter apoptosis stage in HD patients (153, 154). Neurturin selectively protects the striatopallidal neurons expressing glutamic acid decarboxylase and preproenkephalin (155). This suggests that neurturin protects only GABAergic and enkephalinergic neurons that project to the external segment of the globus pallidus [for review, see (143)]. Therefore, these results suggest that neurturin is considered a candidate for the treatment of HD.
Ciliary neurotrophic factor in HD. CNTF is a member of the alpha-helical IL-6 cytokine superfamily with neurotrophic actions in the peripheral and CNS. CNTF stimulates gene expression, cell survival and differentiation in several types of neurons including GABAergic and cholinergic neurons and also plays a role in oligodendrocyte maturation (156, 157). CNTF provides neuroprotection for striatal neurons; it is suggested to be a potential therapeutic agent for HD (158, 159).
Application of CNTF in CNS has been a challenge due to its difficult accessibility into the brain via systemic injection. CNTF is a large protein that does not cross the blood brain barrier. Thus, local delivery using polymer-encapsulated cells engineered to secrete CNTF shows pre-clinical and clinical success for the delivery of this trophic factor to the CNS (159-163). Moreover, the protective effect of encapsulated cells producing human CNTF was successful in degenerating striatal neurons as well as neurons in the cerebral cortex. In HD monkeys, CNTF prevents degeneration of striatal and cortical neurons (163). A neuroprotective effect was observed in HD patients using polymer-encapsulated cells engineered to release human CNTF in the striatum. The clinical phase I studies show a promising neuroprotective effect of CNTF for the treatment of HD patients (160).
CONCLUSION
Although the mechanisms of neurodegeneration in HD remain unclear, mutant huntingtin protein is suggested to alter intracellular trafficking including membrane recycling, clathrin-mediated endocytosis and neuronal transport. Deficit of neurotrophic factor levels in HD mouse models may be linked to direct action of mutant huntingtin protein in the transport of these trophic factors. In addition, mutant huntingtin protein has been demonstrated to interact with cellular transcriptional regulatory proteins, some of which are neurotrophic factors.
Understanding the molecular mechanism of actions of mutant huntingtin protein may underlie expectations for the discovery of drugs that can lead to neurorestoration or neuroprotection in which growth of new axons, dendrites, and synapses might be consequences of functional improvement. These mechanisms might be of great interest for patients who already have the diseases or, in some cases, where the disease is in progression. Neurotrophic factors are considered one of the therapeutic tools for the treatment of HD to overcome neurodegeneration and behavioral abnormalities.
ACKNOWLEDGMENTS
I would like to thank the National Institutes of Health-NIAA for their continuous support (R21AA017735; R21AA016115). I would also like to thank Charisse Montgomery for editing this review article.
ABBREVIATIONS
- HD
Huntington’s disease
- BDNF
Brain-derived neurotrophic factor
- FGF
Fibroblast growth factor
- GDNF
Glial cell line-derived neurotrophic factor
- CNTF
Ciliary neurotrophic factor
- HAP-1
Huntingtin protein-associated protein
- HIP-1
Huntingtin protein-interacting protein 1
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- YAC
Yeast artificial chromosome
- NMDA
N-methyl-D-aspartate receptor
- mGluR5
metabotropic glutamate receptor 5
- Apaf-1
Apoptotic protease activating factor 1
- CREB
cAMP response-element binding protein
- CBP
cAMP binding protein
- SP1
Specificity protein 1
- CtBP
C-terminal binding protein
- CNS
Central nervous system
- NGF
Nerve growth factor
- NT-3
Neurotrophin 3
- Trk
Tyrosine kinase
- TGF-β
Transforming growth factor β
- AAV
Adenoassociated viral
- 3NP
3-nitropropionic acid
REFERENCES
- 1.(HDCRG) HsDCRG. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993 Mar 26;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.McMurray CT. Huntington’s disease: new hope for therapeutics. Trends Neurosci. 2001 Nov;24(11 Suppl):S32–38. doi: 10.1016/s0166-2236(00)01997-4. [DOI] [PubMed] [Google Scholar]
- 3.de la Monte SM, Vonsattel JP, Richardson EP., Jr Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington’s disease. J. Neuropathol Exp. Neurol. 1988 Sep;47(5):516–525. doi: 10.1097/00005072-198809000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, et al. Neuropathological classification of Huntington’s disease. J. Neuropathol Exp. Neurol. 1985 Nov;44(6):559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Vonsattel JP, DiFiglia M. Huntington disease. J. Neuropathol Exp. Neurol. 1998 May;57(5):369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Kumar P, Kalonia H, Kumar A. Huntington’s disease: pathogenesis to animal models. Pharmacol Rep. 2010 Jan-Feb;62(1):1–14. doi: 10.1016/s1734-1140(10)70238-3. [DOI] [PubMed] [Google Scholar]
- 7.Han I, You Y, Kordower JH, Brady ST, et al. Differential vulnerability of neurons in Huntington’s disease: the role of cell type-specific features. J. Neurochem. 2010 Jun;113(5):1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinsen H, Rub U, Bauer M, Ulmar G, et al. Nerve cell loss in the thalamic mediodorsal nucleus in Huntington’s disease. Acta. Neuropathol. 1999 Jun;97(6):613–622. doi: 10.1007/s004010051037. [DOI] [PubMed] [Google Scholar]
- 9.Oyanagi K, Takeda S, Takahashi H, Ohama E, et al. A quantitative investigation of the substantia nigra in Huntington’s disease. Ann. Neurol. 1989 Jul;26(1):13–19. doi: 10.1002/ana.410260103. [DOI] [PubMed] [Google Scholar]
- 10.Alberch J, Perez-Navarro E, Canals JM. Neurotrophic factors in Huntington’s disease. Prog Brain Res. 2004;146:195–229. doi: 10.1016/s0079-6123(03)46014-7. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004 Jul 9;118(1):127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends in biochemical sciences. 2003 Aug;28(8):425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 13.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001 Jul 20;293(5529):493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 14.Zuccato C, Tartari M, Crotti A, Goffredo D, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003 Sep;35(1):76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Lin F, Qin ZH. The role of post-translational modifications of huntingtin in the pathogenesis of Huntington’s disease. Neurosci Bull. 2010 Apr;26(2):153–162. doi: 10.1007/s12264-010-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies SW, Turmaine M, Cozens BA, DiFiglia M, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997 Aug 8;90(3):537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 17.Dragatsis I, Dietrich P, Zeitlin S. Expression of the Huntingtin-associated protein 1 gene in the developing and adult mouse. Neurosci. Lett. 2000 Mar 17;282(1-2):37–40. doi: 10.1016/s0304-3940(00)00872-7. [DOI] [PubMed] [Google Scholar]
- 18.Duyao MP, Auerbach AB, Ryan A, Persichetti F, et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995 Jul 21;269(5222):407–10. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 19.Gusella JF, MacDonald ME. Huntingtin: a single bait hooks many species. Current opinion in neurobiology. 1998 Jun;8(3):425–30. doi: 10.1016/s0959-4388(98)80071-8. [DOI] [PubMed] [Google Scholar]
- 20.Gutekunst CA, Levey AI, Heilman CJ, Whaley WL, et al. Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc. Natl. Acad. Sci. USA. 1995 Sep 12;92(19):8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasir J, Floresco SB, O’Kusky JR, Diewert VM, et al. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995 Jun 2;81(5):811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 22.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, et al. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995 Oct;11(2):155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 23.Petersen A, Mani K, Brundin P. Recent advances on the pathogenesis of Huntington’s disease. Exp. Neurol. 1999 May;157(1):1–18. doi: 10.1006/exnr.1998.7006. [DOI] [PubMed] [Google Scholar]
- 24.Reddy PH, Williams M, Tagle DA. Recent advances in understanding the pathogenesis of Huntington’s disease. Trends Neurosci. 1999 Jun;22(6):248–255. doi: 10.1016/s0166-2236(99)01415-0. [DOI] [PubMed] [Google Scholar]
- 25.Wu LL, Zhou XF. Huntingtin associated protein 1 and its functions. Cell Adh. Migr. 2009 Jan 26;3(1) doi: 10.4161/cam.3.1.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao J, Sharp AH, Wagster MV, Becher M, et al. Expansion of polyglutamine repeat in huntingtin leads to abnormal protein interactions involving calmodulin. Proc. Natl. Acad. Sci. USA. 1996 May 14;93(10):5037–5042. doi: 10.1073/pnas.93.10.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burke JR, Enghild JJ, Martin ME, Jou YS, et al. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat. Med. 1996 Mar;2(3):347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 28.Kalchman MA, Graham RK, Xia G, Koide HB, et al. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J. Biol. Chem. 1996 Aug 9;271(32):19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- 29.Li XJ, Li SH, Sharp AH, Nucifora FC, Jr, et al. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995 Nov 23;378(6555):398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 30.Wanker EE, Rovira C, Scherzinger E, Hasenbank R, et al. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Human molecular genetics. 1997 Mar;6(3):487–495. doi: 10.1093/hmg/6.3.487. [DOI] [PubMed] [Google Scholar]
- 31.Shin JY, Fang ZH, Yu ZX, Wang CE, et al. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. The Journal of cell biology. 2005 Dec 19;171(6):1001–1012. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sack GH., Jr Mitochondrial matters in Huntington disease. J. Bioenerg. Biomembr. 2010 Jun;42(3):189–191. doi: 10.1007/s10863-010-9291-x. [DOI] [PubMed] [Google Scholar]
- 33.Oliveira JM. Nature and cause of mitochondrial dysfunction in Huntington’s disease: focusing on huntingtin and the striatum. J. Neurochem. 2010 Jul;114(1):1–12. doi: 10.1111/j.1471-4159.2010.06741.x. [DOI] [PubMed] [Google Scholar]
- 34.Jin YN, Johnson GV. The interrelationship between mitochondrial dysfunction and transcriptional dysregulation in Huntington disease. J. Bioenerg. Biomembr. 2010 Jun;42(3):199–205. doi: 10.1007/s10863-010-9286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu M, Gash MT, Mann VM, Javoy-Agid F, et al. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann. Neurol. 1996 Mar;39(3):385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 36.Tabrizi SJ, Workman J, Hart PE, Mangiarini L, et al. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann. Neurol. 2000 Jan;47(1):80–86. doi: 10.1002/1531-8249(200001)47:1<80::aid-ana13>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 37.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002 Aug;5(8):731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 38.Milakovic T, Quintanilla RA, Johnson GV. Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: Functional consequences. J. Biol. Chem. 2006 Sep 13; doi: 10.1074/jbc.M603845200. [DOI] [PubMed] [Google Scholar]
- 39.Gutekunst CA, Li SH, Yi H, Ferrante RJ, et al. The cellular and subcellular localization of huntingtin-associated protein 1 (HAP1): comparison with huntingtin in rat and human. J. Neurosci. 1998 Oct 1;18(19):7674–7686. doi: 10.1523/JNEUROSCI.18-19-07674.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desplats PA, Kass KE, Gilmartin T, Stanwood GD, et al. Selective deficits in the expression of striatal-enriched mRNAs in Huntington’s disease. J. Neurochem. 2006 Feb;96(3):743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- 41.Blaustein MP, Lederer WJ. Sodium/calcium exchange: its physiological implications. Physiological reviews. 1999 Jul;79(3):763–854. doi: 10.1152/physrev.1999.79.3.763. [DOI] [PubMed] [Google Scholar]
- 42.Luthi-Carter R, Strand A, Peters NL, Solano SM, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Human molecular genetics. 2000 May 22;9(9):1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 43.Tang TS, Slow E, Lupu V, Stavrovskaya IG, et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2005 Feb 15;102(7):2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J, Tang T, Bezprozvanny I. Evaluation of clinically relevant glutamate pathway inhibitors in in vitro model of Huntington’s disease. Neurosci. Lett. 2006 Sep 5; doi: 10.1016/j.neulet.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 45.Zeron MM, Fernandes HB, Krebs C, Shehadeh J, et al. Potentiation of NMDA receptor-mediated excitotoxicity linked with intrinsic apoptotic pathway in YAC transgenic mouse model of Huntington’s disease. Molecular and cellular neurosciences. 2004 Mar;25(3):469–479. doi: 10.1016/j.mcn.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Behrens PF, Franz P, Woodman B, Lindenberg KS, et al. Impaired glutamate transport and glutamate-glutamine cycling: downstream effects of the Huntington mutation. Brain. 2002 Aug;125(Pt 8):1908–1922. doi: 10.1093/brain/awf180. [DOI] [PubMed] [Google Scholar]
- 47.Estrada-Sanchez AM, Montiel T, Segovia J, Massieu L. Glutamate toxicity in the striatum of the R6/2 Huntington’s disease transgenic mice is age-dependent and correlates with decreased levels of glutamate transporters. Neurobiology of disease. 2009 Apr;34(1):78–86. doi: 10.1016/j.nbd.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Lievens JC, Woodman B, Mahal A, Spasic-Boscovic O, et al. Impaired glutamate uptake in the R6 Huntington’s disease transgenic mice. Neurobiology of disease. 2001 Oct;8(5):807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- 49.Miller BR, Dorner JL, Shou M, Sari Y, et al. Up-regulation of GLT1 expression increases glutamate uptake and attenuates the Huntington’s disease phenotype in the R6/2 mouse. Neuroscience. 2008 Apr 22;153(1):329–337. doi: 10.1016/j.neuroscience.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sari Y, Prieto AL, Barton SJ, Miller BR, et al. Ceftriaxone-induced up-regulation of cortical and striatal GLT1 in the R6/2 model of Huntington’s disease. J. Biomed. Sci. 2010;17:62. doi: 10.1186/1423-0127-17-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bezprozvanny I, Hayden MR. Deranged neuronal calcium signaling and Huntington disease. Biochem Biophys Res Commun. 2004 Oct 1;322(4):1310–1317. doi: 10.1016/j.bbrc.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 52.Tang TS, Guo C, Wang H, Chen X, et al. Neuroprotective effects of inositol 1,4,5-trisphosphate receptor C-terminal fragment in a Huntington’s disease mouse model. J. Neurosci. 2009 Feb 4;29(5):1257–1266. doi: 10.1523/JNEUROSCI.4411-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leist M, Nicotera P. Apoptosis, excitotoxicity, and neuropathology. Experimental cell research. 1998 Mar 15;239(2):183–201. doi: 10.1006/excr.1997.4026. [DOI] [PubMed] [Google Scholar]
- 54.Susin SA, Zamzami N, Castedo M, Hirsch T, et al. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. The Journal of experimental medicine. 1996 Oct 1;184(4):1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998 Aug 28;281(5381):1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 56.Graham RK, Deng Y, Slow EJ, Haigh B, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006 Jun 16;125(6):1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 57.Mende-Mueller LM, Toneff T, Hwang SR, Chesselet MF, et al. Tissue-specific proteolysis of Huntingtin (htt) in human brain: evidence of enhanced levels of N- and C-terminal htt fragments in Huntington’s disease striatum. J. Neurosci. 2001 Mar 15;21(6):1830–1837. doi: 10.1523/JNEUROSCI.21-06-01830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies S, Ramsden DB. Huntington’s disease. Mol. Pathol. 2001 Dec;54(6):409–413. doi: 10.1136/mp.54.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ona VO, Li M, Vonsattel JP, Andrews LJ, et al. Inhibition of caspase-1 slows disease progression in a mouse model of Huntington’s disease. Nature. 1999 May 20;399(6733):263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 60.Wellington CL, Leavitt BR, Hayden MR. Huntington disease: new insights on the role of huntingtin cleavage. J. Neural. Transm. Suppl. 2000;(58):1–17. doi: 10.1007/978-3-7091-6284-2_1. [DOI] [PubMed] [Google Scholar]
- 61.Pattison LR, Kotter MR, Fraga D, Bonelli RM. Apoptotic cascades as possible targets for inhibiting cell death in Huntington’s disease. J. Neurol. 2006 Sep 22; doi: 10.1007/s00415-006-0198-8. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Leavitt BR, van Raamsdonk JM, Dragatsis I, et al. Huntingtin inhibits caspase-3 activation. The EMBO journal. 2006 Dec 13;25(24):5896–5906. doi: 10.1038/sj.emboj.7601445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chopra VS, Metzler M, Rasper DM, Engqvist-Goldstein AE, et al. HIP12 is a non-proapoptotic member of a gene family including HIP1, an interacting protein with huntingtin. Mamm Genome. 2000 Nov;11(11):1006–1015. doi: 10.1007/s003350010195. [DOI] [PubMed] [Google Scholar]
- 64.Kalchman MA, Koide HB, McCutcheon K, Graham RK, et al. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat. Genet. 1997 May;16(1):44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- 65.Modregger J, DiProspero NA, Charles V, Tagle DA, et al. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington’s disease brains. Human molecular genetics. 2002 Oct 1;11(21):2547–2558. doi: 10.1093/hmg/11.21.2547. [DOI] [PubMed] [Google Scholar]
- 66.Seki N, Muramatsu M, Sugano S, Suzuki Y, et al. Cloning, expression analysis, and chromosomal localization of HIP1R, an isolog of huntingtin interacting protein (HIP1) Journal of human genetics. 1998;43(4):268–271. doi: 10.1007/s100380050087. [DOI] [PubMed] [Google Scholar]
- 67.Singaraja RR, Hadano S, Metzler M, Givan S, et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Human molecular genetics. 2002 Nov 1;11(23):2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 68.Sittler A, Walter S, Wedemeyer N, Hasenbank R, et al. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Molecular cell. 1998 Oct;2(4):427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 69.Block-Galarza J, Chase KO, Sapp E, Vaughn KT, et al. Fast transport and retrograde movement of huntingtin and HAP 1 in axons. Neuroreport. 1997 Jul 7;8(9-10):2247–2251. doi: 10.1097/00001756-199707070-00031. [DOI] [PubMed] [Google Scholar]
- 70.Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington’s disease. Fourth in molecular medicine review series. EMBO reports. 2004 Oct;5(10):958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003 May;19(5):233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 72.Dunah AW, Jeong H, Griffin A, Kim YM, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002 Jun 21;296(5576):2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 73.Kegel KB, Meloni AR, Yi Y, Kim YJ, et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J. Biol. Chem. 2002 Mar 1;277(9):7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- 74.McCampbell A, Taylor JP, Taye AA, Robitschek J, et al. CREB-binding protein sequestration by expanded polyglutamine. Human molecular genetics. 2000 Sep 1;9(14):2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 75.Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA. 2000 Jun 6;97(12):6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li SH, Cheng AL, Zhou H, Lam S, et al. Interaction of Huntington disease protein with transcriptional activator Sp1. Molecular and cellular biology. 2002 Mar;22(5):1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 78.Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 2002 May;31(1):47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 79.Glass M, Dragunow M, Faull RL. The pattern of neurodegeneration in Huntington’s disease: a comparative study of cannabinoid, dopamine, adenosine and GABA(A) receptor alterations in the human basal ganglia in Huntington’s disease. Neuroscience. 2000;97(3):505–519. doi: 10.1016/s0306-4522(00)00008-7. [DOI] [PubMed] [Google Scholar]
- 80.Shimohata T, Nakajima T, Yamada M, Uchida C, et al. Expanded poly-glutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet. 2000 Sep;26(1):29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 81.Cornett J, Smith L, Friedman M, Shin JY, et al. Context-dependent dysregulation of transcription by mutant huntingtin. J. Biol. Chem. 2006 Nov 24;281(47):36198–36204. doi: 10.1074/jbc.M607839200. [DOI] [PubMed] [Google Scholar]
- 82.Holbert S, Denghien I, Kiechle T, Rosenblatt A, et al. The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: neuropathologic and genetic evidence for a role in Huntington’s disease pathogenesis. Proc. Natl. Acad. Sci. USA. 2001 Feb 13;98(4):1811–1816. doi: 10.1073/pnas.041566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boutell JM, Thomas P, Neal JW, Weston VJ, et al. Aberrant interactions of transcriptional repressor proteins with the Huntington’s disease gene product, huntingtin. Human molecular genetics. 1999 Sep;8(9):1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- 84.Appel SH. A unifying hypothesis for the cause of amyotrophic lateral sclerosis, parkinsonism, and Alzheimer disease. Ann. Neurol. 1981 Dec;10(6):499–505. doi: 10.1002/ana.410100602. [DOI] [PubMed] [Google Scholar]
- 85.Connor B, Dragunow M. The role of neuronal growth factors in neurodegenerative disorders of the human brain. Brain Res. Rev. 1998 Jun;27(1):1–39. doi: 10.1016/s0165-0173(98)00004-6. [DOI] [PubMed] [Google Scholar]
- 86.Kruttgen A, Saxena S, Evangelopoulos ME, Weis J. Neurotrophins and neurodegenerative diseases: receptors stuck in traffic? J. Neuropathol. Exp. Neurol. 2003 Apr;62(4):340–350. doi: 10.1093/jnen/62.4.340. [DOI] [PubMed] [Google Scholar]
- 87.Murer MG, Yan Q, Raisman-Vozari R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer’s disease and Parkinson’s disease. Prog. Neurobiol. 2001 Jan;63(1):71–124. doi: 10.1016/s0301-0082(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 88.Chao MV. Neurotrophin receptors: a window into neuronal differentiation. Neuron. 1992 Oct;9(4):583–593. doi: 10.1016/0896-6273(92)90023-7. [DOI] [PubMed] [Google Scholar]
- 89.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu. Rev. Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lai KO, Fu WY, Ip FC, Ip NY. Cloning and expression of a novel neurotrophin, NT-7, from carp. Molecular and cellular neurosciences. 1998 May;11(1-2):64–76. doi: 10.1006/mcne.1998.0666. [DOI] [PubMed] [Google Scholar]
- 91.Lewin GR, Barde YA. Physiology of the neurotrophins. Annu. Rev. Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 92.Nilsson AS, Fainzilber M, Falck P, Ibanez CF. Neurotrophin-7: a novel member of the neurotrophin family from the zebrafish. FEBS Lett. 1998 Mar 13;424(3):285–290. doi: 10.1016/s0014-5793(98)00192-6. [DOI] [PubMed] [Google Scholar]
- 93.Alderson RF, Alterman AL, Barde YA, Lindsay RM. Brain-derived neurotrophic factor increases survival and differentiated functions of rat septal cholinergic neurons in culture. Neuron. 1990 Sep;5(3):297–306. doi: 10.1016/0896-6273(90)90166-d. [DOI] [PubMed] [Google Scholar]
- 94.McAllister AK, Katz LC, Lo DC. Neurotrophins and synaptic plasticity. Annu. Rev. Neurosci. 1999;22:295–318. doi: 10.1146/annurev.neuro.22.1.295. [DOI] [PubMed] [Google Scholar]
- 95.Nonomura T, Hatanaka H. Neurotrophic effect of brain-derived neurotrophic factor on basal forebrain cholinergic neurons in culture from postnatal rats. Neurosci Res. 1992 Aug;14(3):226–233. doi: 10.1016/0168-0102(92)90083-o. [DOI] [PubMed] [Google Scholar]
- 96.Glass DJ, Yancopoulos GD. The neurotrophins and their receptors. Trends Cell Biol. 1993 Aug;3(8):262–268. doi: 10.1016/0962-8924(93)90054-5. [DOI] [PubMed] [Google Scholar]
- 97.Chao MV, Hempstead BL. p75 and Trk: a two-receptor system. Trends Neurosci. 1995 Jul;18(7):321–326. [PubMed] [Google Scholar]
- 98.Lin LF, Doherty DH, Lile JD, Bektesh S, et al. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993 May 21;260(5111):1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 99.Cacalano G, Farinas I, Wang LC, Hagler K, et al. GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron. 1998 Jul;21(1):53–62. doi: 10.1016/s0896-6273(00)80514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heuckeroth RO, Enomoto H, Grider JR, Golden JP, et al. Gene targeting reveals a critical role for neurturin in the development and maintenance of enteric, sensory, and parasympathetic neurons. Neuron. 1999 Feb;22(2):253–263. doi: 10.1016/s0896-6273(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 101.Klein RD, Sherman D, Ho WH, Stone D, et al. A GPI-linked protein that interacts with Ret to form a candidate neurturin receptor. Nature. 1997 Jun 12;387(6634):717–721. doi: 10.1038/42722. [DOI] [PubMed] [Google Scholar]
- 102.Adler R, Landa KB, Manthorpe M, Varon S. Cholinergic neuronotrophic factors: intraocular distribution of trophic activity for ciliary neurons. Science. 1979 Jun 29;204(4400):1434–1436. doi: 10.1126/science.451576. [DOI] [PubMed] [Google Scholar]
- 103.Manthorpe M, Davis GE, Varon S. Purified proteins acting on cultured chick embryo ciliary ganglion neurons. Federation proceedings. 1985 Sep;44(12):2753–2759. [PubMed] [Google Scholar]
- 104.Davis S, Aldrich TH, Ip NY, Stahl N, et al. Released form of CNTF receptor alpha component as a soluble mediator of CNTF responses. Science. 1993 Mar 19;259(5102):1736–1739. doi: 10.1126/science.7681218. [DOI] [PubMed] [Google Scholar]
- 105.Davis S, Yancopoulos GD. The molecular biology of the CNTF receptor. Current opinion in cell biology. 1993 Apr;5(2):281–285. doi: 10.1016/0955-0674(93)90117-9. [DOI] [PubMed] [Google Scholar]
- 106.Altar CA, Cai N, Bliven T, Juhasz M, et al. Anterograde transport of brain-derived neurotrophic factor and its role in the brain. Nature. 1997 Oct 23;389(6653):856–860. doi: 10.1038/39885. [DOI] [PubMed] [Google Scholar]
- 107.Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J. Neurosci. 2004 Apr 28;24(17):4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lynch G, Kramar EA, Rex CS, Jia Y, et al. Brain-derived neurotrophic factor restores synaptic plasticity in a knock-in mouse model of Huntington’s disease. J. Neurosci. 2007 Apr 18;27(16):4424–4434. doi: 10.1523/JNEUROSCI.5113-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998 Oct 2;95(1):55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 110.Pineda JR, Canals JM, Bosch M, Adell A, et al. Brain-derived neurotrophic factor modulates dopaminergic deficits in a transgenic mouse model of Huntington’s disease. J. Neurochem. 2005 Jun;93(5):1057–1068. doi: 10.1111/j.1471-4159.2005.03047.x. [DOI] [PubMed] [Google Scholar]
- 111.Ferrer I, Goutan E, Marin C, Rey MJ, et al. Brain-derived neurotrophic factor in Huntington disease. Brain Res. 2000 Jun 2;866(1-2):257–261. doi: 10.1016/s0006-8993(00)02237-x. [DOI] [PubMed] [Google Scholar]
- 112.Duan W, Guo Z, Jiang H, Ware M, et al. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc. Natl. Acad. Sci. USA. 2003 Mar 4;100(5):2911–2916. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gines S, Seong IS, Fossale E, Ivanova E, et al. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington’s disease knock-in mice. Human molecular genetics. 2003 Mar 1;12(5):497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 114.Hermel E, Gafni J, Propp SS, Leavitt BR, et al. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell death and differentiation. 2004 Apr;11(4):424–438. doi: 10.1038/sj.cdd.4401358. [DOI] [PubMed] [Google Scholar]
- 115.Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Human molecular genetics. 2002 Aug 15;11(17):1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 116.Spires TL, Grote HE, Varshney NK, Cordery PM, et al. Environmental enrichment rescues protein deficits in a mouse model of Huntington’s disease, indicating a possible disease mechanism. J. Neurosci. 2004 Mar 3;24(9):2270–2276. doi: 10.1523/JNEUROSCI.1658-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zuccato C, Liber D, Ramos C, Tarditi A, et al. Progressive loss of BDNF in a mouse model of Huntington’s disease and rescue by BDNF delivery. Pharmacol Res. 2005 Aug;52(2):133–139. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 118.Zuccato C, Tartari M, Goffredo D, Cattaneo E, et al. From target identification to drug screening assays for neurodegenerative diseases. Pharmacol Res. 2005 Sep;52(3):245–251. doi: 10.1016/j.phrs.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 119.Canals JM, Pineda JR, Torres-Peraza JF, Bosch M, et al. Brain-derived neurotrophic factor regulates the onset and severity of motor dysfunction associated with enkephalinergic neuronal degeneration in Huntington’s disease. J. Neurosci. 2004 Sep 1;24(35):7727–7739. doi: 10.1523/JNEUROSCI.1197-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu LL, Fan Y, Li S, Li XJ, et al. Huntingtin-associated protein-1 interacts with pro-brain-derived neurotrophic factor and mediates its transport and release. J. Biol. Chem. 2010 Feb 19;285(8):5614–5623. doi: 10.1074/jbc.M109.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pardo R, Molina-Calavita M, Poizat G, Keryer G, et al. pARIS-htt: an optimised expression platform to study huntingtin reveals functional domains required for vesicular trafficking. Mol Brain. 2010;3:17. doi: 10.1186/1756-6606-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gines S, Bosch M, Marco S, Gavalda N, et al. Reduced expression of the TrkB receptor in Huntington’s disease mouse models and in human brain. Eur. J. Neurosci. 2006 Feb;23(3):649–658. doi: 10.1111/j.1460-9568.2006.04590.x. [DOI] [PubMed] [Google Scholar]
- 123.Gines S, Paoletti P, Alberch J. Impaired TrkB-mediated ERK1/2 activation in huntington disease knock-in striatal cells involves reduced p52/p46 Shc expression. J. Biol. Chem. 2010 Jul 9;285(28):21537–21548. doi: 10.1074/jbc.M109.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deogracias R, Espliguero G, Iglesias T, Rodriguez-Pena A. Expression of the neurotrophin receptor trkB is regulated by the cAMP/CREB pathway in neurons. Molecular and cellular neurosciences. 2004 Jul;26(3):470–480. doi: 10.1016/j.mcn.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 125.Sugars KL, Brown R, Cook LJ, Swartz J, et al. Decreased cAMP response element-mediated transcription: an early event in exon 1 and full-length cell models of Huntington’s disease that contributes to polyglutamine pathogenesis. J. Biol. Chem. 2004 Feb 6;279(6):4988–4999. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- 126.Wyttenbach A, Swartz J, Kita H, Thykjaer T, et al. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington’s disease. Human molecular genetics. 2001 Aug 15;10(17):1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 127.Gharami K, Xie Y, An JJ, Tonegawa S, et al. Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington’s disease phenotypes in mice. J. Neurochem. 2008 Apr;105(2):369–379. doi: 10.1111/j.1471-4159.2007.05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bemelmans AP, Horellou P, Pradier L, Brunet I, et al. Brain-derived neurotrophic factor-mediated protection of striatal neurons in an excitotoxic rat model of Huntington’s disease, as demonstrated by adenoviral gene transfer. Hum Gene Ther. 1999 Dec 10;10(18):2987–2997. doi: 10.1089/10430349950016393. [DOI] [PubMed] [Google Scholar]
- 129.Kells AP, Fong DM, Dragunow M, During MJ, et al. AAV-mediated gene delivery of BDNF or GDNF is neuroprotective in a model of Huntington disease. Mol Ther. 2004 May;9(5):682–688. doi: 10.1016/j.ymthe.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 130.Martinez-Serrano A, Bjorklund A. Protection of the neostriatum against excitotoxic damage by neurotrophin-producing, genetically modified neural stem cells. J. Neurosci. 1996 Aug 1;16(15):4604–4616. doi: 10.1523/JNEUROSCI.16-15-04604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Perez-Navarro E, Alberch J, Neveu I, Arenas E. Brain-derived neurotrophic factor, neurotrophin-3 and neurotrophin-4/5 differentially regulate the phenotype and prevent degenerative changes in striatal projection neurons after excitotoxicity in vivo. Neuroscience. 1999;91(4):1257–1264. doi: 10.1016/s0306-4522(98)00723-4. [DOI] [PubMed] [Google Scholar]
- 132.Perez-Navarro E, Canudas AM, Akerund P, Alberch J, et al. Brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4/5 prevent the death of striatal projection neurons in a rodent model of Huntington’s disease. J. Neurochem. 2000 Nov;75(5):2190–2199. doi: 10.1046/j.1471-4159.2000.0752190.x. [DOI] [PubMed] [Google Scholar]
- 133.Ryu JK, Kim J, Cho SJ, Hatori K, et al. Proactive transplantation of human neural stem cells prevents degeneration of striatal neurons in a rat model of Huntington disease. Neurobiology of disease. 2004 Jun;16(1):68–77. doi: 10.1016/j.nbd.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 134.Haque NS, Isacson O. Neurotrophic factors NGF and FGF-2 alter levels of huntingtin (IT15) in striatal neuronal cell cultures. Cell transplantation. 2000 Sep-Oct;9(5):623–627. doi: 10.1177/096368970000900507. [DOI] [PubMed] [Google Scholar]
- 135.Bjugstad KB, Zawada WM, Goodman S, Free CR. IGF-1 and bFGF reduce glutaric acid and 3-hydroxyglutaric acid toxicity in striatal cultures. Journal of inherited metabolic disease. 2001 Nov;24(6):631–647. doi: 10.1023/a:1012706908779. [DOI] [PubMed] [Google Scholar]
- 136.Palmer TD, Ray J, Gage FH. FGF-2-responsive neuronal progenitors reside in proliferative and quiescent regions of the adult rodent brain. Molecular and cellular neurosciences. 1995 Oct;6(5):474–486. doi: 10.1006/mcne.1995.1035. [DOI] [PubMed] [Google Scholar]
- 137.Zhou D, DiFiglia M. Basic fibroblast growth factor enhances the growth of postnatal neostriatal GABAergic neurons in vitro. Exp. Neurol. 1993 Aug;122(2):171–188. doi: 10.1006/exnr.1993.1118. [DOI] [PubMed] [Google Scholar]
- 138.Jin K, LaFevre-Bernt M, Sun Y, Chen S, et al. FGF-2 promotes neurogenesis and neuroprotection and prolongs survival in a transgenic mouse model of Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2005 Dec 13;102(50):18189–18194. doi: 10.1073/pnas.0506375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hoffer BJ, Hoffman A, Bowenkamp K, Huettl P, et al. Glial cell line-derived neurotrophic factor reverses toxin-induced injury to midbrain dopaminergic neurons in vivo. Neurosci. Lett. 1994 Nov 21;182(1):107–111. doi: 10.1016/0304-3940(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 140.Kim BT, Rao VL, Sailor KA, Bowen KK, et al. Protective effects of glial cell line-derived neurotrophic factor on hippocampal neurons after traumatic brain injury in rats. J. Neurosurg. 2001 Oct;95(4):674–679. doi: 10.3171/jns.2001.95.4.0674. [DOI] [PubMed] [Google Scholar]
- 141.Perez-Navarro E, Arenas E, Reiriz J, Calvo N, et al. Glial cell line-derived neurotrophic factor protects striatal calbindin-immunoreactive neurons from excitotoxic damage. Neuroscience. 1996 Nov;75(2):345–352. doi: 10.1016/0306-4522(96)00336-3. [DOI] [PubMed] [Google Scholar]
- 142.Watabe K, Ohashi T, Sakamoto T, Kawazoe Y, et al. Rescue of lesioned adult rat spinal motoneurons by adenoviral gene transfer of glial cell line-derived neurotrophic factor. J. Neurosci Res. 2000 May 15;60(4):511–519. doi: 10.1002/(SICI)1097-4547(20000515)60:4<511::AID-JNR10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 143.Alberch J, Perez-Navarro E, Canals JM. Neuroprotection by neurotrophins and GDNF family members in the excitotoxic model of Huntington’s disease. Brain research bulletin. 2002 Apr;57(6):817–822. doi: 10.1016/s0361-9230(01)00775-4. [DOI] [PubMed] [Google Scholar]
- 144.Ebert AD, Barber AE, Heins BM, Svendsen CN. Ex vivo delivery of GDNF maintains motor function and prevents neuronal loss in a transgenic mouse model of Huntington’s disease. Exp. Neurol. 2010 Jul;224(1):155–162. doi: 10.1016/j.expneurol.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 145.McBride JL, Ramaswamy S, Gasmi M, Bartus RT, et al. Viral delivery of glial cell line-derived neurotrophic factor improves behavior and protects striatal neurons in a mouse model of Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2006 Jun 13;103(24):9345–9350. doi: 10.1073/pnas.0508875103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Popovic N, Maingay M, Kirik D, Brundin P. Lentiviral gene delivery of GDNF into the striatum of R6/2 Huntington mice fails to attenuate behavioral and neuropathological changes. Exp. Neurol. 2005 May;193(1):65–74. doi: 10.1016/j.expneurol.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 147.Gratacos E, Perez-Navarro E, Tolosa E, Arenas E, et al. Neuroprotection of striatal neurons against kainate excitotoxicity by neurotrophins and GDNF family members. J. Neurochem. 2001 Sep;78(6):1287–1296. doi: 10.1046/j.1471-4159.2001.00538.x. [DOI] [PubMed] [Google Scholar]
- 148.Rosenblad C, Kirik D, Devaux B, Moffat B, et al. Protection and regeneration of nigral dopaminergic neurons by neurturin or GDNF in a partial lesion model of Parkinson’s disease after administration into the striatum or the lateral ventricle. Eur. J. Neurosci. 1999 May;11(5):1554–1566. doi: 10.1046/j.1460-9568.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- 149.Ramaswamy S, McBride JL, Herzog CD, Brandon E, et al. Neurturin gene therapy improves motor function and prevents death of striatal neurons in a 3-nitropropionic acid rat model of Huntington’s disease. Neurobiology of disease. 2007 May;26(2):375–384. doi: 10.1016/j.nbd.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 150.Lebherz C, Auricchio A, Maguire AM, Rivera VM, et al. Long-term inducible gene expression in the eye via adeno-associated virus gene transfer in nonhuman primates. Hum Gene Ther. 2005 Feb;16(2):178–186. doi: 10.1089/hum.2005.16.178. [DOI] [PubMed] [Google Scholar]
- 151.Ramaswamy S, McBride JL, Han I, Berry-Kravis EM, et al. Intrastriatal CERE-120 (AAV-Neurturin) protects striatal and cortical neurons and delays motor deficits in a transgenic mouse model of Huntington’s disease. Neurobiology of disease. 2009 Apr;34(1):40–50. doi: 10.1016/j.nbd.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 152.Perez-Navarro E, Akerud P, Marco S, Canals JM, et al. Neurturin protects striatal projection neurons but not interneurons in a rat model of Huntington’s disease. Neuroscience. 2000;98(1):89–96. doi: 10.1016/s0306-4522(00)00074-9. [DOI] [PubMed] [Google Scholar]
- 153.Albin RL, Reiner A, Anderson KD, Dure LSt, et al. Preferential loss of striato-external pallidal projection neurons in presymptomatic Huntington’s disease. Ann. Neurol. 1992 Apr;31(4):425–430. doi: 10.1002/ana.410310412. [DOI] [PubMed] [Google Scholar]
- 154.Reiner A, Albin RL, Anderson KD, D’Amato CJ, et al. Differential loss of striatal projection neurons in Huntington disease. Proc. Natl. Acad. Sci. USA. 1988 Aug;85(15):5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marco S, Perez-Navarro E, Tolosa E, Arenas E, et al. Striatopallidal neurons are selectively protected by neurturin in an excitotoxic model of Huntington’s disease. J. Neurobiol. 2002 Mar;50(4):323–332. doi: 10.1002/neu.10033. [DOI] [PubMed] [Google Scholar]
- 156.Linker RA, Maurer M, Gaupp S, Martini R, et al. CNTF is a major protective factor in demyelinating CNS disease: a neurotrophic cytokine as modulator in neuroinflammation. Nat. Med. 2002 Jun;8(6):620–624. doi: 10.1038/nm0602-620. [DOI] [PubMed] [Google Scholar]
- 157.Stankoff B, Aigrot MS, Noel F, Wattilliaux A, et al. Ciliary neurotrophic factor (CNTF) enhances myelin formation: a novel role for CNTF and CNTF-related molecules. J. Neurosci. 2002 Nov 1;22(21):9221–9217. doi: 10.1523/JNEUROSCI.22-21-09221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Anderson KD, Panayotatos N, Corcoran TL, Lindsay RM, et al. Ciliary neurotrophic factor protects striatal output neurons in an animal model of Huntington disease. Proc. Natl. Acad. Sci. USA. 1996 Jul 9;93(14):7346–7351. doi: 10.1073/pnas.93.14.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Emerich DF, Lindner MD, Winn SR, Chen EY, et al. Implants of encapsulated human CNTF-producing fibroblasts prevent behavioral deficits and striatal degeneration in a rodent model of Huntington’s disease. J. Neurosci. 1996 Aug 15;16(16):5168–5181. doi: 10.1523/JNEUROSCI.16-16-05168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bloch J, Bachoud-Levi AC, Deglon N, Lefaucheur JP, et al. Neuroprotective gene therapy for Huntington’s disease, using polymer-encapsulated cells engineered to secrete human ciliary neurotrophic factor: results of a phase I study. Hum Gene Ther. 2004 Oct;15(10):968–975. doi: 10.1089/hum.2004.15.968. [DOI] [PubMed] [Google Scholar]
- 161.Emerich DF, Thanos CG. Intracompartmental delivery of CNTF as therapy for Huntington’s disease and retinitis pigmentosa. Current gene therapy. 2006 Feb;6(1):147–159. doi: 10.2174/156652306775515547. [DOI] [PubMed] [Google Scholar]
- 162.Emerich DF, Winn SR. Neuroprotective effects of encapsulated CNTF-producing cells in a rodent model of Huntington’s disease are dependent on the proximity of the implant to the lesioned striatum. Cell transplantation. 2004;13(3):253–259. doi: 10.3727/000000004783983981. [DOI] [PubMed] [Google Scholar]
- 163.Emerich DF, Winn SR, Hantraye PM, Peschanski M, et al. Protective effect of encapsulated cells producing neurotrophic factor CNTF in a monkey model of Huntington’s disease. Nature. 1997 Mar 27;386(6623):395–399. doi: 10.1038/386395a0. [DOI] [PubMed] [Google Scholar]