Abstract
High affinity capture agents recognizing biomolecular targets are essential to the performance of many proteomic detection methods. Herein, we report the application of a label-free silicon photonic biomolecular analysis platform for simultaneously determining kinetic association and dissociation constants for two representative protein capture agents: a thrombin-binding DNA aptamer and an anti-thrombin monoclonal antibody. The scalability and inherent multiplexing capability of the technology make it an attractive platform for simultaneously evaluating the binding characteristics of multiple capture agents recognizing the same target antigen, and thus a tool complementary to emerging high-throughput capture agent generation strategies.
Common to most quantitative protein measurement approaches is a requirement for analyte-specific capture agents that recognize, and in many cases localize, the target antigen allowing specific detection via one of many biomolecular transduction technologies.1 Regardless of the eventual detection modality, the physical nature of the interaction between capture agent and target protein often represents the fundamental limitation in assay sensitivity. With the goal of quantitating an ever increasing breadth of the proteome, the development of new and improved capture agents has become an active area of research.2, 3
Monoclonal antibodies are the most commonly used capture agents for protein detection. However, producing monoclonal antibodies is expensive and time- and labor-intensive. 4 Furthermore, the resulting antibodies are often unstable to conditions that compromise protein secondary and tertiary structure. To avoid limitations associated with antibodies, a range of alternative protein capture agent strategies have been proposed, including nucleic acid aptamers,5, 6 multivalent peptide scaffolds,7–9 peptoids,10 and molecularly imprinted polymers,11 amongst others. Many of these alternative strategies improve upon the throughput of agent generation by replacing animal immunization and hybridoma generation with high-throughput methodologies such as ligand evolution (e.g. SELEX) and solid-phase library synthesis. Similarly to combinatorial drug discovery, these methods should be capable of generating libraries of “lead” capture agents against single analytes and the binding characteristics of each member will need to be independently determined. For purposes of assay consistency and antigen conservation, it would be ideal if capture agent evaluation could be carried out in parallel, establishing a need for multiplexable capture agent screening methods.
The most common quantitative metric used to describe the interaction between a capture agent and its target antigen is the equilibrium dissociation constant, KD. Classically, KD is determined via radioimmunoassay,12 but more common now are kinetic methods that allow measurement of association and dissociation rate constants, ka and kd, from which KD is computed (KD=kd/ka). Real-time interaction analysis tools such as surface plasmon resonance (SPR)13 have been successfully applied to binding affinity determination,14 but a need still remains for highly multiplexable capture agent evaluation technologies.
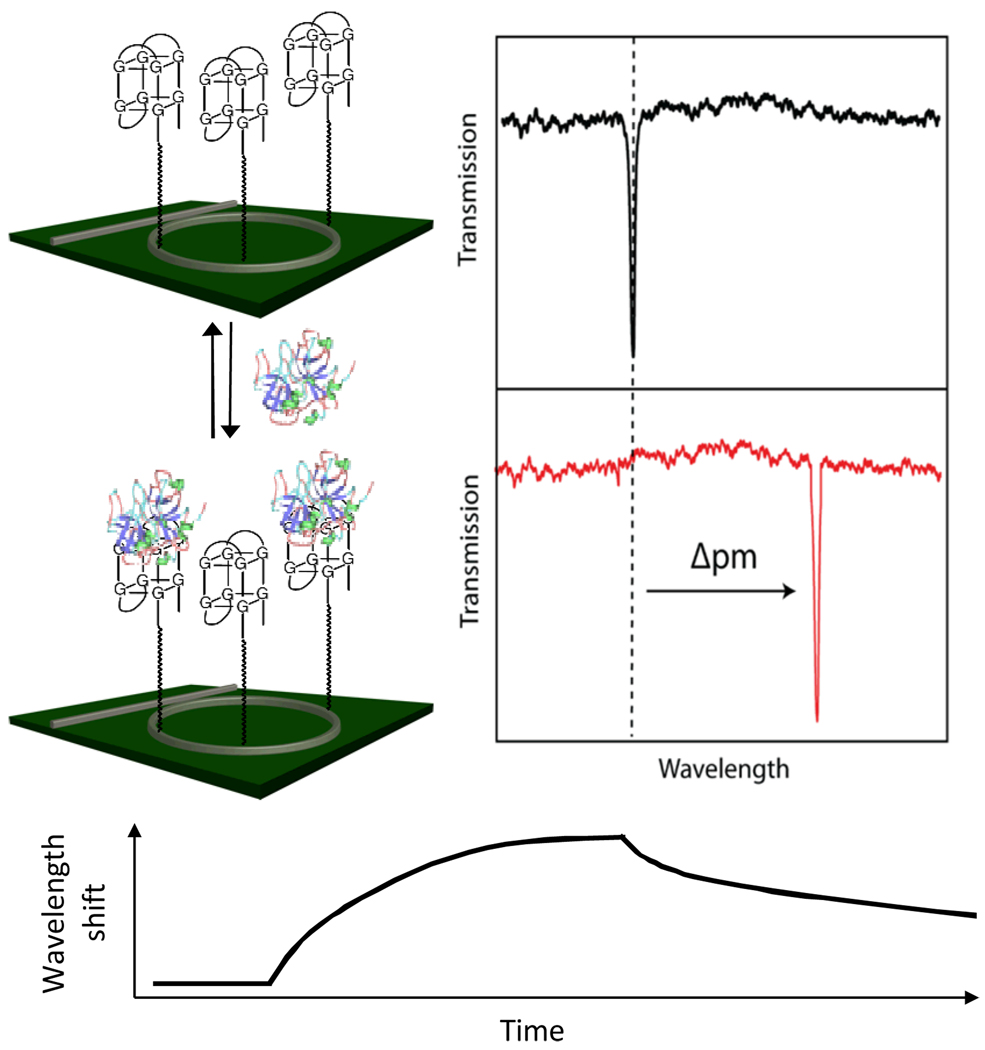
Our group has recently developed a highly multiplexable, real-time biomolecular analysis technology based upon arrays of silicon photonic microring resonators, and have utilized this platform for the quantitative detection of proteins and nucleic acids in single15–17 and multiplexed assays.18, 19 Through optical interference, wavelengths of light that satisfy a resonance condition are strongly localized in the microring and the resulting optical mode samples the proximal refractive index environment. Binding of antigens to capture agents covalently attached to the microring changes the effective refractive index of the optical mode, eliciting a corresponding shift in the resonant wavelengths, as shown in Figure 1. These shifts can be monitored in real time, allowing visualization of biomolecular interaction kinetics.
Fig. 1.
Schematic diagram illustrating the principle of silicon photonic microring resonators presenting aptamer capture probes. Upon target binding, the resonance peak shifts to longer wavelength, a process that can be followed in real-time to enable measurement of binding kinetics.
In order to demonstrate the applicability and versatility of this platform for multiplexed capture agent screening, we wanted to evaluate two different classes of capture agents that both recognize the same target antigen. To accomplish this, we chose to interrogate a monoclonal mouse anti-human thrombin antibody and a 15-nucleotide long aptamer that also recognized human thrombin.20 Aptamers are single-stranded nucleic acids that can bind to their targets with high affinity and specificity,21 and thus have found utility in a range of biomolecular assays.22, 23 Both the antibody and aptamer were covalently immobilized onto different regions of the sensor array and their respective interactions with identical thrombin solutions simultaneously interrogated.
The sensor substrates and read out instrumentation have been described previously.15, 16 A clean sensor chip natively coated with silicon dioxide was functionalized by exposure to 3-N-((6-(N'-isopropylidenehydrazino))-nicotinamide)propyl triethyoxysilane (HyNic silane) to instill an aryl hydrazine moiety capable of reacting with capture agents presenting 4-formylbenzamide (4FB) groups via hydrazone bond formation. Separately, the amine-modified aptamer (5′-NH2 – (CH2)12–TTTTTGGTTGGTGTGGTTGG-3′), control DNA sequence (5′-NH2–(CH2)12–GGTAGTACAGCATATTCGAAAGTGTATAAGATT-3′), and anti-thrombin antibody were each reacted with succinimidyl 4-formylbenzoate (S-4FB) to attach the 4FB reactive group. The sensor chip was then loaded into a custom-made four channel microfluidic device and each of the three 4FB-modified biomolecules, along with bovine serum albumin (BSA), were flowed across different regions of the microring array, allowing simultaneous sensor functionalization. Microrings coated with the control DNA sequence and BSA served as on-chip controls to correct for non-specific adsorption and bulk refractive index changes.
After sensor chip functionalization, the aptamer and antibody were each covalently attached to three microring array elements. Various concentrations of human α-thrombin, solutions ranging from 500 pM to 10 nM, were then flowed over the entire sensor array and the shifts in resonance wavelength were monitored as a function of time as thrombin associated and dissociated from each of the capture agent-modified sensor surfaces. Figure 2 shows real-time resonance wavelength shifts from representative microrings functionalized with the anti-thrombin antibody (a) and thrombin-binding aptamer (b).
Fig. 2.
Time-resolved detection of human thrombin using microrings modified with (a) an antibody and (b) an aptamer. Thrombin solutions were injected t = 5 min and the chamber returned to buffer at t = 17 min. Each measurement was made with three independent microrings; however, data from only a single ring is presented for clarity.
Both capture agents display expected concentration-dependent responses where larger shifts in resonance wavelength are elicited when thrombin solutions having higher concentrations are flowed across the sensor array. Upon switching the solution to buffer (no thrombin), the bound antigen dissociates and the resonance wavelength returns towards the original baseline value. To ensure complete removal of bound thrombin, the sensor surface is chemically regenerated in between exposure to solutions of different concentration. Antibody-antigen complexes are dissociated using glycine buffer (pH = 2.2), while aptamer surfaces are regenerated using proteinase K, which enzymatically consumes any bound thrombin, leaving the covalently bound DNA aptamer available for subsequent binding experiments.
In order to verify the selectivity of capture agents, solutions containing elastase, a serine protease with a molecular weight and an isoelectric point similar to that of thrombin, were flowed across the sensor surface (see Fig. S2). No specific binding response was detected over a relevant concentration range on either the antibody or aptamer-coated microrings. However, at a higher concentration of elastase (50 nM) a negative shift in the resonance wavelength was observed for microrings functionalized with the antibody, an observation that we attribute to the enzymatic degradation of the protein-based capture agent due to the proteolytic activity of elastase. Notably, the negative shift was not observed for the DNA-based aptamer capture agent.
The real-time shifts in resonance wavelength accompanying target binding and unbinding were used to determine the capture agent evaluative metrics, namely the association (ka) and the dissociation rate constants (kd) by assuming a 1:1 binding model.24–26 Both ka and kd were determined by fitting the association component of the real-time binding trace to
where θ is the fractional total occupancy and
Therefore, values of ka and kd were determined by plotting γ as a function of thrombin concentration as shown in Figure 3. For both the anti-thrombin antibody and thrombin-binding aptamer, the plots of θ versus concentration nicely followed a linear relation and ka and kd were extracted as the slope and intercept of the linear fit, respectively. However, when kd is small, the y-intercept values determined by the above approach can be inaccurate.24 Therefore, the kd values were also independently determined by directly fitting the dissociation curves to the simple exponential decay function
Both methods of kd determination give similar values, which are compiled in Table 1 along with values for ka.
Fig. 3.
Plot of γ values as a function of thrombin concentration obtained from the association curves of microrings modified with (a) and antibody and (b) an aptamer. Error bars represent ±1 standard deviation of γ values determined from three independent microrings simultaneously measuring at each concentration.
Table. 1.
Kinetic constants of antibody and aptamer capture agents
| Antibody | Aptamer | |
|---|---|---|
| ka (M−1s−1) a | 4.6 (± 0.1) × 105 | 5.8 (± 0.4) × 105 |
| kd (s−1) a | 1.5 (± 0.01) × 10−3 | 4.7 (± 0.2) × 10−3 |
| KD (M) a | 3.3 (± 0.1) × 10−9 | 8.2 (± 0.6) × 10−9 |
| kd (s−1) b | 1.4 (± 0.3) × 10−3 | 3.5 (± 1.3) × 10−3 |
Rate constants determined from the plot of γ vs thrombin concentration
Rate constants determined from the dissociation curve fittings
Consistent with visual inspection of the real-time binding data in Figure 2, the thrombin-binding aptamer has faster rates of both antigen association and dissociation, when compared to the antibody. Using determined values for ka and kd, KD was calculated (KD=kd/ka) to be in good agreement with published values for both the antibody27 and aptamer.28 When comparing KD values we determine that the antibody is a higher affinity (smaller KD) capture agent, which means that it will give lower limits of detection for most sensor applications.
Since one application of the microring resonator technology is biomolecular detection, we simultaneously investigated the utility of both the antibody and aptamer capture agents for the quantitation of thrombin. Previously we have utilized an initial slope-based quantitation method15, 18, 19 whereby the initial slope of sensor response is plotted versus concentration to give a linear calibration curve. This approach has several advantages over equilibrium methods in that it offers a shorter time to result and broader linear dynamic range. Applied to the detection of thrombin, and using the standard solution data from Figure 2, microrings functionalized with the antibody and aptamer capture agents were both successful in determining the concentration of a blinded thrombin solution, yielding values of 2.5 ± 0.6 nM and 3.8 ± 1.6 nM, respectively, for a solution prepared to be 3.0 nM (see the supporting information for more details). Interestingly, the faster association rate of the aptamer capture agent gives it a steeper calibration plot, i.e. the initial slope is greater for the aptamer, compared to the antibody, for the same concentration. This is significant because it means that the aptamer capture agent actually gives a lower limit of detection, despite having an affinity that is “worse” than its antibody counterpart.
In this paper, we have demonstrated the ability to simultaneously evaluate multiple capture agents against the same target antigen using arrays of silicon photonic microring resonators. In parallel we determined the evaluative metrics of antibody and aptamer capture agents that recognize human thrombin and suggest that, despite having a “worse” affinity, the aptamer agent can be used to quantitate the target antigen with a lower limit of detection. Importantly, we envision that the ability to monitor in parallel the binding kinetics of multiple biomolecular interactions on a scalable and cost-effective platform will be of great utility in evaluating protein binding agents for a range of biomedical applications.
Supplementary Material
Notes and references
- 1.Kodadek T. Chem. Biol. 2001;8:105. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 2.Elia G, Silacci M, Scheurer S, Scheuermann J, Neri D. Trends Biotech. 2002;20:S19. doi: 10.1016/s1471-1931(02)00201-x. [DOI] [PubMed] [Google Scholar]
- 3.Kodadek T, Reddy MM, Olivos HJ, Bachhawat-Sikder K, Alluri PG. Acc. Chem. Res. 2004;37:711. doi: 10.1021/ar030145l. [DOI] [PubMed] [Google Scholar]
- 4.Chiarella P, Fazio VM. Biotechnology Letters. 2008;30:1303. doi: 10.1007/s10529-008-9706-5. [DOI] [PubMed] [Google Scholar]
- 5.Jayasena SD. Clinical Chemistry. 1999;45:1628. [PubMed] [Google Scholar]
- 6.Sefah K, Phillips JA, Xiong X, Meng L, Van Simaeys D, Chen H, Martin J, Tan W. Analyst. 2009;134:1765. doi: 10.1039/b905609m. [DOI] [PubMed] [Google Scholar]
- 7.Agnew HD, Rohde RD, Millward SW, Nag A, Yeon W-S, Hein J, Pitram SM, A.A T, Burns VM, Krom RJ, Fokin VV, Sharpless KB, Heath JR. Angew. Chem. Intl. Ed. 2009;48:4944. doi: 10.1002/anie.200900488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachhawat-Sikder K, Kodadek T. J. Am. Chem. Soc. 2003;125:9550. doi: 10.1021/ja034912n. [DOI] [PubMed] [Google Scholar]
- 9.Williams BAR, Diehnelt CW, Belcher P, Greving M, Woodbury NW, Johnston SA, Chaput JC. J. Am. Chem. Soc. 2009;131:17233. doi: 10.1021/ja9051735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ, Kodadek T. J. Am. Chem. Soc. 2003;125:13995. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 11.Bossi A, Piletsky SA, Piletska EV, Righetti PG, Turner APF. Analytical Chemistry. 2001;73:5281. doi: 10.1021/ac0006526. [DOI] [PubMed] [Google Scholar]
- 12.Berzofsky JA. Clinical Chemistry. 1978;24:419. [PubMed] [Google Scholar]
- 13.Homola J. Chem. Rev. 2008;108:462. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 14.Campbell CT, Kim G. Biomaterials. 2007;28:2380. doi: 10.1016/j.biomaterials.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 15.Washburn AL, Gunn LC, Bailey RC. Anal. Chem. 2009;81:9499. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iqbal M, Gleeson MA, Spaugh B, Tybor F, Gunn WG, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. IEEE J. Sel. Top. Quantum Electron. 2010;16:654. [Google Scholar]
- 17.Luchansky MS, Bailey RC. Anal. Chem. 2010;82:1975. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Anal. Chem. 2010;82:69. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qavi AJ, Bailey RC. Angew Chem Int Ed. 2010;49:1. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 21.Klussmann S. The aptamer handbook: functional oligonucleotides and their applications. 2006 [Google Scholar]
- 22.Cho EJ, Lee JW, Ellington AD. Annual Review of Analytical Chemistry. 2009;2:241. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 23.Liu JW, Cao ZH, Lu Y. Chemical Reviews. 2009;109:1948. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson R, Michaelsson A, Mattsson L. Journal of Immunological Methods. 1991;145:229. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 25.Wegner GJ, Wark AW, Lee HJ, Codner E, Saeki T, Fang SP, Corn RM. Anal. Chem. 2004;76:5677. doi: 10.1021/ac0494275. [DOI] [PubMed] [Google Scholar]
- 26.Morton TA, Myszka DG, Chaiken IM. Analytical Biochemistry. 1995;227:176. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 27.Haematologic Technologies, Inc. http://www.haemtech.com/. [Google Scholar]
- 28.Baldrich E, Restrepo A, O'Sullivan CK. Analytical Chemistry. 2004;76:7053. doi: 10.1021/ac049258o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.