Abstract
Lafora disease is a fatal autosomal recessive form of progressive myoclonus epilepsy. Patients manifest myoclonus and tonic–clonic seizures, visual hallucinations, intellectual, and progressive neurologic deterioration beginning in adolescence. The two genes known to be involved in Lafora disease are EPM2A and NHLRC1 (EPM2B). The EPM2A gene encodes laforin, a dual-specificity protein phosphatase, and the NHLRC1 gene encodes malin, an E3-ubiquitin ligase. The two proteins interact with each other and, as a complex, are thought to regulate glycogen synthesis. Here, we report three Lafora families with two novel pathogenic mutations (C46Y and L261P) and two recurrent mutations (P69A and D146N) in NHLRC1. Investigation of their functional consequences in cultured mammalian cells revealed that malinC46Y, malinP69A, malinD146N, and malinL261P mutants failed to downregulate the level of R5/PTG, a regulatory subunit of protein phosphatase 1 involved in glycogen synthesis. Abnormal accumulation of intracellular glycogen was observed with all malin mutants, reminiscent of the polyglucosan inclusions (Lafora bodies) present in patients with Lafora disease.
Keywords: Epilepsy, Mendelian disorder, Genetics, Autosomal recessive
Introduction
Lafora disease (OMIM 254780) is an autosomal recessive, progressive myoclonus epilepsy that manifests during the first or second decade of life with myoclonic seizures, generalized tonic–clonic seizures, and focal occipital seizures, followed by progressive central nervous system degeneration with severe motor and coordination disability, constant myoclonus, dementia, and, finally, death within 10 years of onset [1]. The pathological hallmark of Lafora disease is the accumulation of polyglucosan (poorly branched and insoluble form of glycogen) inclusion bodies, called Lafora bodies, in the cytoplasm of both neuronal and non-neuronal cells. The diagnosis of the disease relies on clinical findings, presence of Lafora bodies based on axillary skin biopsy, and genetic analysis.
The vast majority of Lafora disease cases (90%) are due to mutations in two genes: EPM2A, which encodes laforin, a member of the dual-specificity protein tyrosine phosphatase family [2, 3], and NHLRC1 (NHL repeat containing 1, EPM2B) [4], which encodes malin, a protein containing a zinc-binding RING finger motif with E3-ubiquitin ligase activity [5, 6]. Malin interacts with and polyubiquitinates laforin, leading to its degradation by the proteasome [5, 6]. Several groups have shown that the laforin–malin complex promotes ubiquitination and proteasome-dependent degradation of proteins involved in the regulation of glycogen biosynthesis, in particular protein targeting to glycogen, R5/PTG, a regulatory subunit of type 1 protein phosphatase [7–9]. An abnormal rise in the cellular levels of R5/PTG, due to loss or alteration of laforin or malin, leads to glycogen accumulation.
We report here the clinical characteristics of three Lafora disease families with four NHLRC1 missense mutations (C46Y, P69A, D146N, and L261P) and provide evidence that all mutations prevented the laforin–malin complex from downregulating R5/PTG-induced glycogen synthesis, thereby leading to glycogen accumulation.
Materials and methods
Families and patients
Twelve unrelated patients affected by Lafora disease were included in the genetic screening. Blood samples were obtained with signed informed consent, and genomic DNA was extracted using standard procedures. We focused on families A, B, and C which were investigated at Pitié-Salpêtrière, Necker-Enfants Malades, and Nancy hospitals, respectively.
NHLRC1 mutational screening
Mutation analysis was performed in the index case of each of the 12 families by direct sequencing of the NHLRC1 single coding exon. Primer sequences are available on request. PCR products were sequenced using the Big Dye Terminator Cycle sequencing kit on a 3710 Applied Biosystems sequencer. Effect of mutations, including conservation of amino acids among species, was evaluated using AlaMut-1.42 prediction software (Interactive Biosoftware, France).
Cell culture and transfection
The full-length human pEGFPC2-malin construct was kindly provided by Dr. S. Ganesh and was used for immunofluorescence experiments. Malin mutants (C46Y, P69A, D146N, and L261P) were generated using the Quickchange®II XL site-directed mutagenesis kit (Stratagene) and sequenced to confirm the correct incorporation of the mutation. To construct pFLAG-malin wild-type and mutant plasmids, BglII/BamHI fragments from pEGFPC2-malin-containing plasmids were subcloned into pFLAG-CMV6 (Sigma). Other plasmids used in this study were pACT2-laforin, pCIneo-laforin, and pCMV-HA-R5/PTG [10]. We used COS-7 cells for immunofluorescence experiments and human embryonic kidney (HEK293) cells for biochemistry experiments and Western blotting. Both cell lines (COS-7 and HEK293) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2 mM of l-glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. All transient transfections were performed using Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen).
Immunofluorescence
COS-7 cells grown on chamber slides were rinsed with PBS and fixed for 20 min in PBS containing 4% paraformaldehyde. After fixation, cells were permeabilized with 0.2% Triton X-100 for 10 min and blocked for 30 min with 10% goat serum. Cells were analyzed using GFP-derived fluorescence for observation of malin and incubated with anti-Myc antibody to label laforin (9E10, Santa Cruz Biotechnology). Finally, the glass slides were washed, air-dried, and mounted with Mowiol. Immunofluorescence images were obtained using a laser scanning confocal microscope (Zeiss 510) with a ×63 objective.
RNA purification and retrotranscription
Total RNAs were extracted from COS-7 cells with RNeasy minicolumns (Qiagen). Quantification was done spectrophotometrically at 260 nm. Up to 5 μg of total RNA from each sample were reverse transcribed for 50 min at 50ºC in a 20-μl reaction volume using 200 units of Superscript® III Reverse Transcriptase (Invitrogen) in the presence of 50 ng random hexamers. Specific primers were designed to specifically amplify a fragment of approximately 100 bp from the 5′ region of the plasmid to the coding sequence of NHLRC1 (5′-CCGGACTCAGATCTCGAGCT-3′ and 5′-CTTCGGCCGCCATGG-3′).
Western blotting
HEK293 cells were lysed 24 h after transfection, as previously described [10]. Total proteins (25 μg) from the soluble fraction of cell lysates were analyzed by SDS-PAGE and Western blotting using appropriate antibodies: mouse monoclonal anti-laforin [10], rabbit polyclonal anti-R5/PTG [10], mouse monoclonal anti-Flag (1/5,000, Sigma), rabbit polyclonal anti-actin (1/4,000, Sigma), rabbit anti-GFP (1/1,000, ab290, Abcam), and monoclonal anti-α-tubulin (1/1,000, Sigma). Immunoreactive bands were visualized using SuperSignal West Dura kit following the manufacturer's instructions (Thermo Scientific). Membranes were revealed on a KODAK Image Station 4000MM Digital Imaging System.
Yeast two-hybrid assay
The yeast two-hybrid system is based on the fact that the protein of interest (laforin) is expressed as a fusion protein to the DNA-binding domain (DBD, here LexA) and the activation domain (AD) is fused to the second protein of interest (malin). Only if laforin and malin physically interact with one another are the DBD and AD brought together to activate expression of the downstream reporter gene (here lac Z which codes for the β-galactosidase).
To construct wild-type and mutant (C46Y, P69A, D146N, and L261P) pBTM116-malin plasmids, EcoRI fragments from pEGFPC2-malin plasmids replaced the EcoRI fragment present in plasmid pBTM116-malin [9]. The yeast CTY10.5d strain was cotransformed with combinations of pACT2-laforin and pBTM116-malin wild-type and mutants. Transformants were grown in selective SC medium, and quantitative β-galactosidase activity was assayed in permeabilized cells and expressed in Miller units.
Glycogen determination
Glycogen determination was carried out by homogenizing transfected HEK293 cells in ice-cold 30% KOH and the extract was then heated at 100ºC for 15 min. Glycogen was then measured as previously described [9]. The amount of glycogen was expressed as the amount of released glucose per milligram of total protein.
Statistical analyses
Values are given as means±standard error of the mean (SEM) of at least three independent experiments. Differences between groups were analyzed by two-tailed Student's t tests. Significance was considered at p < 0.01 and p < 0.001, as indicated in each case.
Results
Clinical description of the families
Family A members originated from a small village in Turkey (Fig. 1a, Table 1). The proband had normal birth and psychomotor development. Epilepsy began at 13 years of age with numerous generalized tonic–clonic seizures, absences with eyelid myoclonia, fragmentary and multifocal, and bilateral massive myoclonus jerks. She never complained of visual hallucinations. Epilepsy was pharmacoresistant to polytherapy with valproate, phenobarbital, and clonazepam. Intellectual decline appeared rapidly after the beginning of the disease. At 27 years, the patient displayed major cognitive disturbances, aphasia, and apraxia. Motor impairment was mainly due to cerebellar symptoms and myoclonus. Axillary skin biopsy was negative. The brother, who was 25 years old, was not examined but displayed similar symptoms starting in adolescence.
Fig. 1.
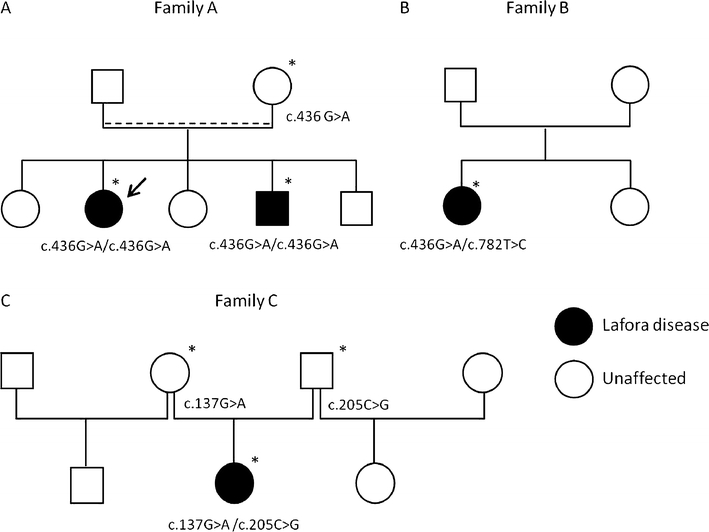
Identification of NHLRC1 mutations. a Segregation of the c.436G>A (p.Asp146Asn/D146N) mutation in family A. Arrow indicates the index case, dashed line indicates probable consanguinity. b Detection of c.782T>C (p.Leu261Pro/L261P) and c.436G>A (p.Asp146Asn/D146N) mutations in patient of family B. c Segregation of c.137G>A (p.Cys46Tyr/C46Y) and c.205C>G (p.Pro69Ala/P69A) mutations in family C. Asterisk indicates DNA sample available for the study
Table 1.
Clinical characteristic of the patients with Lafora disease
| Patient | Sex/Age at time of study or death | Symptoms at onset | Age at onset | Type of seizures | Clinical status at time of study | EEG | MRI | Axillary skin biopsy |
|---|---|---|---|---|---|---|---|---|
| Family A | F/27 | GTCS and absences with eyelid myoclonia, myoclonus | 13 | GTCS, absences | Severe intellectual impairment, ataxia, disabling myoclonus | Slow background, diffuse and frontotemporal epileptic abnormalities | Diffuse cortical atrophy, predominant in temporal regions | Negative for Lafora bodies |
| Photosensitivity | ||||||||
| Family B | F/17 | Absences with eyelid myoclonia, myoclonus | 13 | GTCS, occipital seizures (visual hallucinations during ethosuximide treatment) | Major cognitive deficiency, disabling myoclonus | Generalized discharges of spike waves and polyspikes | Normal | Not done |
| Family C | F/16 | GTCS and myoclonia | 14 | GTCS, SGTCS with visual hallucinations suggestive of occipital seizures | Deceased | Slowed and irregular background, diffuse subclinical spike waves | Normal | Lafora bodies |
GTCS generalized tonic clonic seizures, SGTCS secondarily generalized tonic–clonic seizures
Family B was of Russian origin (Fig. 1b, Table 1). The parents of the proband were not consanguineous. The proband was a 17-year-old girl with normal birth and psychomotor development. The disease began at 13 years of age with brief episodes of loss of contact associated with eyelid myoclonia. She started to present fragmentary and multifocal as well as bilateral massive (both upper limbs) myoclonus jerks, sometimes causing her to fall. She had brief visual hallucinations while being treated with ethosuximide. School difficulties and depressive symptoms progressively appeared. Generalized tonic–clonic seizures on awakening started a few years later. At last follow-up examination, the patient was continuing to suffer from fragmentary and massive myoclonus, occasional tonic–clonic seizures, and major cognitive deficiency.
Family C, of Ukrainian/French origin, was recently described [11] (Fig. 1c, Table 1). This patient had normal birth and psychomotor development. She experienced a first generalized tonic–clonic seizure at 14 years of age. Neuropsychological examination showed a heterogeneous profile with major impairment of nonverbal performance, including impairment of attention, memorization, and visuospatial skills. The patient was pharmacoresistant with fragmentary and bilateral massive myoclonus jerks and frequent loss of contact and photosensitive generalized seizures. Cognitive and motor abilities declined progressively. At age 16, she had a very poor language with complete loss of autonomy, weight loss, and was sleeping most of the day. She died from cardiovascular failure in intensive care for refractory status epilepticus. Axillary skin biopsy showed typical Lafora bodies.
Identification of NHLRC1 mutations
We performed sequence analysis of the NHLRC1 gene in the index cases belonging to a cohort of 12 families with Lafora disease and identified mutations in NHLRC1 in three of them. None of these patients harbored mutations in the coding region of the EPM2A gene. In both affected individuals of family A, we identified the c.436G>A missense mutation at the homozygous state resulting in the previously reported p.Asp146Asn/D146N mutation [4, 12, 13] (Fig. 1a). The patient in family B was a compound heterozygote for two distinct missense mutations: (1) a novel sequence variant c.782T>C resulting in p.Leu261Pro/L261P and (2) a second nucleotide change c.436G>A (D146N; Fig. 1b). The L261P variant has not been described before and most likely represents a novel mutation responsible for Lafora disease since it was not found in 170 Caucasian and 84 North African controls. Since DNA of the parents was not available, we cloned the NHLRC1 ORF from the lymphoblasts of the patient and confirmed that L261P and D146N variants were located on two distinct alleles, suggesting that the parents were heterozygous for each mutation. The patient in family C was a compound heterozygote for two missense mutations: c.137G>A resulting in p.Cys46Tyr/C46Y and c.205C>G resulting in p.Pro69Ala/P69A (Fig. 1c). While P69A results from a recurrent mutational event reported in numerous Lafora disease families [4, 13, 14], C46Y mutation is a recently reported novel mutation [11], which was absent from 110 control individuals of same ethnic origin.
The two mutations C46Y and P69A were located in the RING domain mediating E3 ubiquitin ligase activity, while the other two, D146N and L261P, were located in NHL domains (Fig. 2a). All four amino acid residues affected by the missense mutations identified in the present study were highly conserved across a broad range of different NHLRC1 orthologous proteins, reflecting the evolutionary constraint placed on these residues (Fig. 2b). We therefore considered them to be pathogenic mutations.
Fig. 2.
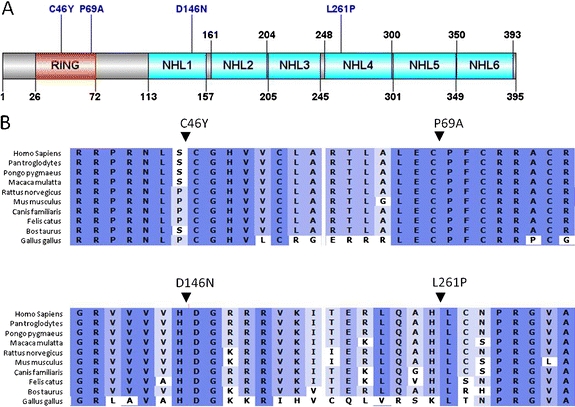
Representation of malin showing mutations. a Schematic representation showing domain organization and location of the mutations of the malin protein. b Multiple protein alignment of malin showing conservation of Cys46, Pro69, Asp146, and Leu261 residues in different vertebrates
Expression levels of malin mutants are similar to wild type
We next analyzed the mRNA and protein profile of malinC46Y, malinP69A, malinD146N, malinL261P, and wild-type malin. COS-7 cells were transiently transfected with equal amounts of plasmids and, 24 h later, RNAs from cell lysates were analyzed by RT-PCR. As shown in Fig. 3, C46Y, P69A, D146N, and L261P disease-causing mutations did not cause major alterations of the malin transcript (upper panel). We also analyzed the protein levels of wild-type and mutant forms of malin by Western blot. The expression constructs encoding GFP-tagged malin and the four mutant forms produced fusion proteins of expected size. As shown, the steady-state levels of expression of malinC46Y, malinP69A, malinD146N, malinL261P, and wild type were similar, suggesting that all mutants had normal conformation and were stable in COS-7 cells (Fig. 3, bottom panels).
Fig. 3.
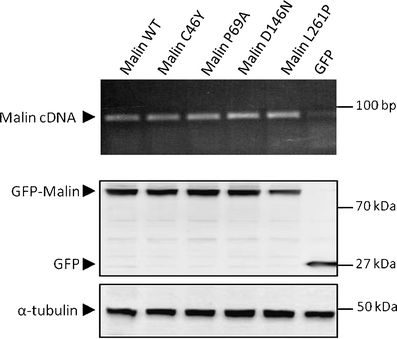
Expression levels of mutants of malin. COS-7 cells were transiently transfected with the wild-type (WT) or indicated mutant GFP-malin-expressing plasmids. Upper panel RT-PCR products of exogenous wild-type and mutants of malin separated on a 1.5% agarose gel. All mRNA transcripts were detected in comparable amounts. Bottom panel detection of wild type and mutants of malin by Western blot with anti-GFP antibody. Equal amounts of proteins were loaded as shown by the α-tubulin control
Subcellular distribution of malin mutants is similar to wild type
We examined the subcellular localization of wild-type and Lafora disease-causing mutant forms of malin in transfected COS-7 cells by fluorescence imaging. Overexpressed GFP-tagged malin was observed in the cytoplasm of cells, predominantly in a diffuse manner (Fig. 4a) but also as a punctate perinuclear staining in ~15% of cells (Fig. 4b), or as a large aggregate in a small proportion of cells (~8%; Fig. 4c). Large aggregates were present in apparently healthy cells, presenting an intact DAPI-labeled nucleus as reported in another study [15]. Expression of GFP-malin at low levels (five times less transfected plasmid) caused a similar proportion of cells containing aggregates, suggesting that it is not a consequence of overexpression (data not shown).
Fig. 4.
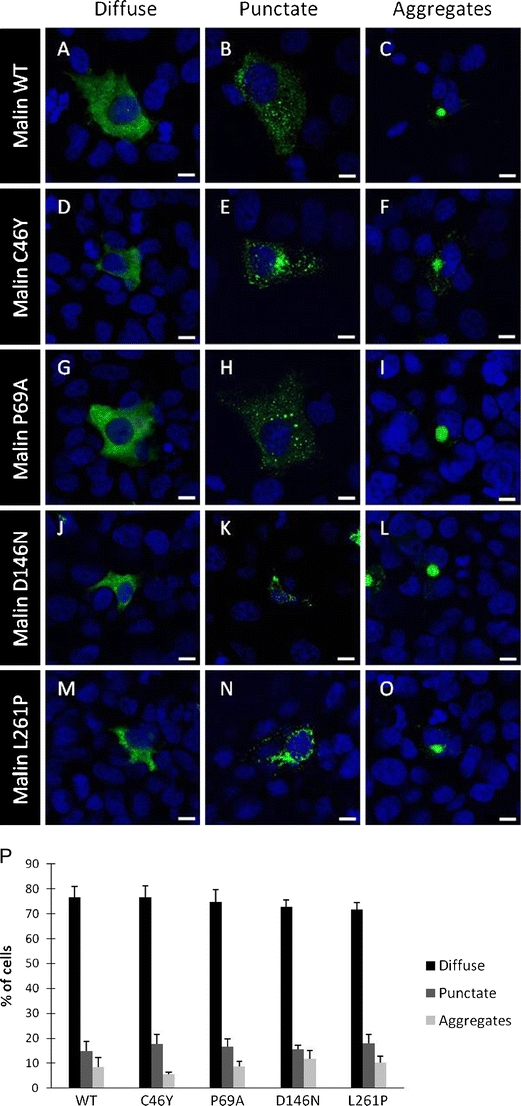
Subcellular distribution of malin mutants. COS-7 cells were transiently transfected with the wild-type (WT) or indicated mutant GFP-malin-expressing plasmids. Malin was detected with a confocal microscope using the GFP-derived fluorescence, and nuclei were stained with DAPI. All malin variants showed a cytoplasmic localization, either as a diffuse or punctate staining or as large aggregates. Most representative localizations of wild-type (WT) malin (a–c), C46Y (d–f), P69A (g–i), D146N (j–l), and L261P (m–o) mutants are shown. Magnification bars, 10 μm (p). Histograms showing the percentage of cells in each experiment (WT, n = 1,140; C46Y, n = 1,247; P69A, n = 1,262; D146N, n = 1,306; L261P, n = 1,145 cells; bars indicate SEM of three independent transfections)
Next, we analyzed the subcellular distribution and aggregate formation of the pathogenic malinC46Y (Fig. 4d–f), malinP69A (Fig. 4g–i), malinD146N (Fig. 4j–l), and malinL261P mutants (Fig. 4m–o). The subcellular distribution and aggregation profile of these mutant proteins were similar to those of wild-type malin. Among three independent experiments, there was no statistical difference in proportion between the number of cells with diffuse versus punctated and aggregated malin between malinC46Y, malinP69A, malinD146N, malinL261P, and wild-type protein (Fig. 4p).
We also examined the colocalization of malin with its interacting partner laforin (Fig. 5). Both wild-type malin and laforin were colocalized in a diffuse and punctate manner in the cytoplasm of the transfected cells (Fig. 5c). All four malin mutants also colocalized with laforin in the cytoplasm of COS-7 cells (Fig. 5f, i, l, o).
Fig. 5.
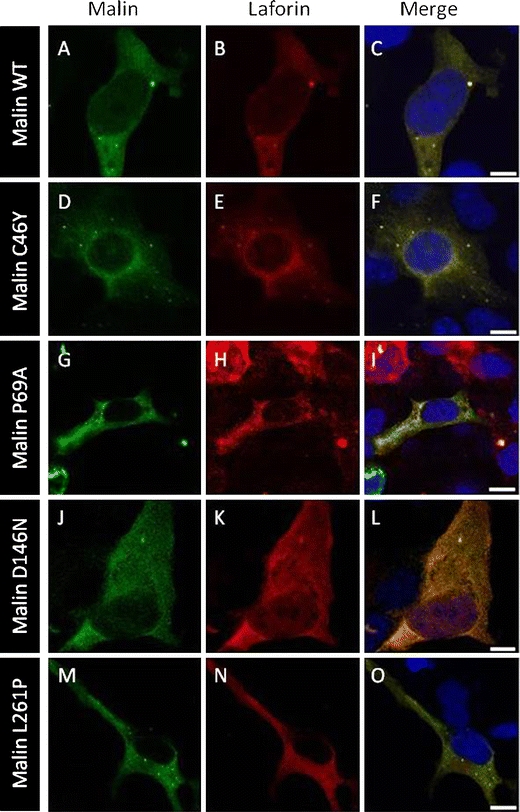
Malin mutants colocalize with laforin. COS-7 cells were transiently cotransfected with the GFP-malin and laforin-Myc plasmids. Direct visualization of GFP-malin (green) and immunofluorescence with anti-Myc (red) was performed. Most representative localizations with wild-type (WT) malin (a–c), C46Y (d–f), P69A (g–i), D146N (j–l), and L261P (m–o) mutants are shown. Magnification bars, 10 μm
Impaired interaction of malin mutants with laforin in yeast two-hybrid assay
We next performed a yeast two-hybrid assay to examine the ability of malin mutants to interact with laforin. With this aim, we constructed pBTM116-malin plasmids (wild-type and C46Y, P69A, D146N, and L261P mutants) that produced LexA–malin fusion proteins. As we previously reported [9], wild-type malin interacted with laforin. However, all four malin mutations caused a severe decrease of interaction with laforin (Fig. 6a), although we observed a residual interaction between laforin and malinP69A and malinD146N. The differences in protein–protein interaction between mutants and wild type were not due to different expression levels of the constructs, since all of them were expressed at similar levels in yeast (Fig. 6b).
Fig. 6.
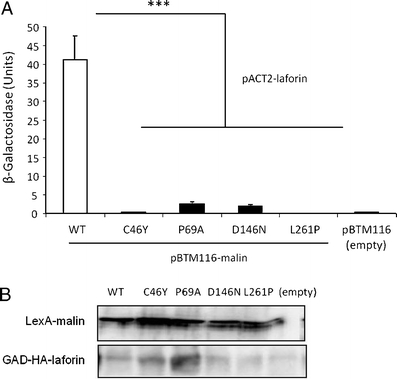
Malin mutations affect yeast two-hybrid interaction with laforin. Yeast CTY10.5d cells were cotransformed with plasmids pBTM116-malin (WT or indicated mutants) or empty vector (pBTM116) and plasmid pACT2-laforin. a Protein interaction was estimated using the yeast two-hybrid system, by measuring the β-galactosidase activity. Values correspond to means from four to six different transformants (bars indicate SEM). b Western blot analysis indicated that all proteins were expressed at similar levels
Downregulation of R5/PTG is impaired by malin mutations
Since the laforin–malin complex downregulates R5/PTG-induced glycogenesis [9], we investigated the ability of disease-causing malin mutations to degrade the R5/PTG protein. HEK293 cells were cotransfected with Flag-malin, pCIneo-laforin, and pCMV-HA-R5/PTG, and proteins were analyzed by Western blot. As expected, coexpression of laforin and wild-type malin with R5/PTG resulted in a marked reduction of R5/PTG levels in comparison with cells that did not overexpress laforin and malin (Fig. 7a). In contrast, the coexpression of laforin with either malinC46Y, malinP69A, malinD146N, or malinL261P caused an accumulation of R5/PTG in the cells (Fig. 7a). As illustrated in Fig. 7b, quantification by densitometry of the relative levels of R5/PTG revealed a statistically significant difference between wild-type and mutant forms of malin. Increased levels of R5/PTG suggest that overproduced malin mutants could not form a functional complex with laforin to mediate ubiquitin-dependent proteasomal degradation of R5/PTG.
Fig. 7.
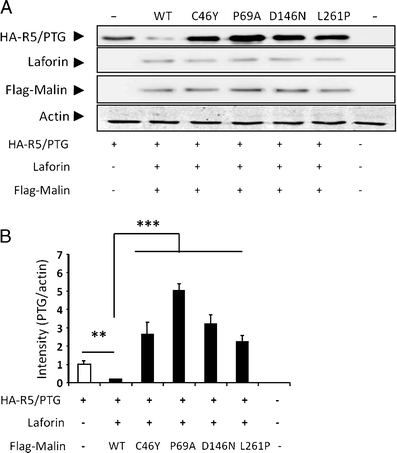
Malin mutations do not downregulate R5/PTG protein levels. HEK293 cells were cotransfected with pCMV-HA-R5/PTG, pCIneo-laforin, and pFLAG-Malin wild-type (WT) or different mutant plasmids. a Twenty-four hours after transfection, cell extracts (25 μg) were analyzed by Western blotting using anti-R5/PTG, anti-laforin, anti-Flag, and anti-actin antibodies. b Relative intensity of the R5/PTG bands was normalized to the actin levels and referred to the levels found in cells only expressing R5/PTG. Bars indicate SEM of three independent experiments: **p < 0.01 and ***p < 0.001
Malin mutations lead to glycogen accumulation
We next measured the impact of malin pathogenic mutations on the glycogenic capacity of R5/PTG protein. The cotransfection of laforin with wild-type malin led to a marked decrease of glycogen levels in HEK293 cells expressing R5/PTG compared to cells overexpressing only R5/PTG (Fig. 8). This result is consistent with the fact that the laforin–malin complex downregulates R5/PTG via a proteasomal degradation pathway. In contrast, malinC46Y, malinP69A, malinD146N, and malinL261P mutants all failed to reduce glycogen levels. As illustrated in Fig. 8, accumulation of glycogen levels was detected in cells expressing malin mutants but not in cells expressing wild-type malin (p < 0.01).
Fig. 8.
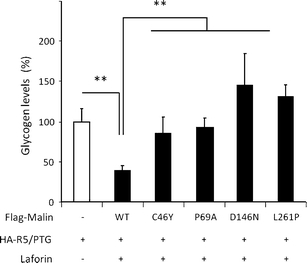
Malin mutations do not downregulate the glycogenic activity of R5/PTG. Glycogen levels were measured in transfected HEK293 cells with pCMV-HA-R5/PTG, pCINEO-laforin, and pFLAG-malin wild-type (WT) or different mutant plasmids. Values were referred to those found in cells expressing only HA-R5/PTG. Bars indicate SEM of three independent experiments. **p < 0.01
Discussion
In this paper, we have described three families with Lafora disease mutated in the NHLRC1 gene and investigated the functional impact of the corresponding mutations. We identified four pathogenic mutations in NHLRC1: the novel mutation L261P, the recently reported mutation C46Y, and two recurrent missense mutations, P69A and D146N. We first aimed to assess by fluorescence imaging the impact of these mutations on the subcellular targeting of GFP-tagged malin in transfected COS-7 cells. In accordance with previous studies [4–6, 16], we observed that the wild-type malin protein displayed a diffuse cytoplasmic expression pattern with small punctate inclusions or large aggregates in some cells. A thorough analysis of the cellular localization of the disease-causing malinC46Y, malinP69A, malinD146N, and malinL261P mutants showed that all presented a similar expression pattern to that of wild-type malin. The effect of Lafora disease-associated malin mutations on cellular localization is known to be heterogeneous since a fraction of NHLRC1 missense mutations alter the subcellular localization of malin, while others have no effect [16, 17]. In our experimental conditions, C46Y, P69A, D146N, and L261P mutations did not impair the subcellular targeting of GFP-tagged malin protein and all colocalized with laforin similarly to wild-type malin, confirming that alteration of the subcellular localization is not a mechanism common to all NHLRC1 mutations. We then assessed by a yeast two-hybrid assay the interaction with laforin and provide evidence that all four mutations affected the physical interaction with laforin, suggesting that mutations might prevent the formation of a laforin–malin complex. Surprisingly, mutations in the RING domain of malin (C46Y and P69A) impaired interaction with laforin, similarly as mutations in the NHL domains (D146N and L261P), probably due to perturbation in the overall conformation of the protein. Since several laforin–malin interaction partners involved in the glycogenesis might be affected by malin mutations, we investigated whether the pathogenic mutations of malin could affect the glycogen synthesis pathway. A key positive regulator of glycogen synthesis is R5/PTG, a subunit of protein phosphatase 1, which is downregulated by the laforin–malin complex via ubiquitination and proteasome-dependent degradation. First, we investigated the capacity of malin mutants to regulate R5/PTG levels in HEK293 cells simultaneously overexpressing laforin, malin, and R5/PTG. Our results revealed that levels of R5/PTG in cells expressing malinC46Y, malinP69A, malinD146N, and malinL261P mutants concomitantly with laforin were increased, consistent with an absence of degradation of R5/PTG. Destabilization of the laforin–malin complex probably precludes the action of malin on its substrate R5/PTG. We next measured the levels of intracellular glycogen and observed that all disease-causing mutations caused a marked increase in glycogen level. Since overexpression of R5/PTG results in glycogen accumulation [9], we speculate that accumulation of glycogen with mutant forms of malin is likely due to the lack of downregulation of R5/PTG levels. However, the impairment of the laforin–malin interaction probably causes the loss of interaction with other partners of the glycogen synthesis pathway, including the muscle glycogen synthase, which could also lead to a glycogen accumulation [7]. Vilchez et al. had reported a similar observation for D146N mutation [7], and we provide evidence here that accumulation of glycogen is likely to be a frequent consequence of NHLRC1 mutations. Epm2b−/− knockout mouse was recently reported to display higher levels of glycogen in skeletal muscle, liver, and brain, which is also consistent with malin having a critical role in the regulation of glycogen metabolism [18, 19]. In the present study, while all four NHLRC1 mutations had similar effects on glycogen levels of transfected cells, they were not all associated with the positive finding of Lafora polyglucosan bodies. Indeed, the skin biopsy of the patient harboring both C46Y and P69A mutations (family C) revealed the presence of Lafora bodies, whereas the patient harboring the D146N mutation (family A) was negative for Lafora bodies. The patient with D146N and L261P mutations (family B) could not be tested (Table 1). Since all mutations led to an in vitro increase of glycogen levels, we can speculate that Lafora bodies may have been missed due to a high false-negative rate of skin biopsy in Lafora disease [11]. This may well have been the case with the patient with the D146N mutation, since Lafora bodies were reported to be present in other patients with the same mutation at the homozygous state [12, 13]. Interestingly, several groups have reported that the D146N mutation in NHLRC1 is associated with a milder clinical course with late age at onset and slow progression of disease [12–14]. We also noticed that the disease course was longer (14 years at the time of the study) in the index case of family A with the D146N mutation at the homozygous state, compared to the patient of family B who was compound heterozygote for the D146N and L261P mutations. A possible explanation for the milder severity of the disease in patients carrying a homozygous malin D146N mutation could be that the enzymatic activity of the D146N variant is still preserved, at least for some substrates, consistent with previous in vitro results indicating that purified malin D146N retained E3-ubiquitin ligase activity [9]. In contrast, the patient in family C had an unusually fast disease progression. Since the P69A mutation, which has been reported in many patients of Southern European descent, was not shown to be associated with an unusual Lafora phenotype [14], we can hypothesize that the C46Y mutation played a role in the rapid disease course of this patient. It would suggest the existence of possible phenotype–genotype correlations for some mutations at the homozygous state. However, this notion will need to be confirmed in a larger number of families.
In conclusion, we have functionally characterized four disease-causing mutations in NHLRC1, two of which were recurrent (P69A and D146N) and two were novel (C46Y and L261P). Our data revealed that the major and common effect of these variants, irrespective of their position in the protein (RING domain or NHL domains), is to lead to an accumulation of intracellular glycogen that could be the cause of the formation of Lafora bodies in patients.
Acknowledgments
We are grateful to the patients and their families. We would like to thank Dr. Aurélien Dauphin for imaging, Dr. Subramaniam Ganesh for providing the pEGFPC2-malin clone, and Dr. Antoinette Gelot for the examination of skin biopsies.
Disclosure
The authors have nothing to disclose and have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Philippe Couarch and Santiago Vernia contributed equally to this study.
References
- 1.Minassian BA. Lafora's disease: towards a clinical, pathologic, and molecular synthesis. Pediatr Neurol. 2001;25:21–29. doi: 10.1016/S0887-8994(00)00276-9. [DOI] [PubMed] [Google Scholar]
- 2.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 3.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum Mol Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 4.Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 5.Gentry MS, Worby CA, Dixon JE. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci USA. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohi H, Ianzano L, Zhao X-C, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet. 2005;14:2727–2736. doi: 10.1093/hmg/ddi306. [DOI] [PubMed] [Google Scholar]
- 7.Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat Neurosci. 2007;10:1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 8.Worby CA, Gentry MS, Dixon JE. Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG) J Biol Chem. 2008;283:4069–4076. doi: 10.1074/jbc.M708712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, et al. Regulation of glycogen synthesis by the laforin–malin complex is modulated by the AMP-activated protein kinase pathway. Hum Mol Genet. 2008;17:667–678. doi: 10.1093/hmg/ddm339. [DOI] [PubMed] [Google Scholar]
- 10.Vernia S, Solaz-Fuster MC, Gimeno-Alcaniz JV, Rubio T, Garcia-Haro L, Foretz M, de Cordoba SR, Sanz P. AMP-activated protein kinase phosphorylates R5/PTG, the glycogen targeting subunit of the R5/PTG-protein phosphatase 1 holoenzyme, and accelerates its down-regulation by the laforin–malin complex. J Biol Chem. 2009;284:8247–8255. doi: 10.1074/jbc.M808492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesca G, Boutry-Kryza N, de Toffol B, Milh M, Steschenko D, Lemesle-Martin M, Maillard L, Foletti G, Rudolf G, Nielsen JE, et al. Novel mutations in EPM2A and NHLRC1 widen the spectrum of Lafora disease. Epilepsia. 2010;51:1691–1698. doi: 10.1111/j.1528-1167.2010.02692.x. [DOI] [PubMed] [Google Scholar]
- 12.Baykan B, Striano P, Gianotti S, Bebek N, Gennaro E, Gurses C, Zara F. Late-onset and slow-progressing Lafora disease in four siblings with EPM2B mutation. Epilepsia. 2005;46:1695–1697. doi: 10.1111/j.1528-1167.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 13.Franceschetti S, Gambardella A, Canafoglia L, Striano P, Lohi H, Gennaro E, Ianzano L, Veggiotti P, Sofia V, Biondi R, et al. Clinical and genetic findings in 26 Italian patients with Lafora disease. Epilepsia. 2006;47:640–643. doi: 10.1111/j.1528-1167.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Abad C, Gomez-Garre P, Gutierrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, Rodriguez de Cordoba S, Serratosa JM. Lafora disease due to EPM2B mutations: a clinical and genetic study. Neurology. 2005;64:982–986. doi: 10.1212/01.WNL.0000154519.10805.F7. [DOI] [PubMed] [Google Scholar]
- 15.Rao SNR, Sharma J, Maity R, Jana NR. Co-chaperone CHIP stabilizes aggregate-prone malin, a ubiquitin ligase mutated in Lafora disease. J Biol Chem. 2010;285:1404–1413. doi: 10.1074/jbc.M109.006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittal S, Dubey D, Yamakawa K, Ganesh S. Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum Mol Genet. 2007;16:753–762. doi: 10.1093/hmg/ddm006. [DOI] [PubMed] [Google Scholar]
- 17.Singh S, Satishchandra P, Shankar SK, Ganesh S. Lafora disease in the Indian population: EPM2A and NHLRC1 gene mutations and their impact on subcellular localization of laforin and malin. Hum Mutat. 2008;29:E1–12. doi: 10.1002/humu.20737. [DOI] [PubMed] [Google Scholar]
- 18.Turnbull J, Wang P, Girard J-M, Ruggieri A, Wang TJ, Draginov AG, Kameka AP, Pencea N, Zhao X, Ackerley CA, et al. Glycogen hyperphosphorylation underlies lafora body formation. Ann Neurol. 2010;68:925–933. doi: 10.1002/ana.22156. [DOI] [PubMed] [Google Scholar]
- 19.Depaoli-Roach AA, Tagliabracci VS, Segvich DM, Meyer CM, Irimia JM, Roach PJ. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to Lafora bodies and the accumulation of insoluble laforin. J Biol Chem. 2010;285:25372–25381. doi: 10.1074/jbc.M110.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]


