Abstract
Proper execution of transcriptional programs is a key requirement of gene expression regulation, demanding accurate control of timing and amplitude. How precisely the transcription machinery fulfills this task is not known. Using an in situ hybridization approach that detects single mRNA molecules, we measured mRNA abundance and transcriptional activity within single Saccharomyces cerevisiae cells. We found that expression levels for particular genes are higher than initially reported and can vary substantially among cells. However, variability for most constitutively expressed genes is unexpectedly small. Combining single-transcript measurements with computational modeling indicates that low expression variation is achieved by transcribing genes using single transcription-initiation events that are clearly separated in time, rather than by transcriptional bursts. In contrast, PDR5, a gene regulated by the transcription coactivator complex SAGA, is expressed using transcription bursts, resulting in larger variation. These data directly demonstrate the existence of multiple expression modes used to modulate the transcriptome.
Regulation of gene expression occurs on multiple levels, beginning with promoter accessibility1. As a key step in gene expression, transcription is probably one of the most complex and tightly regulated processes within the cell, requiring a series of events to occur in a coordinated fashion to initiate mRNA synthesis2. Chromatin rearrangement makes promoters accessible for sequence-specific transcription factors that mediate the assembly of coactivators, additional regulatory factors, the basal transcription machinery and finally RNA polymerase II resulting in initiation2–5. Once promoter complexes are assembled, the interaction of transcription factors with DNA keeps the gene active, probably by recruiting polymerases to a preassembled transcription complex. The stability of promoter complexes and their assembly efficiency will therefore influence the amplitude of a transcription response2–7. Different trans-acting factors and promoter elements including the TATA box have been shown to be important to stabilize promoter complexes and allow efficient transcription, for example, by rapid re-initiation on an assembled promoter complex3,6,8,9.
As is true for most biological processes, the different steps leading to transcription are subject to stochastic fluctuations10. A gene will not be expressed identically in two cells, even if they are grown under the same conditions. Such fluctuations should optimally be minimal, because many proteins such as transcription or splicing factors require well-defined concentrations. High-throughput analyses in yeast showed that protein variation for most genes is low11. However, in the yeast Saccharomyces cerevisiae, most mRNAs are present in low abundance; 80% of the transcriptome, including many essential genes, are expressed at less than two copies per cell12. Therefore, high mRNA expression variation would be likely to lead to a situation where many cells are depleted of essential mRNAs, making it difficult to keep protein levels constant. How the cell keeps this variation low is not known.
This question has been difficult to address owing to technical limitations. Classical ensemble methodologies such as northern blots and reverse-transcription PCR (RT-PCR) are unsuitable for the study of single-cell variability. Most single-cell studies have measured gene expression variation using green fluorescent protein (GFP) reporters to monitor the variability of protein concentrations13,14. However, by measuring protein concentration, they could only determine the combined result of transcription and translation, not the direct output of transcription since the mRNA itself was not measured. To understand how cells mediate mRNA expression and how this results in expression variation requires single-cell analysis with single-mRNA resolution.
Few studies have used single-molecule techniques to understand gene expression kinetics. Fluorescence in situ hybridization (FISH) suggested that genes in mammalian cells are expressed as ‘bursts’ of transcription: infrequent periods of transcriptional activity that produce many transcripts within a short time15. Such transcription bursts were shown to lead to large variability in mRNA numbers15. Using different techniques, transcriptional bursting has been described for many genes and has become the prominent model for gene expression14,16–20. Transcriptional bursting has also been observed in bacteria, although in this system bursting was much weaker and measured only on an inducible gene21. However, bursting with its consequential large mRNA variation does not explain the low-noise characteristics found for most genes in yeast. To measure variation precisely and the underlying transcriptional activity and expression levels, we have derived a single-molecule counting approach that allows us to enumerate every single mRNA and nascent transcript from a given gene within a cell. The approach is nondisruptive and simple, is applicable to any endogenous gene and does not require any genetic manipulation.
We have used single molecule–sensitivity FISH to determine the exact number of mRNAs that are present in individual S. cerevisiae cells for different genes while characterizing the transcriptional status in the same cell by enumerating the number of nascent transcripts. By using these numbers in a mathematical modeling approach that constrains the probable outcomes, we were able to determine kinetic parameters that mediate the expression of these genes. We show that expression of genes in yeast can be achieved by single, noncorrelated transcription-initiation events, in contrast to what occurs in higher eukaryotes. However, we also find that some genes can show bursting expression as well.
RESULTS
Single mRNA–sensitivity FISH to analyze gene expression
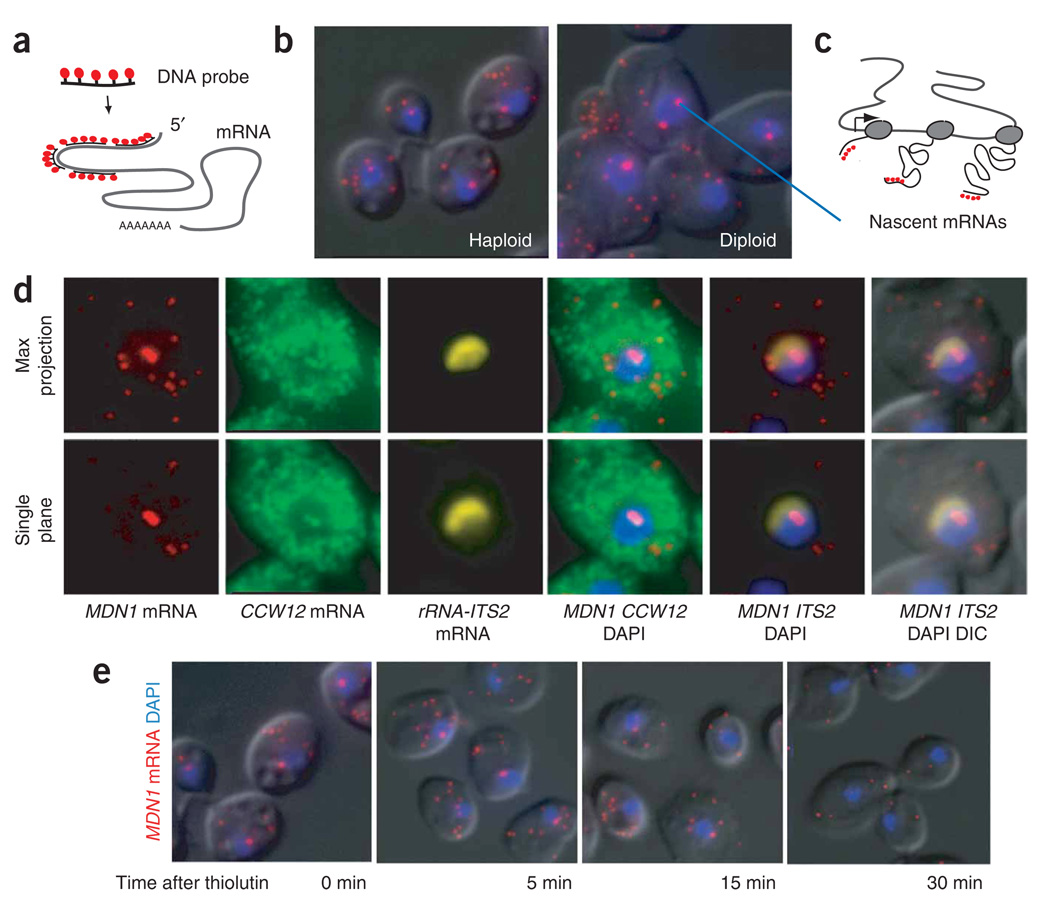
To achieve single-transcript resolution, we adapted a FISH technique previously described in mammalian cells22. The protocol uses multiple oligodeoxynucleotides, each labeled with five fluorescent dyes, creating a sufficient signal-to-noise ratio to allow single-mRNA detection (Fig. 1a). To validate the approach in paraformaldehyde-fixed yeast, we hybridized a mixture of four DNA probes complementary to the MDN1 gene (Fig. 1b). MDN1, the largest gene in yeast (14.7 kb) is an essential, constitutively expressed gene involved in preribosomal processing and reportedly expressed at one mRNA copy per cell12,23. Probes were designed to hybridize to the 5′ end of the gene to allow the detection of an mRNA from the very beginning of its synthesis, when it is still associated with the site of transcription.
Figure 1.
Single mRNA–sensitivity FISH. (a) Schematic diagram of the FISH protocol. A mixture of four 50-nt DNA oligonucleotides, each labeled with five fluorescent dyes, is hybridized to paraformaldehyde-fixed yeast cells to obtain a single-transcript resolution. (b) Single-mRNA FISH for MDN1 mRNA. Single mRNAs are detected in the cytoplasm, with a higher intensity spot in the nucleus. Haploid and diploid yeast cells are shown. Probes hybridize to the 5′ end of the mRNA. MDNI mRNA, red; DAPI, blue; superimposed on the differential interference contrast (DIC) image. (c) Cartoon showing how the number of nascent mRNAs at the site of transcription is used to determine the polymerase loading on a gene when using FISH probes that hybridize to the 5′ end of the gene. (d) Nascent transcripts of neighboring genes colocalize at the site of transcription. Diploid cells are hybridized with probes against MDN1 (labeled with cy3) and CCW12 (labeled with cy3.5). The nucleolus is stained with probes against the ITS2 spacer of the ribosomal RNA precursor (labeled with Cy5). Maximum projection of a three-dimensional data set and single plane containing the transcription sites is shown. (e) Nascent-transcript detection requires ongoing transcription. Cells were fixed 0, 5, 15 and 30 min after addition of the transcription inhibitor thiolutin (4ug ml−1) to the media. FISH was carried out using probes to MDN1 mRNA as shown in b. Representative cells are shown for each time point.
We acquired three-dimensional data sets and reduced them to two-dimensional images to facilitate data analysis. The fluorescent in situ probes appeared as multiple diffraction-limited spots within the cytoplasm of individual yeast cells; this is similar to what has been seen in mammalian cells, where they were shown to represent single mRNAs15,22 (Fig. 1b). Higher-intensity spots were found in the nucleus, colocalizing with the DAPI signal, and were likely to represent the assembly of multiple nascent transcripts associated with the MDN1 gene (Fig. 1c). Consistently, a single higher-intensity nuclear spot is found in haploid cells, whereas two are present in diploid strains. Nascent transcripts of neighboring genes should colocalize within these nuclear spots.
CCW12 is a short but actively transcribed gene starting 6,000 bp upstream from the MDN1 promoter. Signals for CCW12 and MDN1 mRNA colocalized in the nucleus, indicating that the nuclear spots represent sites of transcription (Fig. 1d). Notably, although frequently transcribed, only nascent RNA for CCW12 was found in the nucleus, indicating that export of these mRNAs after their release from the site of transcription is rapid. As expected, sites of transcription disappear with treatment by the transcription inhibitor thiolution, followed by the reduction of cytoplasmic mRNAs (Fig. 1e).
Different studies have shown that many genes in yeast associate with the nuclear periphery when they are transcribed24,25. Notably, although we often found MDN1 transcription sites at the border of the DAPI stain, they did not localize to the nuclear periphery but to the region between the nucleoplasm and the nucleolus. This is likely to be caused by the proximity of the MDN1 and CCW12 genes to the ribosomal RNA genes located only about 90 kb further upstream (Fig. 1d).
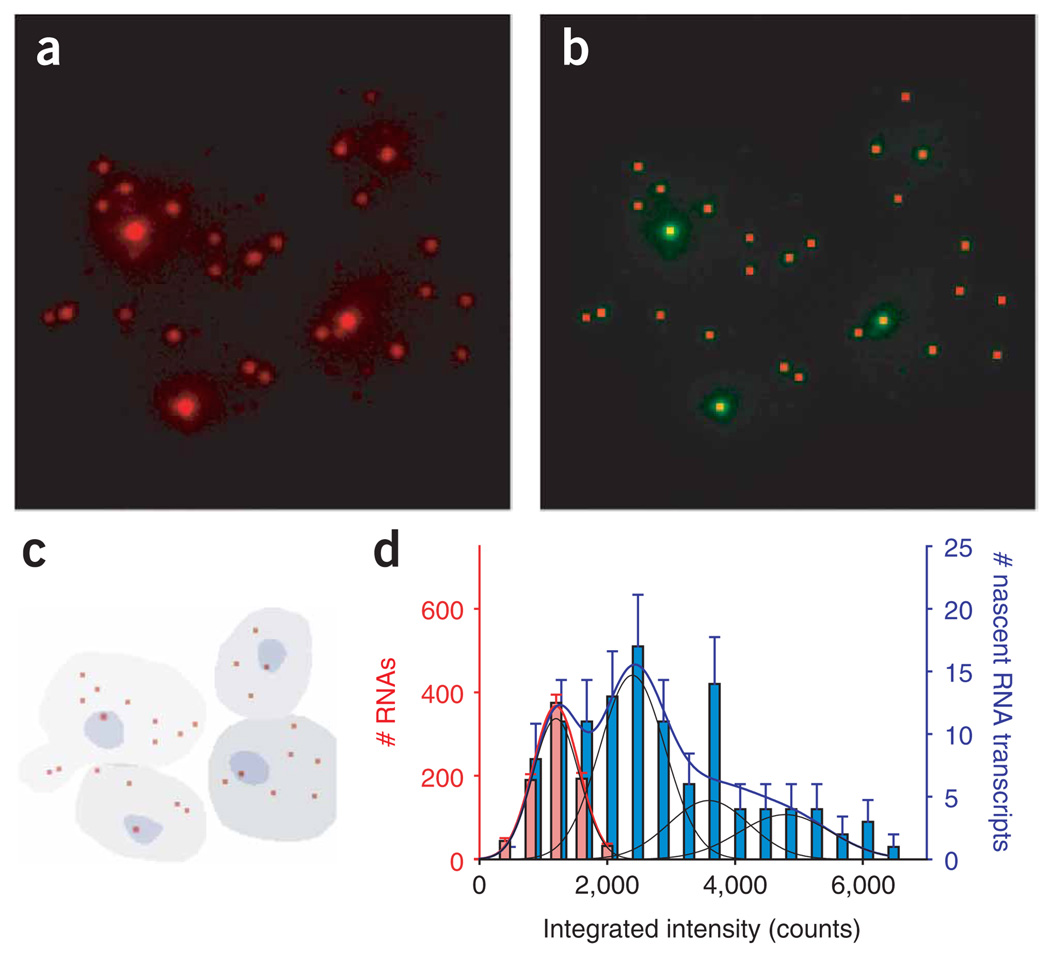
To demonstrate that the detected signals correspond to single mRNAs and not to multiple mRNAs clustering in a diffraction-limited spot, we quantified the signal intensities of the cytoplasmic and nuclear signals using a spot-detection program that detects and quantifies the signal intensities for each spot26 (Fig. 2a–c). The signal intensities of the cytoplasmic spots show a uniform distribution and can be fitted to a single Gaussian curve, as expected for the detection of single mRNAs (Fig. 2d). Consistently, the intensity of these single mRNAs hybridized to four oligonucleotide probes equals four times the intensity of a single probe (Supplementary Fig. 1 online). The intensity distribution for spots in the nucleus can be fitted to a superposition of Gaussian distributions corresponding to the assembly of multiple nascent transcripts associated with the MDN1 gene (Fig. 1c,d). This provides a direct measure of how many mRNAs are being transcribed. Therefore, this methodology allowed us to determine two essential parameters defining gene expression: the ‘expression state’, the total number of mRNAs per cell; and the ‘transcriptional status’, an instantaneous measure of the number of nascent transcripts on a gene. Notably, this analysis addressed endogenous RNA as close to a physiological state as was experimentally possible, as genetic modifications were not required.
Figure 2.
Quantitative single-molecule, single-cell gene expression analysis. (a,b) A spot-detection algorithm detects and quantifies FISH signals. Red dots in b show signals identified by the spot-detection software from the raw signals in a. (c) The nucleus and cytoplasm were segmented using a hand-drawn mask (cellular boundaries) and DAPI thresholding (nucleus). (d) Histogram of cytoplasmic (left axis, red bars) and nuclear (right axis, blue bars) signal intensities of MDN1 signals from multiple fields, determined using the spot-detection algorithm. The cytoplasmic mRNA intensities fit to a Gaussian distribution (red line), and the mean is used as the brightness of a single transcript. The nuclear signal intensities (assembly of nascent mRNAs associated with the gene) fit with multiple Gaussian distributions (blue line), where the mean of each Gaussian distribution is an integer multiple of the single-peak intensity. The width of each peak also scales with the mean, as expected for Poisson noise in spot localization26. The individual contributions to the composite fit are shown in black. Error bars indicate s.e.m.
Expression variation of constitutively active genes
We then analyzed the expression of one of the most common classes of genes, the housekeeping genes. The extent of RNA variation for these genes is not known, although protein variation has been the subject of many studies14.
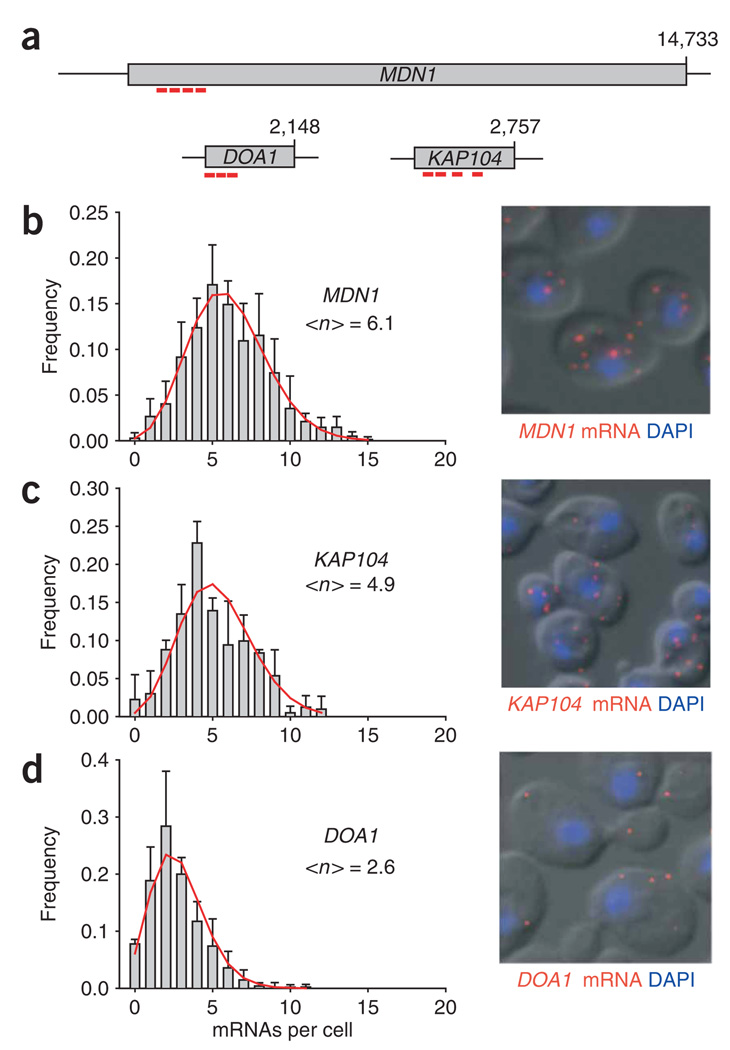
To address this question directly, we analyzed the expression profiles of three unrelated, constitutively expressed genes—MDN1, KAP104 and DOA1—involved in such diverse functions as ribosome biogenesis, ubiquitin-mediated protein degradation and nucleocytoplasmic transport. All genes have been indicated to be expressed at one copy per cell12. The three genes show similar expression profiles, suggesting a common mode of expression, and several characteristics are immediately evident (Fig. 3). First, expression levels were higher than previously estimated. On average, cells contained three to six times the number of mRNAs as had been measured by microarray. This observation corrects the long-standing assumption that most mRNAs in yeast are expressed at only one or two copies per cell and that many genes are transcribed only once during a cell cycle12,27,28. Second, few cells were devoid of mRNA for any of the genes tested. Even for DOA1, which is expressed at the lowest levels, only about 8% of the cells lacked DOA1 mRNA, indicating that cells have evolved a transcriptional behavior to maintain a basal level of expression. Third, the expression levels varied among individual cells in the population. For example MDN1 mRNA was expressed from 1 to 15 mRNAs per cell with a mean of 6.1. Finally, expression variation for housekeeping genes fell within a narrow range that can be described by a Poisson distribution, suggesting that the variation might be explained by uncoordinated transcription initiation.
Figure 3.
Expression profiles of constitutively active genes. (a) Cartoon showing the position of the FISH probes used according to their target region on the corresponding mRNAs. (b–d) mRNA expression profiles of different yeast genes shown in a were determined using FISH. Frequency (y axis) of mRNA numbers (x axis) per cell determined for MDN1, KAP104 and DOA1 are shown. <n> shows the average number of mRNAs per cell. Red lines in b–d show fits describing the expression kinetics (see text). Error bars indicate s.e.m. Representative FISH images (mRNA, red; DAPI, blue) superimposed on the differential interference contrast (DIC) are shown on the right.
Modeling expression kinetics from mRNA-abundance data
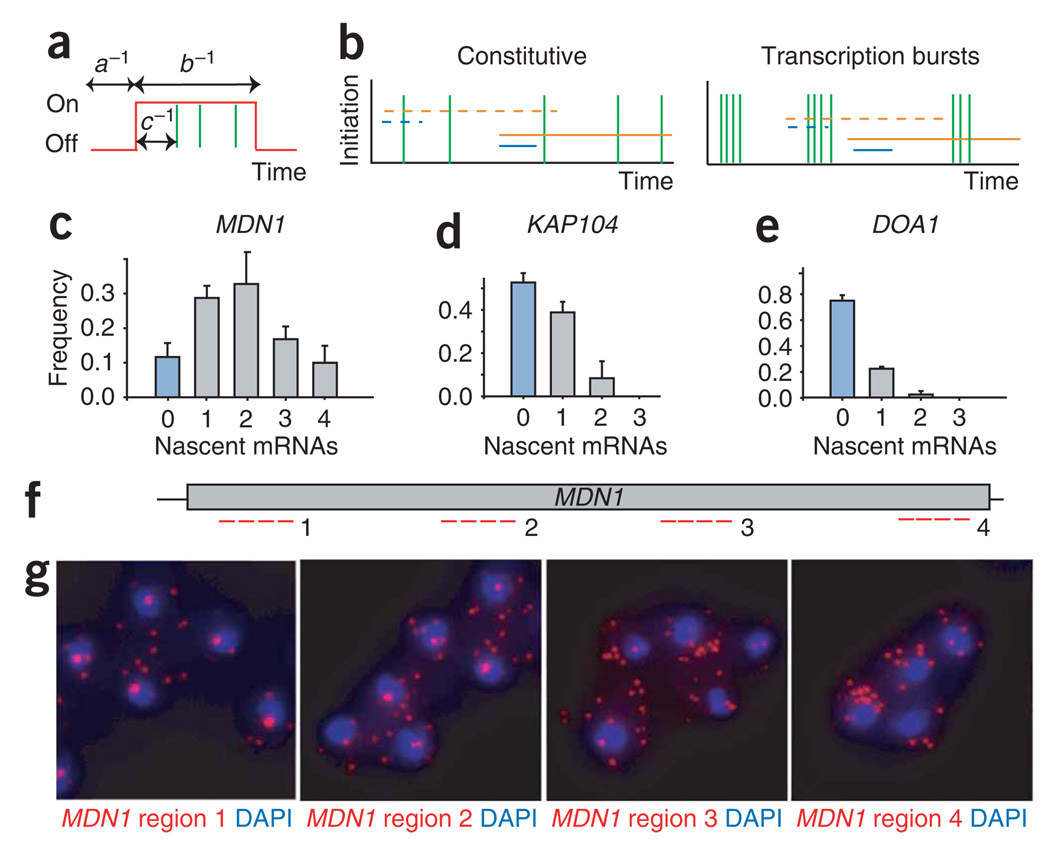
To obtain a general understanding of the kinetic parameters leading to the observed mRNA distributions, we performed simulations using a mathematical framework based on a gene-activation and -inactivation model15,29. In this model, a gene alternates between an active ‘on’ and an inactive ‘off ’ state (Fig. 4a). The three variable parameters that describe the distribution of mRNA in the cytosol are the rate for switching to an on state (parameter a; Fig. 4a), the rate for switching to an off state (parameter b; Fig. 4a) and the rate of transcription while in the on state (parameter c; Fig. 4a). The transcripts accumulate in the cytoplasm where they are degraded at a fixed, specific rate (parameter d)12. Notably, this mathematical framework allowed us to distinguish between two transcriptional modes suggested to mediate mRNA expression: ‘bursts’ (infrequent on states producing multiple transcripts rapidly), or the ‘constitutive’ mode (initiation distributed in time; Fig. 4b). Simulations have shown that these two modes can lead to distinctly different mRNA distributions14. However, our simulations of RNA abundance alone resulted in poorly constrained transcriptional models that did not differentiate between a bursting model (c/b > 1) and a nonbursting model (c/b ≤1). Recent theoretical studies similarly have suggested that cytoplasmic distributions alone do not allow the full description of expression modes30.
Figure 4.
Transcriptional loading determines the mode of transcription. (a) Gene-activation and -inactivation model used to simulate the expression kinetics. The gene transitions to the on state (red line, high) with rate a and to the off state (red line, low) with rate b. The initiation rate from the on state is c, and initiation events are denoted by a vertical green hash mark. The average time intervals are a−1, b−1 and c−1, respectively. (b) Alternative transcription modes. In the constitutive transcription mode, individual initiation events are clearly separated in time, whereas for transcription bursts multiple transcripts are produced within short time intervals followed by long periods of transcription inactivity. Initiation of a single transcript is shown as a green vertical line. Red and blue lines indicate the time the polymerase needs to synthesize an mRNA and is therefore equal to the time an mRNA stays at the site of transcription. On a long gene (orange), constitutive and bursting transcription can lead to similar distributions. On short genes (blue), bursting and constitutive expression lead to different distributions. Full and broken lines show two time points when cells are fixed. (c–e) Transcription status profiles of MDN1, KAP104 and DOA1 determined using FISH. The frequency (y axis) of the number of nascent transcripts (x axis) per cell is shown. The fraction of cells not containing an active site of transcription are highlighted in blue. Error bars indicate s.e.m. (f) Position of FISH probes to different region on the MDN1 gene. (g) Polymerases do not cluster on the MDN1 gene. MDN1 mRNA FISH using cy3-labeled probes to regions 1, 2, 3 and 4 on MDN1. RNA, red; DNA stained with DAPI, blue.
Measuring polymerase loading to determine expression kinetics
To obtain an additional kinetic parameter allowing a better description of the expression kinetics, we determined the temporal spacing of individual transcription-initiation events by measuring the number of active polymerases at a gene. We achieved this by determining the number of nascent mRNAs at the site of transcription (Figs. 1c, 2d and 4b). For example, a transcription site containing multiple nascent mRNAs indicates that several transcripts were initiated within the time interval it takes to synthesize a complete transcript. This synthesis time (τ) depends on the length of the gene.
The transcriptional status is shown in Figure 4c–e and 4g. Nascent mRNAs of the DOA1 gene were detected in about 20% of the cells, and cells transcribing DOA1 contain only a single nascent mRNA (Fig. 4e). Assuming that RNA polymerase II elongates at 2 kb min−1, the synthesis of its 2.2 kb transcript will last at least 1 min31. Therefore, in a cell containing a single nascent DOA1 mRNA, at least 1 min has passed after the initiation of the previous transcript. Thus, determining the number of nascent mRNAs at a site of transcription acts a direct measure for initiation. The KAP104 gene shows a similar polymerase-loading distribution to DOA1, indicating that individual initiation events are well separated in time (Fig. 4d). However MDN1, the longest gene investigated in this study, shows a transcriptional profile with up to four nascent mRNAs at the gene (Fig. 4c). This could have resulted from a transcriptional burst where several transcripts were initiated in rapid succession. If this were the case, we would expect to observe a cluster of up to four nascent chains somewhere in the gene. Therefore, multiple nascent transcripts should be detected using FISH probes against a 3′ subregion within the gene, as there is a probability that the polymerases would have progressed into this region together. However, multiple nascent RNAs were observed only when using FISH probes that hybridized to 5′ regions, but never with probes that hybridized closer to the 3′ end of the gene, suggesting that clustering of polymerases does not occur on MDN1 and that correlated initiations do not occur (Fig. 4f,g). Taken together, these data indicate that, for constitutively active genes, individual initiation events are spaced minutes apart.
Modeling MDN1 expression kinetics
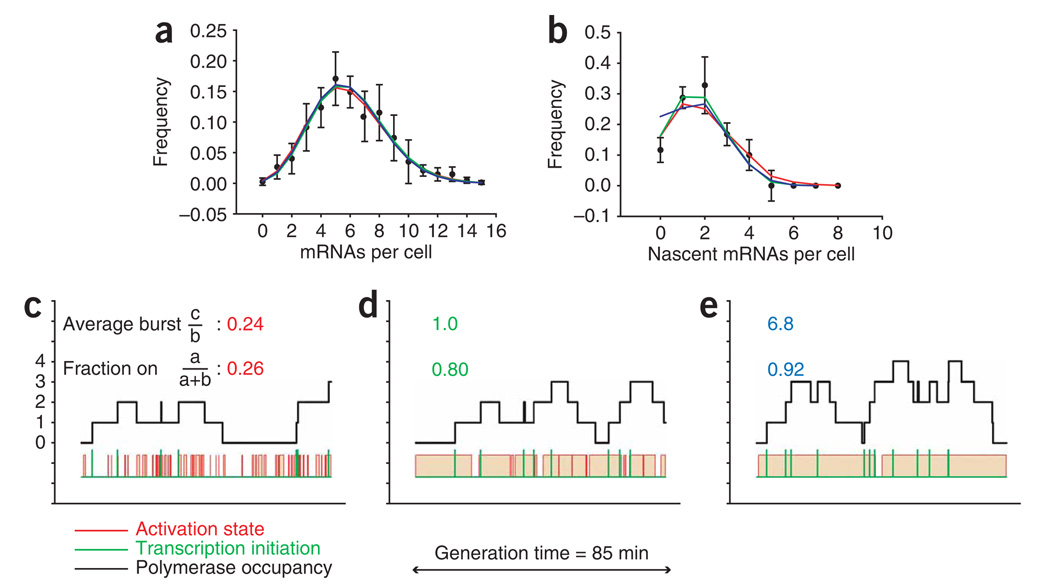
To test this hypothesis, we modeled the polymerase-loading data using the activation-inactivation model. The variables a, b and c were defined as previously and an additional parameter, τ, the time a nascent transcript is associated with the gene, was introduced. Figure 5a–e (see also Supplementary Table 1 online) shows three examples of models that fit the measured MDN1 data equally well for both the distribution of total mRNA (Fig. 5a, χ2 < 2.43) and the nascent chains (Fig. 5b, χ2 < 9.15). Representative Monte Carlo time traces are shown in Figure 5c–e. In the first model, the gene is on 26% of the time, and 0.24 transcripts are produced on average from each active state (Fig. 5a,b, red curve, and 5c). This represents the extreme limit of nonbursting transcription, where some active states are too short to even allow transcription initiation (c/b ≪ 1). An intermediate case occurs when the on state is exactly as long as the average time between transcription-initiation events (c/b = 1) (Fig. 5a,b, green curve, and 5d), and in this case the gene is on 80% of the time. Finally, there is the case where the gene is practically always on (Fig. 5a,b, blue curve, and 5e); here, the burst size is substantial (c/b ≫ 1), with each active period producing around seven transcripts. These models result in statistically similar distributions, and all three describe the measured data within the variation. Furthermore, the polymerase occupancy (Fig. 5b–e, black lines) is not noticeably different for the three models.
Figure 5.
Modeling MDN1 expression kinetics. (a,b) mRNA abundance (, a) and nascent transcripts (, b) for MDN1 fit with a model based on the scheme in a. Three different scenarios with different values for a, b and c are shown (red, green and blue curves, respectively). Error bars indicate s.e.m. (c–e) Representative Monte Carlo time traces of transcription, where c (red), d (green) and e (blue) show a different set of rate constants a, b and c, corresponding to the curves in a and b. The black curve is the polymerase-occupancy level on the gene; the red curve is the on/off state of the gene; the green curve marks initiation events. The average burst size and fraction of time spent in the on state are shown above each time trace.
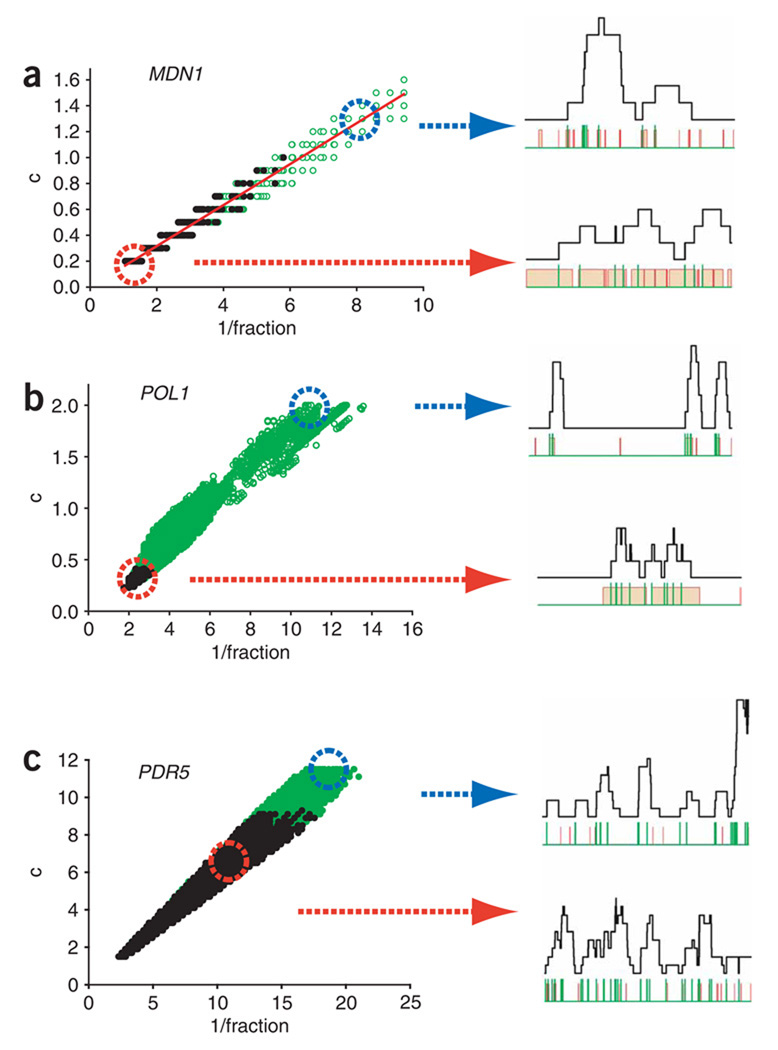
The difference between models is due only to which rate constant is limiting, suggesting that c/b by itself is not a sufficient determination of bursting. For example, when using only the value c/b > 1 as the definition of bursting, scenario 3 with c/b of 6.8 would suggest a bursting expression for MDN1. However, the gene is on for almost the entire generation time, and initiation events are spaced minutes apart, hardly consistent with bursting. Therefore, a better way to describe the expression modes of these genes is needed. To obtain a fully inclusive picture of the parameter space that describes the experimental data, we considered a locus of points that fits both the mRNA abundance and the nascent-chain data (Fig. 7a). When initiation rate (c) is plotted against fraction−1 ((a + b)/a), the acceptable (χ2 < 25.99; see Supplementary Tables 2 and 3 online) models cluster around a line. The slope of this line is defined by ac/(a + b), and this value provides an effective transcription rate (that is, the initiation frequency in the on state multiplied by the fractional time spent in the on state) that is necessary to balance the degradation in steady state. The locus of points is an unambiguous description of the possible modes of transcription and shows a continuum of kinetic modes without relying on the arbitrary binary classification of ‘bursting’ or ‘nonbursting’. To the right of the graph are models where the fraction of time the gene spends in the on state is low, and the initiation rate is high (bursting limit); to the left, the fraction of time the gene spends in the on state is high, and initiation is low (nonbursting limit). In addition, the models that fit the RNA abundance alone (Fig. 7a, open green circles, ), are further restricted to models that fit both RNA abundance and nascent-chain data (Fig. 7a, black circles, ). Monte Carlo traces from those fits taken from the nonbursting end of the graph (Fig. 7a, dashed red circle) that fits the nascent-chain data, and traces from the bursting end of the graph (Fig. 7a, dashed blue circle) that does not fit the nascent-chain data ( ), clearly demonstrate the importance of determining the nascent-chain loading. Notably, only scenarios with low initiation frequencies fit the data.
Figure 7.
Transcription kinetics of endogenous yeast genes. (a) The combinations of transcription rate constants that result in statistically significant models for MDN1 are shown as circles designating a particular value of a, b and c (min−1), with τ implicit. 1/fraction = (a + b)/a. Models that fit the mRNA abundance only are shown in open green circles; models that fit both the mRNA abundance and the nascent-mRNA loading are shown in closed black circles (χ2 significance level = 0.10). Simulated Monte Carlo time traces for MDN1 transcription using the parameters corresponding to the parameters used for the regions in red and blue circles are shown on the right. The black line shows the occupancy level of the gene; the red line shows the activation state of the gene (high = active, low = inactive); the vertical green lines mark single initiation events. (b,c) Parameters describing the expression kinetics of POL1 and PDR5.
Expression kinetics of a cell cycle–regulated gene
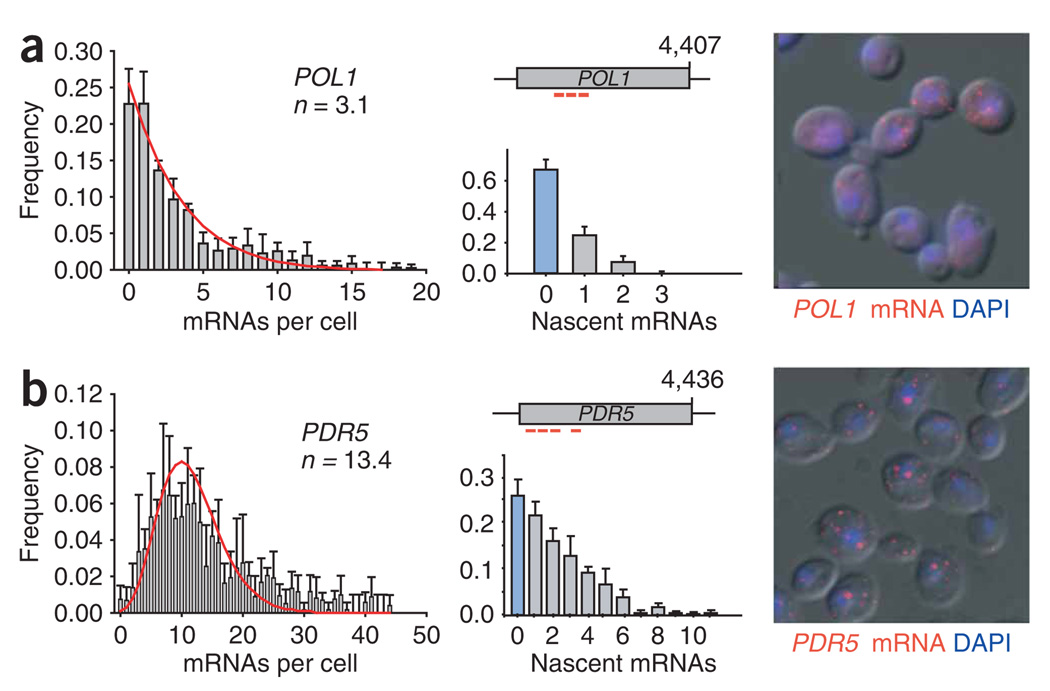
For comparison, we extended this analysis to a gene that is not constitutively expressed, the POL1 gene, which encodes DNA polymerase I and is expressed during part of the G1 and S phases32. As expected, the expression profile for this gene was different, with many cells not expressing POL1 mRNA or having a single mRNA in the cytoplasm (Fig. 6a). However, in cells containing active sites of transcription, nascent mRNA distributions resemble constitutively active genes: only one and rarely two nascent mRNAs were found associated with the POL1 gene, suggesting a more ‘constitutive’ mode of transcription in the on state, but no transcription bursts. When evaluated using the mathematical framework, the POL1 data showed low initiation frequency, during a prolonged on state that occurs infrequently during the generation time, suggestive of a portion of the cell cycle (Fig. 7b). The bursting limit can be ruled out by an inadequate fit to the nascent-chain data. Hence, the part of the cell cycle in which POL1 is expressed is long enough to permit uncorrelated initiations.
Figure 6.
Expression profiles of a cell cycle–regulated and a SAGA-controlled gene. mRNA expression and transcription status profiles of POL1 (a) and PDR5 (b), as determined using FISH. Frequency (y axis) of the number of cytoplasmic mRNAs (left) and nascent transcripts (below middle) (x axis) per cell are shown. <n> shows the average number of mRNAs per cell. Fractions of cells not containing an active site of transcription are highlighted in blue. Error bars indicate s.e.m. Above middle, position of FISH probes. Representative FISH images (mRNA, red; DAPI, blue) superimposed on the differential interference contrast (DIC) are shown on the right.
Bursting expression of PDR5
In contrast to what we observed in yeast, genes in higher eukaryotes are reported to show transcription bursts15,17,18,22. We investigated whether bursting genes might also exist in yeast. Analyses on the yeast HIS3 promoter have suggested that, depending on the conservation of the TATA element, expression could be achieved by a constitutive or inducible transcription mode8,33,34. It was then shown that the presence of a consensus TATA box leads to robust transcription mediated by transcription re-initiation, a process that could be the cause of transcriptional bursting6,8,9. Measurements of protein variation in yeast identified a subset of genes whose expression showed substantially higher variation than found for most of the proteome, suggesting that they might be regulated differently11. Many of these genes were regulated by the transcriptional coactivator SAGA (Spt–Ada–Gcn5–acetyl transferase complex) and contain conserved TATA boxes11,35. We therefore determined the mRNA distribution and transcriptional status of the TATA-containing, SAGA-regulated PDR5 gene. The mRNA distribution for PDR5 was much wider than the constitutive genes (Fig. 6b). Nascent-transcript analysis showed that about 50% of cells contained no or only a single nascent PDR5 transcript, whereas the remaining cells showed up to 11 nascent transcripts, indicating the presence of transcription bursting (Fig. 6b, below middle). Simulating the PDR5 distributions showed that the expression kinetics fit a bursting mode (Fig. 7c). Thus, the SAGA-regulated PDR5 gene shows a transcriptional mode that is comparable to those observed in higher eukaryotes.
Defining constitutive expression
We have described different expression modes in S. cerevisae in which bursting and constitutive, or nonbursting, are limiting descriptive classifications when bursting is defined only as the ratio of the initiation frequency and the on state of a promoter (c/b). The kinetic modes are determined by different rates of gene activation and inactivation and the initiation frequency. The physical meaning of the gene activation and inactivation parameters for transcription can be partially assessed by considering scenarios that apply to all genes. The expression states of the constitutive genes (Fig. 3b–d, left, red curves) can all be fit with a single set of gene activation and inactivation parameters (a,b) and a variable initiation rate (c). The average off time (1/a) for this particular model is 1.4 min; the average on time (1/b) is 8.7 min (87% on time). Using this scenario, the average number of transcripts produced during each on state is 1.4 transcripts. Using only c/b > 1 to define bursting, these genes would show a weak bursting expression. However, considering the short off times and the low initiation frequency, the individual initiations are spaced by minutes, making the term bursting inaccurate. These data suggest that a promoter stays in an open state long enough to initiate one or two transcripts. Mechanistically, this observation indicates that, after the assembly of a transcription competent complex at a promoter, at most only one or two transcripts are produced before the complex falls apart and the complex must be reassembled on a promoter that is still accessible. This scenario might be different for bursting genes such as PDR5, where factors such as SAGA might stabilize promoter complexes and allow multiple initiations from a single complex assembly.
Extracting polymerase speed and termination time
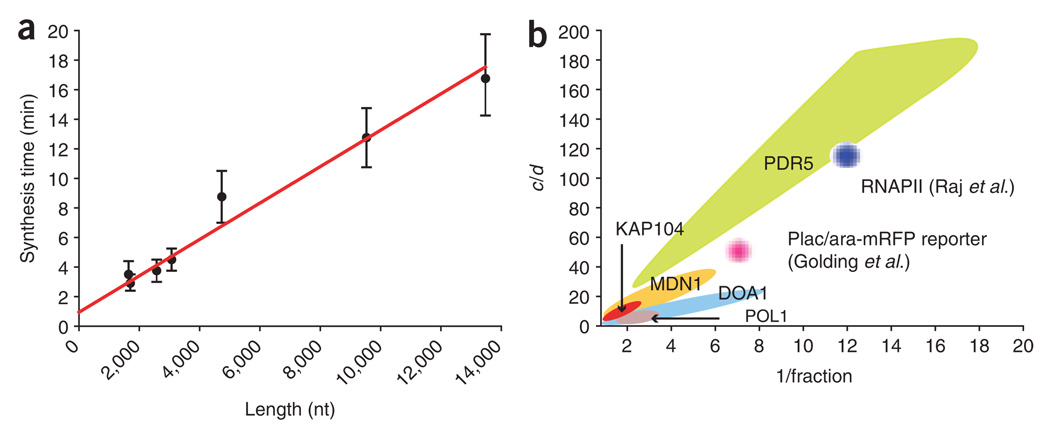
The distribution of nascent chains further implies a synthesis time uniquely determined from the fit. If plotted against the effective length of the gene, the inverse slope provides the average speed of RNA polymerase: 0.81 ± 0.07 kb minute−1 (Fig. 8a). In addition, the y-intercept corresponds to a termination time of 56 ± 20 s. The elongation speed is slower than the elongation speed measured from a Gal promoter–driven gene, measured at 2 kb min−1 (ref. 31). A velocity of 2 kb min−1, however, does not fit our data, suggesting that different elongation speeds exist for different classes of gene. Different elongation speeds have been measured in various organisms and on different genes, ranging from 0.7 kb min−1 to 4.4 kb min−1 (refs. 31,36–41). One reason for the differences in elongation speed might be that polymerases on strongly transcribed genes, such as Gal-induced genes, are more processive because the chromatin is more open compared to sporadically transcribed genes42,43. Additionally, elevated polymerase densities on highly transcribed genes might increase polymerase velocity, as shown in bacteria40.
Figure 8.
Extracting kinetic data from fixed-cell analysis. (a) Determining polymerase speed from FISH data. The synthesis time (τ) is plotted against the length of the gene. The length is determined from the position of the FISH probe nearest the 3′ end of the gene. Error bars indicate s.d. determined from the model by allowing τ to vary for a fixed set of a, b and c parameters. The slope of the line gives (polymerase speed)−1, resulting in a speed of 0.80 ± 0.07 kb min−1; the y intercept corresponds to a termination time of 56 ± 20 s. The individual data points correspond to the previously described genes (KAP104, DOA1, MDN1, POL1 and PDR5) and multiple regions of MDN1. (b) The parameters space for endogenous gene transcription. The statistically significant models for each gene are presented as in Figure 7. The y axis is the initiation rate constant c normalized by the mRNA decay constant d, which allows for comparison between genes. 1/fraction = (a + b)/a. For the genes studied in this report (MDN1, KAP104, DOA1, POL1, PDR5), the colored regions represent the actual parameter space for a, b and c. For the genes described in previous reports15,21, the full parameter space was not reported. The approximate value of a, b and c is based on the findings of these authors (Raj et al.: c/d ~ 120; 1/f ~ 12. Golding et al.: c/d ~ 50; 1/f ~7), but the physical extent of these regions as depicted in b is only for graphic display.
DISCUSSION
We have analyzed the expression behavior of endogenous genes in yeast using single-molecule analysis. For the first time, we have determined the exact number of mRNAs expressed in a single cell and used this information to model the expression kinetics for these genes. The key for these analyses was combining the number of cytoplasmic mRNAs present with the transcriptional status for each of the genes.
The ability to use cells without the need for any genetic modification is one main advantage of FISH. Cells are simply fixed, hybridized and analyzed. By this method, many cells can be analyzed and used for mathematical modeling. Additionally, placing FISH probes at different positions along the mRNA can be used to define the spacing between individual transcription-initiation events or to produce ‘footprints’ of polymerases on a gene (Fig. 4f,g). Expanding this analysis by interrogating multiple genes simultaneously in the same cell will allow not only the dissection of single genes but also the study of co-regulatory networks and provide an important tool for systems analysis.
Our observation that mRNA abundance for most genes was higher than previously suggested was surprising, as these numbers were obtained by different hybridization techniques and are commonly used in the literature12,28,44,45, although higher numbers have been suggested previously for a small subset of genes46. The main reason for the discrepancy may lie in the normalization factor used by these studies, wherein it was assumed that a yeast cell expresses 15,000 mRNAs per cell. As shown in Supplementary Table 4 online, the genes used in this study show a three- to six-fold higher expression than that determined previously12. This would correct the number of transcripts to around 60,000 mRNAs per cell and indicates that the yeast transcriptome is more active than initially thought. This number also fits measurements suggesting that about 85% of the 200,000 yeast ribosomes are associated with mRNAs at an average ribosome density of 1 ribosome per 154 nt47,48. Our observations also illustrate the utility of tools that enable the absolute quantification of gene expression, independently of ensemble measurements that use calibration and normalization factors.
Analyzing the expression of constitutively active genes revealed that mRNA variation is low, almost to a level that would be expected from pure Poisson noise. Although theoretical work has shown that different expression modes can lead to similar distributions30, we show that expression is achieved by single, temporally well-separated initiation events, but not by transcription bursts. Even the cell cycle–regulated POL1 gene is expressed in a similar manner to a constitutive gene during its active period. With respect to promoter kinetics, this indicates that the assembly of an entire transcription complex usually leads to the initiation of a single transcript before the complex falls apart.
Recent work suggests that transcription complexes in general might not be as stable as thought. Even if transcription factors interact stably with their specific binding sites in vitro, the residence time of many transcription factors at promoters in vivo is short49,50. However, for some classes of genes and promoters, factors that stabilize promoter complexes might allow the production of multiple mRNAs from a preassembled and stabilized complex. Transcription re-initiation has long been assumed to be required for efficient transcription from a promoter3,51,52. Transcription bursts found for the PDR5 gene or for genes in higher eukaryotes might depend on factors allowing transcription re-initiation. Many genes in yeast showing high expression variation in protein levels are regulated by SAGA and contain a well-conserved TATA box, which is unusual for genes in yeast. Notably, it had previously been suggested that more stable binding of a TBP–TFIID complex, caused by the conserved TATA box, leads to re-initation–competent complexes, thereby causing transcriptional bursting6,9. Consistent with this, mutating the TATA box in yeast has been shown to affect expression and reduce protein variation8,16,53.
Figure 8b shows the parameter space for each gene tested, with the initiation rate c normalized by the mRNA decay rate d. Although some genes (MDN1, DOA1) have a less-restricted parameter space than others (POL1, KAP104), these genes all overlap in the nonbursting limit, whereas PDR5 is much less restricted. RNA polymerase II in mammalian cells and a bursting, artificial gene in bacteria are shown for comparison (the parameter space depicted for these two genes is only schematic). So, why has yeast but not higher eukaryotes chosen a constitutive expression mode for housekeeping genes? The possible explanation might lie in the fact that yeast is a rapidly dividing single cell. In higher eukaryotes, although mRNA variation is high owing to transcriptional bursting, the final protein variation is relatively low because mRNA noise is damped out by long mRNA and protein half-lives15. In yeast, however, such buffering is not possible, as the average protein half-life is short and only twice as long as the average mRNA half-life54,55. Maintaining constant expression is therefore better achieved by nonbursting, low-variation expression that constantly produces new proteins. Constant protein production is achieved by efficient translation, as most mRNAs (>70%) in a yeast cell are also polysome associated47. However, in some cases, when fast responses are more important than precise control of transcriptional amplitudes, for example, during stress responses, bursting expression might be beneficial56. Notably, bursting as well as constitutive RNA expression have been described in bacteria21,57.
It is reasonable to speculate that the simple structure of yeast promoters, when compared to promoters in higher eukaryotes makes it easier to assemble transcription complexes for single initiation events. Promoters are often only a few hundred base pairs long and consist mainly of the histone-free region just upstream of the transcription start site58,59. Opening promoters and assembling a transcription-competent complex is likely to require much more effort for the cell in higher eukaryotes, so it might be advantageous to transcribe multiple mRNAs once a complex is assembled, especially as higher total mRNA numbers are required as well60. However, genes may exist in higher eukaryotes that are expressed in a less bursting and more constitutive manner. Future studies will show how other eukaryotes have evolved their modes of transcription and whether higher eukaryotes use transcription bursting only to express their transcriptome or if constitutive expression also exists. Single-molecule approaches such as that presented here will be essential to understand the kinetics of gene expression.
METHODS
In situ probes
Probes were designed, synthesized and labeled using cyanine dyes cy3, cy3.5 and cy5 (GE healthcare, #PA23001, PA23501, PA25001) as described previously18. RNA probes were generally 50 nt long and contained four or five amino-modified nucleotides (amino-allyl T). The free amines were chemically coupled to fluorophores after synthesis. Probes used in this study are listed in Supplementatary Table 5 online.
Fluorescence in situ hybridization
Yeast cells (haploid BY4741 or diploid w303) were grown in YPD media at 30 °C to an optical density at 600 nm (OD600) of 0.8, and fixed by adding 32% (v/v) paraformaldehyde directly to the media to a final concentration of 4% (v/v) for 45 min at room temperature (20–25 °C). The cell wall was digested with lyticase (Sigma #L2524), cells were attached to poly-l-lysine (Sigma #P8920)–coated coverslips and stored in 70% (v/v) ethanol at −20 °C. Before hybridization, cells were rehydrated twice in 2× SSC for 5 min and once in 40% (v/v) formamide and 2× SSC (5 min). Coverslips were inverted onto 20 µl of hybridization solution containing 0.5 ng of labeled DNA probe (typically three or four DNA probes per gene) in 50% (v/v) formamide, 2× SSC, 1 mg ml−1 BSA, 10 mM VRC (NEB #S1402S), 5 mM NaHPO4, pH7.5, 0.5 mg ml−1 Escherichia coli tRNA and 0.5 mg ml−1 single-stranded DNA and hybridized overnight at 37 °C. Coverslips were washed twice with 40% (v/v) formamide and 2× SSC at 37 °C for 15 min, once in 2× SSC and 0.1% (v/v) Triton X-100 at room temperature for 15 min and once with 1× SSC at room temperature for 15 min, stained with DAPI and mounted with ProLong Gold antifade reagent (Invitrogen # P36930).
Image acquisition
Images were acquired with an BX61 epi-fluorescence microscope (Olympus) with an internal focus motor and an Olympus UPlanApo 100×, 1.35 numerical aperture oil-immersion objective using an X-Cite 120 PC (EXFO) light source for fluorescence illumination and Uniblitz shutters (Vincent Associates). Differential interference contrast (DIC) was generated using an Olympus U-DICTHC Nomarski prism. Digital images were acquired using a CoolSNAP HQ camera (Photometrics) as stacks of 30 images taken with a Z step size of 0.2 µ m using IPLab software (Windows v3, BD Biosciences) and filter sets 31000 (DAPI), 41001 (FITC), SP-102v1 (Cy3), SP-103v1 (Cy3.5) and CP-104 (Cy5) (Chroma Technology).
Data analysis
RNA counting and nascent-chain determination. Three-dimensional data sets were reduced to a two-dimensional image by maximum Z projection using IPLab software. Spot detection was based on a two-dimensional Gaussian mask algorithm described previously26 and was implemented with custom-made software for the IDL platform (ITT Visual Information Solutions). Single-transcript intensity was defined as the integrated intensity determined from the Gaussian mask algorithm. The number of nascent transcripts at the site of transcription was obtained by dividing the spot intensity of the transcription side by the single-transcript intensity, and rounding up or down to the nearest whole number. Cell segmentation was achieved by a hand-drawn mask using a custom-made script in IPLab. Nuclear segmentation was done by DAPI thresholding using an IPLab script. To obtain the single-cell, single-transcript expression profiles, data from spot detection, nuclear and cell segmentation were combined using custom-made software in IDL, computing total mRNA per cell and the number of nascent transcripts per cell. To obtain mRNA distributions, we used data sets from at least three independent experiments containing more than 80 cells.
Histogram of fluorescence in situ hybridization intensity
The intensity histogram of the cytoplasmic mRNAs (Fig. 2d, red curve) is fit to a single-peak Gaussian distribution:
where A is the amplitude, x0 is the center intensity and m is a gain factor that relates the intensity of the peak in counts to the width. The variance thus has the form σ2 = mx0, which is the variance of a Poisson distribution multiplied by the gain factor to convert counts to photons.
The intensity histogram of the nascent mRNAs in the nucleus is fit to a multiple-peak Gaussian distribution:
where the additional parameters are now the relative amplitudes B, C and D. Using the amplitudes of the fit, the weighted nascent chain occupancy for MDN1 is 1.60. Using simple rounding to the nearest integer value, occupancy in each nucleus gives a mean of 1.77.
Numerical modeling
The theoretical model for mRNA abundance is based on the Markovian model of Peccoud and Ycart as implemented by Raj and coworkers15,29. The analytical form derived by Raj and co-workers for the steady-state solution is:
where a, b, c and d are as defined in the text, N is the number of mRNA transcripts and 1F1 is the confluent hypergeometric function of the first kind. We note that d is a fixed value taken from the literature12. To calculate the distribution of nascent chains, we use a Monte Carlo simulation. For a given set of a, b and c parameters, the gene transitions to an on state with an exponentially distributed waiting time a−1 and remains in the on state for an exponentially distributed waiting time b−1. From the on state, initiation events follow a gamma distribution with mean waiting time c−1 (first initiation: gamma = 1; second initiation: gamma = 2, and so on). Once a polymerase has been initiated, it remains on the gene for a fixed synthesis time τ. The occupancy level therefore reflects the frequency of initiation and the synthesis time. Each time trace is 85 min long, corresponding to the generation time of yeast under these conditions. The number of total time traces (that is, cells) was chosen such that the distribution of nascent chains converged (typically 1,000). Thus, for a given set of a, b and c parameters, one has the analytical calculation of the mRNA distribution; for those same a, b and c parameters and also τ, one has the nascent-mRNA distribution determined from the Monte Carlo calculation.
Model parameters (a, b, c, τ) are varied concurrently to generate a complete map of phase space. Models are evaluated at the P = 0.10 level with a χ2 test. Specific numerical χ2 values, corresponding to the number of data points for each gene, are presented in Supplementary Table 2. Acceptable models are those that fit both the mRNA abundance distribution and the nascent-chain distribution. In general, we find that the nascent-chain distribution results in a more restrictive phase space than the mRNA abundance, as reflected in Figure 7.
The polymerase velocity was obtained by determining the best-fit line for the synthesis time (τ) using a single set of parameters a, b and c that fit the mRNA distributions of all the constitutive genes (parameters described in the section “Defining constitutive expression” 1/a = 1.4, 1/b = 8.7) and another single set of parameters for PDR5 (1/a = 2.3, 1/b = 0.2). The synthesis time τ is varied until the minimum χ2 for the nascent-chain distribution is found. The error bars represent the 95% confidence level. The velocity is determined from the slope of the line where synthesis time is plotted against length.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Burke and S.M. Shenoy for writing scripts for data analysis, and J.R. Warner, E.D. Siggia and M. Keogh for helpful discussions. This work was supported by the US National Institutes of Health (R.H.S.).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONTRIBUTIONS
D.Z. initiated the project and performed the experimental work. D.Z. and D.R.L. analyzed the data. D.R.L. wrote the spot-detection program and performed the numerical modeling. R.H.S. supervised the project. D.Z., D.R.L. and R.H.S. wrote the paper.
References
- 1.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 2.Thomas MC, Chiang C-M. Thegeneral transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 3.Dieci G, Sentenac A. Detours and shortcuts to transcription reinitiation. Trends Biochem. Sci. 2003;28:202–209. doi: 10.1016/S0968-0004(03)00054-9. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 6.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 7.Darzacq X, Singer RH. The dynamic range of transcription. Mol. Cell. 2008;30:545–546. doi: 10.1016/j.molcel.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer V, Struhl K. Mechanism of differential utilization of the His3 TR and TC TATA elements. Mol. Cell. Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yean D, Gralla J. Transcription reinitiation rate: a special role for the TATA box. Mol. Cell. Biol. 1997;17:3809–3816. doi: 10.1128/mcb.17.7.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 11.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 12.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 13.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann BB, van Oudenaarden A. Stochastic gene expression: from single molecules to the proteome. Curr. Opin. Genet. Dev. 2007;17:107–112. doi: 10.1016/j.gde.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blake WJ, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 19.Newlands S, et al. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross IL, Browne CM, Hume DA. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol. Cell Biol. 1994;72:177–185. doi: 10.1038/icb.1994.26. [DOI] [PubMed] [Google Scholar]
- 21.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 23.Bassler J, et al. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 24.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 25.Casolari JM, et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 26.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bon M, McGowan SJ, Cook PR. Many expressed genes in bacteria and yeast are transcribed only once per cell cycle. FASEB J. 2006;20:1721–1723. doi: 10.1096/fj.06-6087fje. [DOI] [PubMed] [Google Scholar]
- 28.Velculescu VE, et al. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 29.Peccoud J, Ycart B. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 1995;48:222–234. [Google Scholar]
- 30.Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–343. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- 31.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 32.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W, Struhl K. Saturation mutagenesis of a yeast his3 “TATA element”: genetic evidence for a specific TATA-binding protein. Proc. Natl. Acad. Sci. USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol. Cell. Biol. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huisinga KL, Pugh BFA. Genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 36.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards AM, Kane CM, Young RA, Kornberg RD. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 38.O’Brien T, Lis JT. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ucker DS, Yamamoto KR. Early events in the stimulation of mammary tumor virus RNA synthesis by glucocorticoids. Novel assays of transcription rates. J. Biol. Chem. 1984;259:7416–7420. [PubMed] [Google Scholar]
- 40.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 41.Boireau S, et al. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 44.Hereford LM, Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977;10:453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- 45.Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 46.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 49.Karpova TS, et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- 50.McNally JG, et al. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 51.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 52.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 53.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, et al. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guido NJ, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 57.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 58.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velculescu VE, et al. Analysis of human transcriptomes. Nat. Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.