Abstract
Protein Inhibitor of Activated Signal Transducer and Activators of Transcription 3 (PIAS3) is a molecule that regulates STAT3 and has antiproliferative properties. Glioblastoma and squamous cell lung cancer lack PIAS3 expression. To test the hypothesis that PIAS3 transcriptional effects are STAT3-independent, we developed models for STAT3 knockdown and PIAS3 overexpression. PIAS3 expression results in a distinct transcriptional profile that does not occur with STAT3 knockdown. We identify novel transcription factor binding partners for PIAS3 including ETS, EGR1, NR1I2 and GATA1. PIAS3 binds to these factors and regulates their transcriptional effects resulting in alterations in canonical pathways including Wnt/β-catenin signaling and functions such as cell death and proliferation. A model is proposed by which PIAS3 effects EGR1 regulated pathways.
Key words: PIAS3, STAT3, lung cancer, signal transduction, epidermal growth factor, transcription factor, EGR
Introduction
Lung cancer is the leading cause of cancer-related death in both the United States and the world. In 2007, 213,380 new lung cancer cases were seen in the United States with 160,390 deaths related to this disease.1 The devastating 5-year relative survival rates for this disease in the United States including all stages is approximately 15.0%.1 Most of the promise for this disease during the last decade relates to the introduction of novel targeted therapeutic agents such as those targeting the epidermal growth factor receptor (EGFR).2 However, these interventions have only minimally decreased overall mortality rates.
Signal Transducer and Activators of Transcription (STAT) are important cytoplasmic proteins that act as transcription factors to regulate gene expression. STAT proteins, especially STAT3, are important in the development and progression of cancers by either preventing apoptosis or promoting proliferation.3 STAT3 is a key molecule in maintaining a transformed phenotype and inhibition of STAT3 has become a potential target for drug development in cancer.4 Indeed, blockade of STAT3 results in extensive apoptosis of NSCLC cells,5and combined inhibition of EGFR and STAT3 can have synergistic anti-proliferative effects.6
PIAS3 (protein inhibitor of activated STAT3), was originally identified as a specific inhibitor of the STAT3 signaling pathway.7A growing body of evidence has shown that the PIAS family (PIAS1, PIAS3, PIASx and PIASy) regulates a variety of cytokine and growth factor signaling pathways and transcription factors, mainly through interaction with STAT proteins but also NFκB and SMADs, through protein-protein interactions.8 It has also been demonstrated that the PIAS family has a conserved ring finger-like zinc binding domain that is involved in SUMO-E3-ligase activity and regulates activity of transcription factors such as SMAD4 9 through sumoylation. The PIAS family is generally associated with transcriptional repression through different mechanisms. For example, PIAS1 blocks DNA-binding by NFκB (nuclear factor κB) p65 and inhibits NFκB-mediated gene activation.10 PIASy interacts with HDACs (histone deacetylases) and represses the transcriptional activity of AR (androgen receptor),11 and SMAD3 by recruiting HDAC.12 In the case of PIAS3, however, little information is available on how PIAS3 may exert anti-proliferative properties.
Recently PIAS3 has emerged as a protein with putative tumor suppressor function with lack of expression of PIAS3 in glioblastoma multiforme13 and a consequent unregulated increase in phosphorylation of STAT3. We have also identified a lack of PIAS3 protein expression in the majority of human squamous cell carcinomas of the lung.14 We have recently demonstrated that PIAS3 binds to STAT3 within minutes of ligand-induced activation and phosphorylation of tyrosine 705 of STAT3, and that PIAS3 negatively regulates STAT3 transcriptional activity in a concentration dependent manner.15 In addition, we have identified a novel mechanism by which PIAS3 may effect STAT3 signaling, by decreasing STAT3 phosphorylation.15 PIAS3 decreases lung cancer growth and increases the antitumor effects of EGFR inhibitors.16 It is, however, unknown if PIAS3 works uniquely by interacting with STAT3 or other mechanisms of action exist.
Our present study convincingly demonstrates that overexpression of PIAS3 results in a downstream transcriptional profile that is not seen with STAT3 knockdown. This indicates a STAT3 independent activity and indeed we show that PIAS3 interacts with a several transcription factors including: ETS, EGR, NR1I2 and GATA1. We demonstrate both in vitro and in vivo binding of PIAS3 to these novel transcription factors as we all the promoter of their target genes. EGR1 is functionally regulated by PIAS3 and PIAS3 overexpression results in alterations in multiple canonical pathways including the Wnt/β-catenin and non-small cell lung cancer signaling as well as cellular functions such as cell death and proliferation.
Results
Upregulation of PIAS3 expression and downregulation of STAT3 in A549 cells.
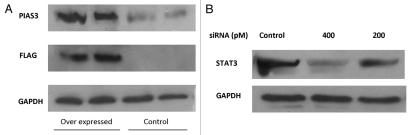
The main goal of the studies presented here is to evaluate the differences in functional effects of overexpression of PIAS3 on the one hand and downregulation of STAT3 on the other hand. To achieve this goal, we transfected A549 cells with a pCMV5 vector encoding human PIAS3 expressing gene and siRNA targeting STAT3. PIAS3 overexpression and STAT3 knockdown were confirmed by immunoblotting with anti-PIAS3, anti-FLAG and anti-STAT3 antibodies respectively (Fig. 1A and B). We were able to achieve a high degree of PIAS3 overexpression in PIAS3 transfected cells (transfection efficiency 30–40%) and were also able to achieve a significant reduction in STAT3 with targeted siRNA (>90% knockdown). The RNA extracted from transfected cells, unstimulated or stimulated with EGF was subsequently used for expression profile analysis utilizing a cDNA microarray. RNA from the different experimental groups was processed and hybridized to Affymetrix U133A expression micro-arrays on which 22,000 transcripts and ESTs are displayed, representing approximately 14,500 unique human genes.
Figure 1.
Confirmation of (A) PIAS3 overexpression: A549 cells were transfected with a PIAS3 expressing construct. Controls refer to trasnfection with an empy vector not containing the PIAS3. The protein gel blot shows a significant increase in pIas3 expression as well as the FLaG protein. GAPDH was used as a loading control. (B) STAT3 downregulation by siRNa: A549 cells were transfected with siRNA targeting STAT3. protein gel blotting shows a significant decrease in STAT3 expression. Controls refer to transfection with mismatched siRNA. GAPDH was used as a loading control.
PIAS3 upregulation and STAT3 knockdown have different downstream transcriptional effects.
Our hypothesis proposed that PIAS3 upregulation and STAT3 downregulation may have different downstream transcriptional effects indicating that PIAS3 exerts functions independent of the negative regulatory of PIAS3 on STAT3. Table 1 depicts the genes that are downregulated by PIAS3 overexpression (but not by STAT3 knockdown) using at least a 4-fold change as cutoff. A total of 37 genes are identified. Many genes are associated with the Wnt/β-catenin signaling, including heparin sulfate proteoglycan,17 transcription factor 4 18 and nucleophosmin.19 Another large category is cell adhesion molecules including CDH8, multimerin 1, CDH6 and fibronectin. Another notable gene with wide implications for cancer progression and metastases is SPARC (osteonectin).20Table 2 demonstrates genes that are upregulated by PIAS3 over-expression (but not STAT3 knockdown) using a 10-fold increase as cutoff. A total of 61 genes are identified. The genes altered include many genes involved in interferon and cytokine signaling. Using 2.5-fold changes as a cutoff STAT3 knockdown downregulated 23 genes and upregulated 7 genes not seen with PIAS3 upregulation (data not shown). As a whole, genes affected by the overexpression of PIAS3 are more numerous than that of STAT3 knockdown. This suggests that PIAS3 has more diverse biological activity independent of its regulation of STATs transcriptional activity.
Table 1.
Genes that are downregulated by pIas3 overexpression (but not by sTaT3 knockdown) using at least a 4-fold change as cutoff
| Symbol | Entrez gene name | Affymetrix/Entrez gene | Fold change |
| CDH8 | cadherin 8, type 2 | 217574_at | −25.99 |
| ATG16L1 | ATG16 autophagy related 16-like 1 (S. cerevisiae) | 220521_s_at | −22.63 |
| AKAP13 | A kinase (PRKA) anchor protein 13 | 213516_at | −21.11 |
| TAGLN | transgelin | 205547_s_at | −18.38 |
| GLRB | glycine receptor, β | 205280_at | −17.15 |
| IL32 | interleukin 32 | 203828_s_at | −17.15 |
| CEP135 | centrosomal protein 135 kDa | 207286_at | −14.93 |
| MMRN1 | multimerin 1 | 205612_at | −14.93 |
| HSPG2 | heparan sulfate proteoglycan 2 | 201654_s_at | −12.13 |
| KLHL4 | kelch-like 4 (Drosophila) | 214591_at | −12.13 |
| GOLGA4 | golgi autoantigen, golgin subfamily a, 4 | 215203_at | −9.85 |
| SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | 200665_s_at | −9.85 |
| CDH6 | cadherin 6, type 2, K-cadherin (fetal kidney) | 205532_s_at | −9.19 |
| TCF4 | transcription factor 4 | 212385_at | −9.19 |
| C4BPB | complement component 4 binding protein, β | 208209_s_at | −8.57 |
| AOX1 | aldehyde oxidase 1 | 205082_s_at | −8 |
| LYPD1 | LY6/PLAUR domain containing 1 | 212909_at | −7.46 |
| MBD6 | methyl-CpG binding domain protein 6 | 214384_s_at | −7.46 |
| MTMR8 | myotubularin related protein 8 | 220537_at | −6.06 |
| NRG1 | neuregulin 1 | 208241_at | −5.66 |
| EPHA5 | EPH receptor A5 | 215664_s_at | −5.28 |
| N4BP2L2 | NEDD4 binding protein 2-like 2 | 214753_at | −5.28 |
| DCLK1 | doublecortin-like kinase 1 | 205399_at | −4.92 |
| FLRT2 | fibronectin leucine rich transmembrane protein 2 | 204359_at | −4.92 |
| SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 213543_at | −4.92 |
| AKT2 | v-akt murine thymoma viral oncogene homolog 2 | 211453_s_at | −4.59 |
| MED13L | mediator complex subunit 13-like | 216109_at | −4.59 |
| PDLIM7 | PDZ and LIM domain 7 (enigma) | 203370_s_at | −4.59 |
| NR5A2 | nuclear receptor subfamily 5, group A, member 2 | 208337_s_at | −4.29 |
| PDGFC | platelet derived growth factor C | 218718_at | −4.29 |
| DIO2 | deiodinase, iodothyronine, type II | 203700_s_at | −4 |
| ETV1 | ets variant 1 | 221911_at | −4 |
| GLS | glutaminase | 203159_at | −4 |
| NPM3 | nucleophosmin/nucleoplasmin 3 | 205129_at | −4 |
| SMA4 | glucuronidase, beta pseudogene | 215599_at | −4 |
| TTLL3 | tubulin tyrosine ligase-like family, member 3 | 217818_s_at | −4 |
Table 2.
Genes that are upregulated by PIAS3 overexpression (but not STAT3 knockdown) using a 10-fold increase as cutoff
| Symbol | Entrez gene name | Affymetrix/Entrez gene | Fold change |
| APOL1 | apolipoprotein L, 1 | 209546_s_at | 11.31 |
| AQP3 | aquaporin 3 (Gill blood group) | 39248_at | 12.13 |
| CST1 | cystatin SN | 206224_at | 12.13 |
| GBP1 | guanylate binding protein 1, interferon-inducible, 67 kDa | 202270_at | 12.13 |
| IRF9 | interferon regulatory factor 9 | 203882_at | 12.13 |
| SP110 | SP110 nuclear body protein | 208392_x_at | 12.13 |
| TRPM4 | transient receptor potential cation channel, subfamily M, member 4 | 219360_s_at | 12.13 |
| ANKRD26 | ankyrin repeat domain 26 | 205705_at | 13.00 |
| TMOD2 | tropomodulin 2 (neuronal) | 219701_at | 13.00 |
| C18ORF1 | chromosome 18 open reading frame 1 | 207996_s_at | 13.93 |
| DDX60 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 | 218986_s_at | 13.93 |
| FCGBP | Fc fragment of IgG binding protein | 203240_at | 13.93 |
| IFIT5 | interferon-induced protein with tetratricopeptide repeats 5 | 203596_s_at | 13.93 |
| LRRTM4 | leucine rich repeat transmembrane neuronal 4 | 220345_at | 13.93 |
| NR4A1 | nuclear receptor subfamily 4, group A, member 1 | 202340_x_at | 13.93 |
| SAMD9 | sterile alpha motif domain containing 9 | 219691_at | 13.93 |
| CCDC69 | coiled-coil domain containing 69 | 212886_at | 14.93 |
| FAM63B | family with sequence similarity 63, member B | 37802_r_at | 14.93 |
| PAQR6 | progestin and adipoQ receptor family member VI | 219236_at | 14.93 |
| FGB | fibrinogen β chain | 204988_at | 16.00 |
| IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | 217502_at | 16.00 |
| PDE1A | phosphodiesterase 1A, calmodulin-dependent | 208396_s_at | 16.00 |
| CYP4F11 | cytochrome P450, family 4, subfamily F, polypeptide 11 | 206153_at | 17.15 |
| IFI16 | interferon, gamma-inducible protein 16 | 208966_x_at | 17.15 |
| S100P | S100 calcium binding protein P | 204351_at | 17.15 |
| OASL | 2′-5′-oligoadenylate synthetase-like | 205660_at | 18.38 |
| IRF7 | interferon regulatory factor 7 | 208436_s_at | 19.70 |
| TNFSF10 | tumor necrosis factor (ligand) superfamily, member 10 | 202688_at | 19.70 |
| ESM1 | endothelial cell-specific molecule 1 | 208394_x_at | 21.11 |
| FGA | fibrinogen alpha chain | 205649_s_at | 21.11 |
| IGHM | immunoglobulin heavy constant mu | 216558_x_at | 21.11 |
| NPHP1 | nephronophthisis 1 (juvenile) | 206285_at | 21.11 |
| LAMP3 | lysosomal-associated membrane protein 3 | 205569_at | 22.63 |
| TNFRSF11B | tumor necrosis factor receptor superfamily, member 11b | 204932_at | 22.63 |
| CES1 | carboxylesterase 1 (monocyte/macrophage serine esterase 1) | 209616_s_at | 24.25 |
| PCSK5 | proprotein convertase subtilisin/kexin type 5 | 213652_at | 24.25 |
| SERPINB3 | serpin peptidase inhibitor, clade B (ovalbumin), member 3 | 209720_s_at | 25.99 |
| HERC5 | hect domain and RLD 5 | 219863_at | 27.86 |
| IFI6 | interferon, alpha-inducible protein 6 | 204415_at | 27.86 |
| TRIM22 | tripartite motif-containing 22 | 213293_s_at | 27.86 |
| DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 218943_s_at | 29.86 |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | 204747_at | 29.86 |
| RSAD2 | radical S-adenosyl methionine domain containing 2 | 213797_at | 29.86 |
| FXYD2 | FXYD domain containing ion transport regulator 2 | 217091_at | 34.30 |
| TM4SF20 | transmembrane 4 L six family member 20 | 220639_at | 34.30 |
| CDH17 | cadherin 17, LI cadherin (liver-intestine) | 209847_at | 36.76 |
| MUC5AC | mucin 5AC, oligomeric mucus/gel-forming | 214303_x_at | 36.76 |
| ISG20 | interferon stimulated exonuclease gene 20 kDa | 204698_at | 39.40 |
| OAS2 | 2′-5′-oligoadenylate synthetase 2, 69/71 kDa | 204972_at | 39.40 |
| IFIH1 | interferon induced with helicase C domain 1 | 219209_at | 42.22 |
| IFI44 | interferon-induced protein 44 | 214453_s_at | 45.25 |
| OAS1 | 2′,5′-oligoadenylate synthetase 1, 40/46 kDa | 205552_s_at | 45.25 |
| ANK3 | ankyrin 3, node of Ranvier (ankyrin G) | 206385_s_at | 51.98 |
| ISG15 | ISG15 ubiquitin-like modifier | 205483_s_at | 55.72 |
| MX1 | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | 202086_at | 55.72 |
| CCL5 | chemokine (C-C motif) ligand 5 | 1405_i_at | 59.71 |
| IFITM1 | interferon induced transmembrane protein 1 (9–27) | 201601_x_at | 90.51 |
| IFI44L | interferon-induced protein 44-like | 204439_at | 103.97 |
| BST2 | bone marrow stromal cell antigen 2 | 201641_at | 119.43 |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | 203153_at | 256.00 |
| IFI27 | interferon, alpha-inducible protein 27 | 202411_at | 362.04 |
Pathway analysis.
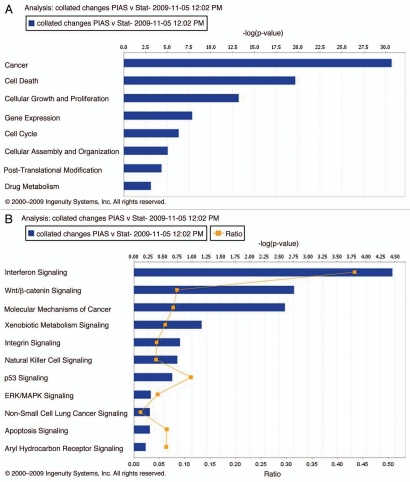
Genes with altered expression by PIAS3 overexpression were analyzed by IPA software. The analysis of the affected functional categories (highest significance) is presented in Figure 2A. The highest scoring category of the genes affected by PIAS3 overexpression was under the heading “cancer.” This comprised 368 out of 1,462 genes analyzed. The next leading function that came out of this analysis included 258 genes involved in cell death and cellular proliferation. The -logp value score of 10.5 indicates a high amount of significance associated with the regulation of these genes. The significant canonical pathways associated with the transcriptional changes associated with PIAS3 overexpression are interferon signaling, Wnt/β-catenin signaling, xenobiotic signaling, natural killer cell signaling, p53 signaling, apoptosis and arylhydrocarbon signaling, of which interferon, Wnt/β-catenin and p53 signaling showed high -log p value scores (Fig. 2B). A high score is indicative of the statistical strength of association of the regulated genes with a particular pathway or function.
Figure 2.
Functions affected by genes altered by PIAS3 overexpression (A). Bars represent -log (p value) for disproportionate representation of affected genes out of the total number of genes in the selected function/disease category. Canonical pathways affected by genes altered by PIAS3 (B). Bars represent -log (p value) for disproportionate representation of affected genes in the selected pathway; the yellow line represents the ratio of affected genes to the total number of genes in a pathway.
PIAS3 differential effect on gene expression is most likely related to its effects on other important transcription factors.
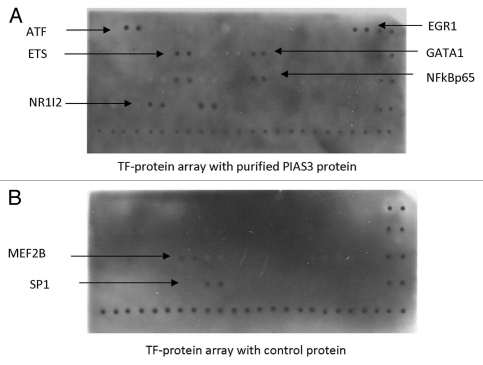
We sought to find other previously not described transcription factors that PIAS3 may regulate. To this effect we purified high amounts of recombinant PIAS3 protein using pGEX-4T-1 expression plasmid. To identify the specific binding partners of PIAS3 we used a Transcription Factor (TF) Protein array, which would identify potential binding partners for PIAS3. The membrane was spotted with different transcription factor proteins which are expressed from full length TF cDNAs. The interaction between PIAS3 protein and other transcription factor proteins was detected by exposing the membrane to a chemiluminescence imaging system. Among the TFs spotted on the membrane, six TFs showed binding with recombinant PIAS3 protein. These transcription factors are ATF1, ETS, EGR1, NR1I2, GATA1 and NFκBp65. Out of these transcription factors ETS, EGR1, NR1I2 and GATA1 are novel binding partners of PIAS3 whereas binding with ATF and NFκBp65 has previously been reported (Fig. 3).
Figure 3.
PIAS3 binding to various transcription factors using a transcription factor protein array. EGR1, ETS, GATA1, NR1I2 transcription factors are identified as novel binding partners for PIAS3. ATF and NFκBp65 have previously been described as binding to PIAS3 protein. (A) PIAS3 purified protein obtained by introduction of PIAS3 expression plasmid into E. coli followed by purification and (B) control protein obtained by introduction of an empty plasmid into E. coli followed by protein purification, were both incubated with the TranSignal TF Protein Array membrane for 2 h and detected by PIAS3 monoclonal antibody (Santa Cruz) and anti-mouse Ig HRP conjugate (Amersham). Images were acquired by exposing the membranes using Hyperfilm (TM) ECL.
Ligand-dependent binding of PIAS3 to novel transcription factor binding partners.
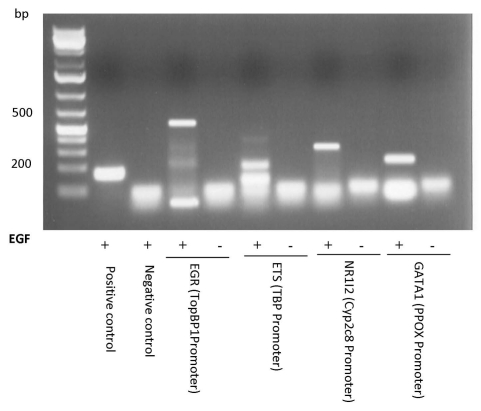
In order to determine if cancer cells show evidence of PIAS3 binding to these new putative transcription factors we performed ChIP studies. A549 cells were either unstimulated or stimulated with EGF and incubated with 1% formaldehyde to crosslink protein to DNA. The cells were lysed, nuclei were prepared and chromatin was sheared to approximately 1 kb. PIAS3 and its associated DNA were immunoprecipitated using an anti-PIAS3 antibody. Crosslinks were reversed and the success of each immunoprecipitation was examined by PCR analysis using primers specific to a known consensus binding site of transcription factors: EGR1 (TopBP1 Promoter), ETS (TBP Promoter), NR1I2 (CYP2C8 Promoter) and GATA1 (PPOX Promoter) which we had demonstrated binding to PIAS3 in the TF-protein array. PCR amplification revealed that PIAS3 binds to all four transcription factors and to the TF DNA binding sites. Of interest is that this binding only occurs upon exposure to EGF (Fig. 4).
Figure 4.
Demonstration of PIAS3 binding by ChIP assay to promoter regions of targets for the newly described binding partners for PIAS3: EGR1, ETS, NR1I2 and GATA1. ChIP was performed using anti-PIAS3 and normal mouse IgG (negative control) antibodies; cMyc promoter was amplified as a positive control (as a downstream target of STAT3/PIAS3 complex). ChIP assays were repeated twice with similar outcomes. PCR was performed using specific primers for the transcription factors promoter region. ChIP assay identifies PIAS3 binding to targets of EGR, ETS, NR1I2 and GATA1 transcription factors.
Demonstration of PIAS3 binding to promoters of EGR1, ETS and NR1I2 through a novel transcription factor promoter tiling array.
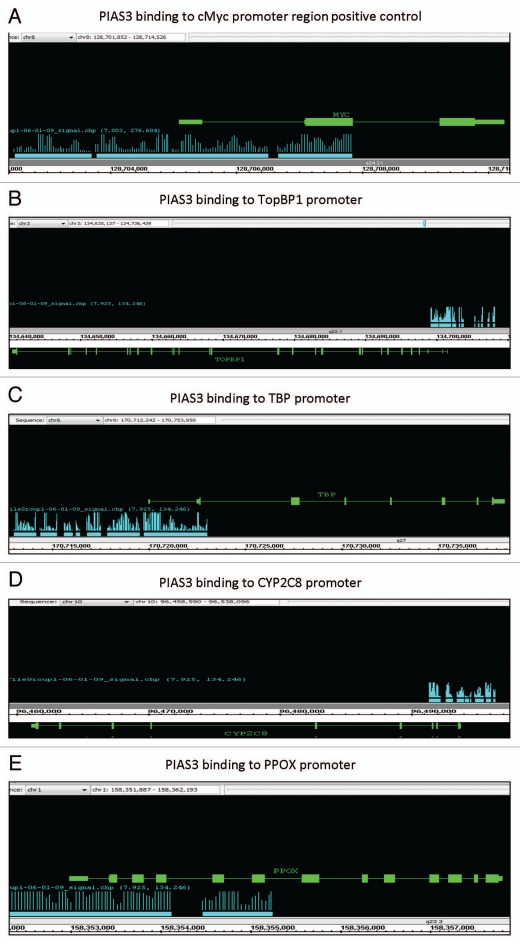
We performed ChIP-on-chip promoter tiling arrays using DNA obtained from the ChIP analysis described above, by using a PIAS3 specific monoclonal antibody. This experiment allowed investigation of interacting binding sites of PIAS3 on a genome-wide basis. Our goal of ChIP-on-chip is to locate ligand-induced PIAS3 DNA binding sites within the promoter region of genes. We discovered over 25 PIAS3 binding sites on each chromosome. All four novel transcription factor binding partners for PIAS3 (EGR1, ETS, NR1I2, GATA1) were evaluated by searching for their target gene binding sites. This included: EGR1 binding site at the TopBP1 Promoter; ETS binding site at the TBP Promoter; NR1I2 binding at the CYP2C8 Promoter; and GATA1 binding site at the PPOX Promoter. cMyc promoter site, which is a known binding site for STAT3 is also demonstrated as a control (Fig. 5).
Figure 5.
Binding of PIAS3 to the promoter region of target genes: Using a promoter tiling array we looked for areas of PIAS3 binding to various genes that are targets of the newly described binding partners of PIAS3. Blue bars indicate areas of PIAS3 binding along the genes of interest (green lines). (A) cMyc promoter (target of PIAS3/STAT3 comples, used as positive control); (B) TopBP1 promoter (target of EGR1); (C) TBP promoter (target of ETS); (D) CYP2C8 promoter (target of NR1I2); and (E) PPOX promoter (target of GATA1).
EGF stimulation results in binding of PIAS3 to EGR1-DNA complex.
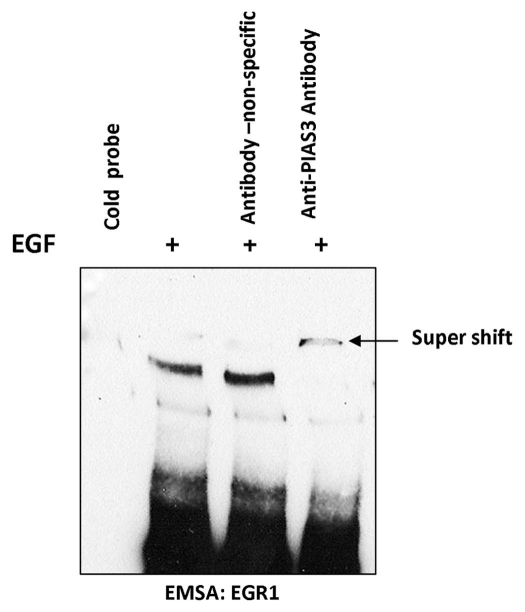
EMSA/super shift analysis was used to confirm the identity of the EGR1 DNA binding to PIAS3. Using A549 cells from which serum was withdrawn for 24 h, and either unstimulated or stimulated with EGF, nuclear extracts were prepared and EMSA was performed with the EGR1 transcription factor probe, which specifically binds EGR1 proteins with high affinity. As shown in Figure 6, EGF stimulation confirms binding of EGR1 to its consensus sequence. PIAS3 association with this complex is confirmed by supershifting with an anti-PIAS3 antibody.
Demonstration of functional and concentration-dependent effect of PIAS3 on EGR1 transcriptional activity.
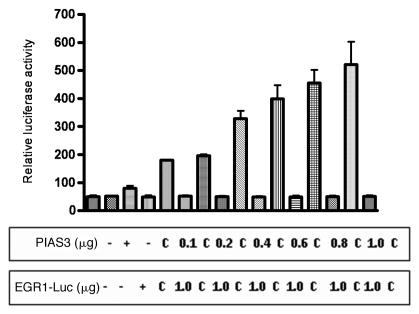
To analyze the functional regulatory affects of PIAS3 on EGR1 transcriptional activity we co-transfected the A549 cell line with vectors containing EGR1, PIAS3 and a vector containing the luciferase reporter gene under the transcriptional control of EGR1. In the absence of EGF, the luciferase activity is minimal. Co-transfection with PIAS3 expression construct results in a significant increase in luciferase expression and is PIAS3 concentration dependent (p < 0.0001) (Fig. 7). These data indicate that PIAS3 has functional effects on EGR1 transcriptional activity.
Figure 7.
PIAS3 has functional effects on EGR1 transcriptional activity. In order to elucidate if PIAS3, in addition to binding, has functional effects on EGR1, A549 cells were transfected with vectors containing PIAS3, EGR1 and a vector containing the luciferase reporter gene under the transcriptional control of EGR1 (EGR1-LUC). After 48 h post-transfection, cells were harvested and relative luciferase activity was measured. The columns denote relative luciferase activity from triplicate experiments. This figure shows that EGR1 transcriptional activity is PIAS3 dose-dependent, with increasing doses of PIAS3 resulting in increased EGR1 binding to its consensus sequence and luciferase activity. The columns with “c” indicate the controls, were instead of PIAS3 expression construct, an empty vector was used.
Network analysis.
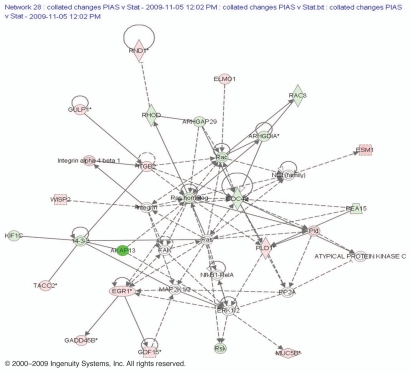
An EGR1 based network was created assembling the combined network from upregulated and downregulated genes after PIAS3 overexpression (Fig. 8). This network provides a hypothesis on how PIAS3 may regulate EGR1. The presence of transcriptional regulator EGR1 suggests cancer suppressor activity.1 Within the network associated with EGR1 we found several genes for which the highest score included gene products associated with GDF15 that belongs to the transforming growth factor b super-family. We also found WNT1-inducible-signaling pathway protein 2, a protein that in humans is encoded by the WISP2 gene. Proteins related to angiogenesis included the endothelial cell-specific molecule 1 (ESM1), a secreted protein which is mainly expressed in the endothelial cells in human lung and kidney tissues. This network also contained groups of interacting proteins that included GADD45A and NFκB, both transcription factors that mediate the transcription of proteins involved in cell survival and proliferation. PIAS3 overexpression results in downregulation of certain genes that are indicated in green color in this EGR1 associated network including the RhoGAP family proteins which are negative regulators of Rho family GTPases. This network also identified Rac, a transcription factor that binds to serum response elements found in the promoters of many growth factor-regulated genes. Thus the network associated with EGR1 related genes indicates that PIAS3 overexpression causes upregulation of the genes responsible for the apoptosis and cell maintenance while downregulated genes are responsible for cell progression and adhesion. This network also contained groups of interacting proteins that included the transcription factor that mediates the transcription of proteins involved in cell survival, proliferation and inflammatory responses.
Figure 8.
Network contains downregulated genes (green) and upregulated genes (red) identified around EGR1 gene. Top functions of the genes were involved in cell survival, proliferation, inflammatory responses and tumor suppression. Genes were identified as differentially expressed in our experiment and were integrated into the computationally generated networks on the basis of the evidence stored in the IPA knowledge memory of available scientific data indicating a relevance to this network. The node shapes denote enzymes, phosphatases, kinases, peptidases, G-protein coupled receptors, transmembrane receptors, cytokines, growth factors, ion channels, transporters, translation factors, nuclear receptors, transcription factors and others.
Discussion
PIAS3 belongs to a multigene family which was first identified as a transcriptional repressor of activated STAT3.7 This transcriptional repressive activity of STAT3 is thought to be the mechanism by which PIAS3 overexpression leads to growth inhibition in multiple lung cancer cell lines.16 However other functions for PIAS3 have been recently described. For example, PIAS3 interacts with p65 subunit of NFκB induced by TNFα or IL-121 and to MITF resulting in its decreased transcriptional activity.22 However neither MITF nor NFκB plays an important role in lung cancer pathogenesis. In order to determine if PIAS3 uniquely affects STAT3 targeted genes in lung cancer we performed either a small interference -mediated knockdown of STAT3 or an overexpression of PIAS3 and did a transcriptional profiling of the downstream modified genes. Surprisingly, PIAS3 modified a significantly greater number of genes as compared to STAT3 knockdown.
The mechanism by which PIAS3 alters significantly more genes than those of STAT3 mediated events is likely due to its promiscuous function. Indeed, we demonstrate four new binding partners for PIAS3: ETS, EGR1, NR1I2 and GATA1. All four binding partners play important roles in cancer development. This interaction was confirmed in our experiments by protein-protein binding, ChIP and a novel tiling microarray. The functional effects of this binding have been demonstrated by the capacity for PIAS3 to increase EGR1 transcriptional activity.
Transcriptional profiling indicated that a significant number of genes altered are related to angiogenesis. This is in line with the demonstration of PIAS3-ETS (v-ets avian erythroblastosis virus E26 oncogene) interaction. ETS serves as a key transcription factor for extracellular matrix remodeling in angiogenesis.23 Our finding that PIAS3 alters genes related to xenobiotic metabolism and CYP3A4 expression is in line with our demonstration of PIAS3 binding to NR1I2. Indeed NR1I2, is a nuclear receptor whose primary function is in the detoxification and clearance of foreign substances from the body and is a direct transcriptional regulator of the cytochrome P450 gene, CYP3A4.24 Their other binding partner for PIAS3 that we identified is GATA1 which is an important transcription factor for the differentiation of the erythroid and megakaryocytic cell lineages. Its relevance in solid tumors is less clear but it plays a role in acute megakaryocytic leukemias.25
The demonstration of PIAS3 interaction EGR1 and changes in apoptotic related genes is a novel finding and suggests a proapoptotic effect for PIAS3. Indeed transcriptional profiling shows upregulation of a number of proapoptotic genes. EGR1 is induced very early in the apoptotic process,26 where it mediates the activation of downstream regulatory genes such as p53.27 EGR1 has been found to be decreased or undetectable in human breast28 and non-small cell lung tumors29 as well as in an array of tumor cell lines.29–31 We further assessed the functional effect of binding of PIAS3 to EGR1 using a luciferase reporter assay. We found that PIAS3 increases the transcriptional promoter activity of the human EGR1 gene, resulting in upregulation of the EGR1 pathway.
A networking analysis was performed using EGR1 to gain insight into pathways that PIAS3 may modulate. Transcriptional regulation and protein-protein interactions among these identified PIAS3 targets form a network which is biologically relevant for processes related to cell cycle and tumor suppression. The central molecules of the EGR1-based network are Wnt1, GDF15, GADD45, NFκB, RhoGAP, CDC4 and Rac amongst others. They are important signaling molecules involved in multiple cellular processes such as cell cycle control, proliferation and differentiation. These results suggest a dynamic model in which PIAS3 and its target genes are integral in cancer-related biological pathways such as cell cycle regulation and control. Because the IPA software used in the study established potential gene interaction networks based solely on empirical knowledge extracted from existing published literature, it is possible that PIAS3 affects other cancer-related pathways as well. The Wnt/βcatenin signaling pathway emerged as a key canonical pathway altered by PIAS3 overexpression and the IPA network analysis again identified Wnt1 as a key protein in this network. Wnt-mediated signaling has been identified as a critical signaling pathway in non-small cell lung cancer.32
As several cancers have demonstrated decrease or loss of PIAS3 expression,13,14,16 restoration of PIAS3 may be a mechanism where one could achieve therapeutic cancer growth control. Our data convincingly shows that in addition to STAT3 mediated effects PIAS3 has a vast STAT3-independent effect by binding to a number of transcription factors with downstream alterations in apoptosis, angiogenesis and a number of signaling pathways.
Material and Methods
Cell lines.
Human pulmonary epithelial cell line A549 cells were purchased from American Type Culture Collection (Manassas, VA). Cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM)/HF12 supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, Hyclone, South Logan, UT), 50 unit/ml penicillin and 50 µg/ml streptomycin in a 5% CO2 humidified incubator at 37°C. Cell number and viability were assessed by Trypan Blue (Sigma) dye exclusion using a hemacytometer.
STAT3 siRNA transfection.
A549 cells were grown in 60-mm plates to 40% confluence before transfection. Media was aspirated from the cells, which were washed twice with sterile phosphate-buffered saline. Five mL of fresh medium containing 10% serum and without antibiotics was added to each plate. Twenty microliters of 10-µM STAT3 siRNA (sc-29493) (Cell Signaling Technology, Danvers, MA) were added to 300 µL of siRNA Transfection Medium (Santa Cruz Biotechnology Inc., Santa Cruz, CA), mixed gently, kept at room temperature for 20 min and added drop-wise to the plates with gentle rocking. After incubation for 24 h at 37°C, the transfection media was removed and the cells were transfected again, following the same protocol. After another 48 h, the cells were harvested and total RNA or protein was extracted for analysis.
Transient transfection of PIAS3.
A549 cells were seeded at 1 × 105 cells per well in 6-well plates. HD FuGENE (Roche Applied Science, Indianapolis, IN) was used as a transfection reagent to transfect the cells with either plasmid pCMV5 or pPIAS3-CMV5 in DMEM/HF12 media without serum and antibiotics. After 5 h incubation, media was replaced with DMEM/HF12 media containing fetal bovine serum (10%) and antibiotics. Following 48 h incubation, cells were stimulated with 50 ng/mL EGF for 15 min, cells were washed with cold PBS, lysed with passive lysis buffer (Promega, Madison, WI) and then centrifuged at 12,000x g for 4 min. The supernatant was collected and stored at −80°C.
Gene transcriptional analysis.
Subconfluent A549 cells were seeded in 75-cm2 flasks for 12 h in media supplemented with 10% FBS. Cells were transfected separately with pCMV5 vector encoding human PIAS3 expressing gene and siRNA against STAT3, respectively. At 48 h of transfection cells were stimulated with EGF or remained unstimulated, then total RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA quality was evaluated using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Double-stranded cDNA was synthesized using SuperScript II Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA). Phase Lock Gel, phenol/chloroform extraction and ethanol precipitation were employed to purify the resultant cDNA. cRNA was synthesized using the GeneChip IVT Labeling Kit (Affymetrix, Santa Clara, CA). In vitro transcription was carried out for 4 h at 37°C, using biotinylated ribonucleotides (Enzo Diagnostics, Farmingdale, NY) for labeling. Labeled cRNA was purified with an RNeasy kit (Qiagen) and fragmented in fragmentation buffer for 30 min at 94°C. Fragmented cRNA was hybridized to human U133A GeneChip microarrays (Affymetrix) in a rotating hybridization oven for 16 h at 45°C. Staining was performed on Affymetrix fluidics station utilizing streptavidin/phycoerythrin conjugate Molecular Probes, Invitrogen Life Science), followed by biotinylated antibody to streptavidin (Vector Laboratories, Burlingame, CA) and finally via a second streptavidin/phycoerythrin conjugate. Stained micro-arrays were scanned on a Hewlett Packard GeneArray Scanner and data was compiled with Affymetrix Microarray Suite 5.0 software. Each sample was repeated in three independent cultures, each of which was subjected to three independent hybridization reactions.
Gene expression analysis.
Intensity data from each of the triplicate micro-arrays was uploaded to GeneTraffic version 2.8 (Iobion, Informatics, La Jolla, CA). Raw data was normalized using Robust Multichip Analysis and p values were calculated using an F-class ratio with variance stabilization, which served as a measure of the difference between the normalized intensityvalue means of various sample groups relative to the variability of intensity values of each replicate within a given treatment group. Significantly regulated gene lists (p < 0.05, absolute fold change greater than 1.2) were then manually clustered based upon known cellular function.
Ingenuity pathway analysis.
The lists of significantly regulated genes obtained from Affymetrix microarrays were subjected to Ingenuity pathway analysis (IPA). The enriched datasets were used for performing “core analysis” function. The involvement of significantly regulated genes in well characterized pathways and functions were analyzed. The significance values associated with pathways and functions (-log p value) were calculated using Fisher's exact t-test. These values indicated the significance of association of differentially regulated genes with specific pathways/functions. The ratio values were calculated based on the number of molecules in a given pathway that meet cutoff criteria, divided by total number of molecules that make up that pathway. The involvement of EGR1 in different signaling networks was analyzed using “build pathway” function and using our own database of significantly regulated genes.
Immunoblotting and antibodies.
From culture cells were scraped, washed with 1x PBS in different time intervals and lysed in lysis buffer. Protein concentrations were estimated using Bradford reagent (Biorad). Equal amount of protein was loaded for immunoblotting. Following SDS-PAGE, resolved proteins were electro-blotted on PVDF membrane (Millipore, Bedford, MA). The membrane was blocked overnight in PBS containing 0.1% Tween-20 (PBST) and 3% BSA. The membrane was then probed with primary antibody in PBST for 2 h at room temperature or overnight at 4°C followed by three 10 min PBST washes at room temperature. Incubation with the secondary antibody was done for 1 h, then three 10-min PBST washes were given prior to chemiluminiscence detection using ECL substrate (Amersham, GE Healthcare Life Sciences, Piscataway, NJ). All antibodies for protein gel blots were obtained from Santa Cruz Biotechnology, Inc.
Production of recombinant protein.
The gene encoding human PIAS3 was subcloned into NotI restriction enzyme site of pGEX-4T-1 expression plasmid. The recombinant clone was verified by DNA sequencing. Recombinant protein production was achieved by introduction of the expression plasmid into Escherichia coli strain BL21(DE3) (Invitrogen Corp., Carlsbad, CA) by transformation. Recombinant E. coli BL21Star (DE3) strain, carrying the plasmids pGEX4T-1-GST or pGEX4T-1-PIAS3-GST were grown in LB medium to an OD600 of 0.6–0.8 and induced with 1 mM IPTG for up to 5 h. Aliquots were taken at 0, 1, 2 and 5 h of incubation, harvested by centrifugation and resuspended in lysis buffer containing 50 mM NaH2PO4 pH 8.0, 300 mM NaCl, frozen at −80°C and treated with 1 mg/ml lysozyme. The cells, after incubation at 4°C for 2 h, were sonicated and cleared by centrifugation. The supernatant was filtered through 0.2-µm hydrophilic sterile filters (Fisher Labosi, Elancourt, France). Total protein concentration was measured by using the Bradford method with BSA as the calibration standard.
Transcription factor-protein array.
The TranSignal Transcription Factor Protein array version IV membrane (Panomics, Inc., Redwood City, CA) was blocked with 1x blocking buffer and incubated for 2 h at room temperature on a shaker. The bacterial extract, containing overexpressed protein to a final concentration of 60 µg/ml was diluted in 4 ml of 1x blocking buffer I. The membrane was incubated with the diluted bacterial extract at room temperature for 2 h with gentle shaking. The membrane was washed twice with 1x Wash Buffer for 5 min each wash at room temperature. The membrane was incubated with 1x blocking buffer II containing primary antibody 1:500 concentration for 2 h at room temperature. The membrane was washed twice with 1x Wash Buffer for 5 min each wash. Then the membrane was incubated with the secondary antibody diluted 1:10,000 in 1x blocking buffer II for 1 h at room temperature. The membrane was washed with 1x Wash Buffer for 5 min each wash at room temperature. The detection solution was prepared combining 250 µl of each detection buffer A and B and placed protein-side-up on the membrane and incubated for 5 min at room temperature. The membrane was exposed to a chemiluminescence's imaging system.
Chromatin immunoprecipitation (ChIP).
The ChIP assay was performed using the Simple ChIP Enzymatic Chromatin IP kit (Cell Signaling Technology, Danvers, MA) according to the manufacturer's instructions. Briefly, 2 × 106 cells were treated with 1% formaldehyde for 10 min at 37°C. The cells were harvested, suspended with SDS lysis buffer [1% SDS, 10 mM EDTA and 50 mM Tris/HCl (pH 8.3)] and incubated on ice for 10 min. Lysates were sonicated and debris was removed from the samples by centrifugation for 10 min at 10,000 g. An aliquot of each chromatin solution (40 µl) was set aside and designated as the input fraction. Supernatants were diluted 10-fold in immunoprecipitation buffer and precleared with Protein A agarose beads. The precleared chromatin solutions were incubated with the relevant antibodies to PIAS3 and positive and negative antibody controls separately for 16 h at 4°C. The immune complexes were then collected with the addition of Sepharose A/G agarose beads, followed by several washes with appropriate buffers, according to the manufacturer's instructions. Each sample was eluted with elution buffer by at 65°C for 2 h. Chromatin-associated proteins were digested with proteinase K (10 mg/ml) and the immunoprecipitated DNA was recovered by phenol/chloroform extraction and ethanol precipitation and analyzed by PCR. The primers used for ChIP were as follows: EGR1 sense 5′-CAC GTA CTC CTC TGT T-3′ and antisense 5′-AGA CAC TGT ACA AGG-3′, product size was 500 bp designed from TOPBP1 promoter binding region; ETS sense 5′-GCG GTG CCG GAA GTA GTC-3′ and antisense 5′-GGC AGC AGC GTC TAT CTC C-3′ product size was 100 bp designed from TBP promoter binding region; NR1I2 sense 5′-tgg ccc ttt cct taa tag tgc t-3′ and antisense 5′-ACC GGG ATT TGA GCA GAG A-3′ product size was 298 bp, designed from CYP2C8 promoter binding region; and GATA1 sense 5′-GGA GTA GCG GAT TTG AAG CA-3′ antisense 5′-TCA CCC ACA ATA GGT AGG GAT-3′, product size was 214 bp designed from PPOX promoter binding region.
Binding microarray analysis.
Human Promoter 1.0R Array, contain more than four million probes corresponding to 25,500 promoter regions. The 25-mer probes (with inter-probe gaps of 10 bp) are tiled over regions spanning from 7.5 kb upstream to 2.5 kb downstream of each transcription start site. The assay starts with ChIP using procedures optimized by Genpathway to give maximum sensitivity and minimal background binding. The assay also includes initial qualification of the factor-specific antibody of interest (or use of a qualified antibody) and validation of the chromatin prior to the primary assay. ChIP DNA preparations are obtained using the specific antibody and control ChIP preparations consisting of either control antibodyimmunoprecipitated DNA or Input (unprecipitated) DNA are generated. The DNA preps, obtained from the above ChIP experiment was further amplified by whole genome amplification kit (WGA) are labeled and hybridized to the arrays at Case Comprehensive Cancer Center Gene Expression Core facility. Raw data from the scans are analyzed using Affymetrix® Tiling Analysis Software (TAS) and the results are viewed both in Affymetrix' Integrated Genome Browser (IGB) and compiled in tables with additional useful information using Genpathway's proprietary ChIP Analysis Software (ChAS).
Electrophoretic mobility shift assay (EMSA) and supershift assay.
Nuclear lysates were prepared using the Panomics kit. Five to ten micrograms of nuclear protein was incubated at 15°C for 30 min with transcription factor probe, EGR1, which specifically binds EGR proteins with high affinity (EMSA “Gel Shift” Kit, Panomics, Inc., Redwood City, CA). Samples were then run on a 7.5% precast acrylamide gel (BioRad, Invitrogen, Carlsbad, CA) and transferred to a nylon membrane. Bound oligos were immobilized by baking the membrane at 85°C or by cross-linking the membrane in a UV Crosslinker for 3 min followed by blocking and staining of the membrane using a Streptavidin-HRP conjugate. Substrate solutions included in the Panomics kit were used for detection and the membrane was exposed to Hyperfilm. Supershift assays were performed using the same procedure, but included the addition of 2 µg anti-PIAS3 antibody, during initial incubation with EGR1 nuclear extracts.
Transient transfection and luciferase assay.
A549 cells were seeded at 1 × 105 per well in 6-well plates. The HD FuGENE (Roche) was used as a transfection reagent to cotransfect the cells with luciferase reporter construct pEGR1-Luc or pCMV5-Luc alone as a nonspecific control and with pCMV5 PIAS3 expression construct or pCMV5 alone as a control. The cells were incubated in DMEM/HF12 medium for 48 h, treated or not with 20 ng/mL EGF for 15 min; cells were washed with cold PBS, lysed with passive lyses buffer (Promega) and then centrifuged at 12,000x g for 4 min. The supernatant was collected and stored at −80°C until assessment of luciferase activity. Luminescence was read in a Berthold luminometer (Lumat LB9501) after briefly mixing the supernatant (20 µL) with 100 µL of firefly luciferase assay substrate solution. The activity of luciferase was normalized to protein concentrations in lysate. Transfections were repeated at least three times and the relative changes are presented as mean ± SE.
Figure 6.
EGF induces PIAS3-EGR1 complex binding to EGR1 consensus sequence. After EGF stimulation, nuclear extracts were made from A549 cells. The extracts were mixed with the EGR1 DNA binding consensus sequence and EMSA performed. Supershift using an anti-PIA3 antibody was seen indicating PIAS3 is part of the complex. With a non-specific antibody no supershift is seen.
Acknowledgments
Supported by K23 CA109348 from the National Institutes of Health and by a support grant from the University Hospitals Ireland Cancer Center.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statisticsm 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Petri ET, Halmos B, Boggon TJ. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J Clin Oncol. 2008;26:1742–1751. doi: 10.1200/JCO.2007.12.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 4.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 5.Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22:4150–4165. doi: 10.1038/sj.onc.1206479. [DOI] [PubMed] [Google Scholar]
- 6.Dowlati A, Nethery D, Kern JA. Combined inhibition of epidermal growth factor receptor and JAK/STAT pathways results in greater growth inhibition in vitro than single agent therapy. Mol Cancer Ther. 2004;3:459–463. [PubMed] [Google Scholar]
- 7.Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 8.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 9.Ohshima T, Shimotohno K. Transforming growth factor-beta-mediated signaling via the p38 MAP kinase pathway activates Smad-dependent transcription through SUMO-1 modification of Smad4. J Biol Chem. 2003;278:50833–50842. doi: 10.1074/jbc.M307533200. [DOI] [PubMed] [Google Scholar]
- 10.Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, et al. Negative regulation of NFkappaB signaling by PIAS1. Mol Cell Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross M, Yang R, Top I, Gasper C, Shuai K. PIASy-mediated repression of the androgen receptor is independent of sumoylation. Oncogene. 2004;23:3059–3066. doi: 10.1038/sj.onc.1207443. [DOI] [PubMed] [Google Scholar]
- 12.Long J, Wang G, Matsuura I, He D, Liu F. Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3) Proc Natl Acad Sci USA. 2004;101:99–104. doi: 10.1073/pnas.0307598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brantley EC, Nabors LB, Gillespie GY, Choi YH, Palmer CA, Harrison K, et al. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin Cancer Res. 2008;14:4694–4704. doi: 10.1158/1078-0432.CCR-08-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kluge A, Dabir S, Vlassenbroeck I, Eisenberg R, Dowlati A. Protein inhibitor of activated STAT3 expression in lung cancer. Mol Oncol. 2011;5:256–264. doi: 10.1016/j.molonc.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabir S, Kluge A, Dowlati A. The association and nuclear translocation of the PIAS3-STAT3 complex is ligand and time dependent. Mol Cancer Res. 2009;7:1854–1860. doi: 10.1158/1541-7786.MCR-09-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kluge A, Dabir S, Kern J, Nethery D, Halmos B, Ma P, et al. Cooperative interaction between protein inhibitor of activated signal transducer and activator of transcription-3 with epidermal growth factor receptor blockade in lung cancer. Int J Cancer. 2009;125:1728–1734. doi: 10.1002/ijc.24553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell MP, Fiori JL, Kershner EK, Frank BP, Indig FE, Taub DD, et al. Heparan sulfate proteoglycan modulation of Wnt5A signal transduction in metastatic melanoma cells. J Biol Chem. 2009;284:28704–28712. doi: 10.1074/jbc.M109.028498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei W, Chua MS, Grepper S, So S. Small molecule antagonists of Tcf4/beta-catenin complex inhibit the growth of HCC cells in vitro and in vivo. Int J Cancer. 2010;126:2426–2436. doi: 10.1002/ijc.24810. [DOI] [PubMed] [Google Scholar]
- 19.Willert J, Epping M, Pollack JR, Brown PO, Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark CJ, Sage EH. A prototypic matricellular protein in the tumor microenvironment—where there's SPARC, there's fire. J Cell Biochem. 2008;104:721–732. doi: 10.1002/jcb.21688. [DOI] [PubMed] [Google Scholar]
- 21.Jang HD, Yoon K, Shin YJ, Kim J, Lee SY. PIAS3 suppresses NFkappaB-mediated transcription by interacting with the p65/RelA subunit. J Biol Chem. 2004;279:24873–24880. doi: 10.1074/jbc.M313018200. [DOI] [PubMed] [Google Scholar]
- 22.Levy C, Nechushtan H, Razin E. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J Biol Chem. 2002;277:1962–1966. doi: 10.1074/jbc.M109236200. [DOI] [PubMed] [Google Scholar]
- 23.Fujimoto J, Aoki I, Toyoki H, Khatun S, Tamaya T. Clinical implications of expression of ETS-1 related to angiogenesis in uterine cervical cancers. Ann Oncol. 2002;13:1598–1604. doi: 10.1093/annonc/mdf248. [DOI] [PubMed] [Google Scholar]
- 24.Watkins RE, Noble SM, Redinbo MR. Structural insights into the promiscuity and function of the human pregnane X receptor. Curr Opin Drug Discov Devel. 2002;5:150–158. [PubMed] [Google Scholar]
- 25.Rainis L, Bercovich D, Strehl S, Teigler-Schlegel A, Stark B, Trka J, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood. 2003;102:981–986. doi: 10.1182/blood-2002-11-3599. [DOI] [PubMed] [Google Scholar]
- 26.Muthukkumar S, Nair P, Sells SF, Maddiwar NG, Jacob RJ, Rangnekar VM. Role of EGR-1 in thapsigargin-inducible apoptosis in the melanoma cell line A375-C6. Mol Cell Biol. 1995;15:6262–6272. doi: 10.1128/mcb.15.11.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair P, Muthukkumar S, Sells SF, Han SS, Sukhatme VP, Rangnekar VM. Early growth response-1-dependent apoptosis is mediated by p53. J Biol Chem. 1997;272:20131–20138. doi: 10.1074/jbc.272.32.20131. [DOI] [PubMed] [Google Scholar]
- 28.Huang RP, Fan Y, de Belle I, Niemeyer C, Gottardis MM, Mercola D, et al. Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int J Cancer. 1997;72:102–109. doi: 10.1002/(sici)1097-0215(19970703)72:1<102::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 29.Levin WJ, Press MF, Gaynor RB, Sukhatme VP, Boone TC, Reissmann PT, et al. Expression patterns of immediate early transcription factors in human non-small cell lung cancer. The Lung Cancer Study Group. Oncogene. 1995;11:1261–1269. [PubMed] [Google Scholar]
- 30.Calogero A, Cuomo L, D'Onofrio M, de Grazia U, Spinsanti P, Mercola D, et al. Expression of Egr-1 correlates with the transformed phenotype and the type of viral latency in EBV genome positive lymphoid cell lines. Oncogene. 1996;13:2105–2112. [PubMed] [Google Scholar]
- 31.Liu C, Yao J, Mercola D, Adamson E. The transcription factor EGR-1 directly transactivates the fibronectin gene and enhances attachment of human glioblastoma cell line U251. J Biol Chem. 2000;275:20315–20323. doi: 10.1074/jbc.M909046199. [DOI] [PubMed] [Google Scholar]
- 32.You L, He B, Xu Z, Uematsu K, Mazieres J, Mikami I, et al. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004;23:6170–6174. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]