Abstract
Metagenomic library was constructed from a deep-sea sediment sample and screened for lipolytic activity. Open-reading frames of six positive clones showed only 33–58% amino acid identities to the known proteins. One of them was assigned to a new group while others were grouped into Families I and V or EstD Family. By employing a combination of approaches such as removing the signal sequence, coexpression of chaperone genes, and low temperature induction, we obtained five soluble recombinant proteins in Escherichia coli. The purified enzymes had optimum temperatures of 30–35°C and the cold-activity property. Among them, one enzyme showed lipase activity by preferentially hydrolyzing p-nitrophenyl palmitate and p-nitrophenyl stearate and high salt resistance with up to 4 M NaCl. Our research demonstrates the feasibility of developing novel lipolytic enzymes from marine environments by the combination of functional metagenomic approach and protein expression technology.
1. Introduction
Lipolytic enzymes such as esterases and lipases belong to the class of carboxylic ester hydrolases that catalyze both the hydrolysis and synthesis of ester bonds. Lipolytic enzymes have been classified into eight families based on the conserved sequence motifs and biological properties [1]. They share a characteristic α/β hydrolase fold in the three-dimensional structure, but show differences in substrate preferences. Esterases (EC 3.1.1.1) hydrolyze water-soluble or emulsified esters with short-chain carboxylic acids (≤10 carbon atoms), while lipases (EC 3.1.1.3) prefer long-chain acylglycerides (≥10 carbon atoms) [2]. Esterases and lipases have a wide range of biotechnological applications, such as organic chemical processing, detergent formulation, synthesis of biosurfactants, the oleochemical industry, the dairy industry, the agrochemical industry, paper manufacturing, nutrition, cosmetics, and pharmaceutical processing [3–6]. Therefore, the identification of novel esterases/lipases will be a useful tactic for finding novel biocatalysts.
The metagenomic approach, direct cloning of genomes of all microorganisms present in a given habitat, can be accessing the potential of nonculturable microorganisms [7–10]. In detail, metagenomic libraries were constructed from DNA of diverse environmental samples in cloning vectors including cosmid, fosmid and bacterial artificial chromosome (BAC) [11–13] and host strains. Two major strategies have been pursued to identify novel biocatalysts or genes with new functions for biotechnological applications. The first approach uses function-based screenings of metagenomic DNA libraries, and the second one includes sequence-based searches [14–16]. Through functional screens of metagenomic libraries, several genes encoding lipolytic enzymes have been previously identified from various environmental samples [17–21].
We have been applying metagenomic approach to search for new lipolytic enzymes from marine environmental samples such as deep-sea and arctic seashore sediments, which possess untouched and potential resources [22–24]. Several novel esterases/lipases have been identified with unique properties including cold activity. In this study, we used another sediment core sample of deep sea which had been explored in our previous researches and could unveil the existence of novel lipolytic enzymes. Here, we describe the identification of novel lipolytic enzyme-encoding genes from metagenomic library, the enhancement of soluble protein expression, and the biochemical characterization of the purified enzymes.
2. Materials and Methods
2.1. Strains, Library Construction, and Screening Escherichia coli
DH5α (Stratagene, La Jolla, CA, USA), EPI300-T1R (Epicentre, Madison, WI, USA), and BL21(DE3) (Novagen, Madison, WI, USA) were used as host strains for cloning and expression. pBluescript SK- (Stratagene), pET-24a(+) vector (Novagen), and fosmid vector (Epicentre) were used as vectors.
2.2. Metagenomic Library Construction and Screening for Lipolytic Clones
Deep-sea sediment sample was collected from the southern clam beds area around the summit of Edison Seamount in the New Ireland Fore-arc near Papua New Guinea (3°89′S/152°49′E; depth 1,440 m). DNA from the sediment sample was extracted based on a previously described method [25] with minor modifications. After extraction, the DNA was further purified by gel electrophoresis in a 1% low-melting-temperature agarose gel (FMC Bioproducts, Rockland, ME) containing 1% polyvinyl polypyrrolidone (Sigma, St. Louis, MO). Gel electrophoresis was performed at 35 V for 13 h, and DNA fragments of approximately 40 to 50 kb were then isolated from the gel. The isolated DNA was end-repaired with End-It DNA End-Repair kit (Epicentre, Madison, WI), which caused the DNA to be blunt ended and 5′-phosphorylated. The blunt-ended DNA was ligated into a pCC1FOS vector (Epicentre, Madison, WI). Lambda packaging extracts were added to ligations, and infection of phage T1-resistant EPI300-T1R cells was performed according to the manufacturer's instructions. The E. coli transformants were transferred to 96-well microtiter plates and stored at −80°C. To screen for esterase/lipase activity, the transformants were plated on Luria-Bertani (LB) agar plates containing 12.5 μg/mL of chloramphenicol and 1% tributyrin as a substrate. Colonies were incubated for one day at 37°C and subsequently incubated for a week at 4°C. Candidates surrounded by a clear halo on the plate were selected. The positive clones were reconfirmed and subcloned.
Fosmid clones showing lipolytic activity on the tributyrin agar plate were inoculated into 200 mL of LB broth containing 12.5 μg/mL of chloramphenicol. After overnight incubation at 37°C, the cells were harvested by centrifugation at 5,000 ×g for 15 min and washed twice with distilled water. The fosmid DNA was purified using the alkaline lysis method [26] with minor modifications and was randomly sheared by nebulization according to the manufacturer's instruction (Invitrogen, Carlsbad, CA). After nebulization, DNA fragments of 2 to 4 kb were isolated from a 0.6% low-melting-temperature agarose (FMC Bioproducts, Rockland, ME) gel and end-repaired to generate blunt ends. The blunt-ended DNA was ligated into the EcoRV site of pBluescript SK(−) (Stratagene, La Jolla, CA), and the ligations were introduced into E. coli DH5α cells. The E. coli transformants were plated onto LB agar plates containing 100 μg/mL of ampicillin and 1% tributyrin. After incubation at 37°C for 24 h, colonies surrounded by a clear halo were selected. Nucleotide sequencing was performed with the automated sequencer (ABI3100) using the BigDye terminator kit (PE Applied Biosystems, Foster City, CA). The DNA sequence was determined by primer walking in both directions and assembled using the ContigExpress program of the Vector NTI suite 7 software package (InforMax, North Bethesda, Md.). The open reading frame (ORF) was detected using the ORF search tool provided by the National Center for Biotechnology Information (NCBI). Sequence homology searches were performed with the basic local alignment search tool (BLAST) program [27]. Signal sequence search was performed with the SignalP 3.0 program [28]. Multiple alignments between protein sequences were performed with the ClustalW program [29]. The phylogenetic tree was constructed by the neighbor-joining method [30] using the Molecular Evolutionary Genetics Analysis 4.1 software (MEGA, version 4.1) [31].
2.3. Overexpression and Purification of the Lipolytic Enzyme-Encoding Genes
The lipolytic enzyme genes were amplified by PCR with primer pairs, and the amplified DNA fragments were inserted into the pET-24a(+) expression vector (Table 1). Three recombinant plasmids including genes of EM3L1, EM3L2, and EM3L3 were transformed into E. coli BL21 (DE3) cells while three recombinant plasmids including genes of EM3L4, EM3L6, and EM3L7 were transformed into E. coli BL21 (DE3) expressing molecular chaperones GroEL-GroES with pGro7 (Takara, Kyoto, Japan) and the transformants were inoculated into LB medium containing 20 μg/mL of chloramphenicol and 50 μg/mL of kanamycin for plasmid selection and 0.5 mg/mL L-arabinose for induction of chaperone expression. A transformants were cultivated at 37°C, and 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce gene expression when the optical density at 600 nm reached 0.4. After incubation for 16 h at 16°C, the cells were harvested by centrifugation (6,000 ×g, 20 min, 4°C) and resuspended in 50 mM Tris-HCl buffer (pH 8.0) containing 0.1 M KCl and 10% glycerol. The cells were disrupted by sonication and centrifuged (20,000 ×g, 1 h, 4°C). The resulting supernatants were applied to a column of TALON metal-affinity resin (BD Biosciences Clontech, Palo Alto, CA) and washed with 10 mM imidazole (Sigma, St. Louis, MO) in 50 mM Tris-HCl buffer (pH 8.0) containing 0.1 M KCl and 10% glycerol, and the enzymes were eluted with 300 mM imidazole in the buffer. The protein concentration was measured by the Bradford method using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin as a standard [32]. The purity of the protein was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions as described method [33].
Table 1.
Primers used in the cloning of lipolytic enzyme-encoding genes.
| Gene | Primer* |
|---|---|
| EM3L1 | 5′-TTTGGAGGCATATGCTTATCCCTTCCGATGGTCTGGAAC-3′ |
| 5′-CTGGTGTCCTCGAGGTTTGCCAAAAAACGGCCGTATATC-3′ | |
| EM3L2 | 5′-TGATAAATCATATGAACAGAATGACAATAGGCTTCTC-3′ |
| 5′-CCTTTTAGCTCGAGATTATTGTCTTGCAACCAGGATT- 3′ | |
| EM3L3 | 5′-TAGCTGCACATATGATCCTTTTTAGCTTGGTCGGTGTC-3′ |
| 5′-AAAGTTGGCTCGAGTGTCTTAATCCCAAGAAAATTCAAG-3′ | |
| EM3L4 | 5′-GAGAAGTGAACCGGGGGACATATGACTGGTAGAATTG-3′ |
| 5′-CAAAAGCGCCCCTCGAGCGGCTGCTCCTCC-3′ | |
| EM3L6 | 5′-AGGAGAAGCATATGAAATGCATCCCATCAGACGGCC-3′ |
| 5′-GTTGCAGACTCGAGATCATTATTCGAGCCTAATTCCTC-3′ | |
| EM3L7 | 5′-CGGAGTGAAAGCATATGACTTATCCGATTGTGCTC-3′ |
| 5′-GTATTACCTCTCGAGGTTAAGGTAGTTGTTCCGCGATTC-3′ |
*Underlined bases in the primers indicate the restriction enzyme recognition site (NdeI/XhoI).
2.4. Characterization of Lipolytic Enzymes
Esterase and lipase activities were measured by a spectrophotometric method using p-nitrophenyl butyrate and p-nitrophenyl palmitate (Sigma, St. Louis, MO) as the substrate, respectively. The reaction mixture contained p-nitrophenyl butyrate in acetonitrile, Tris-HCl buffer, and the enzyme solution. p-nitrophenyl palmitate solutions were mixed with Tris-HCl buffer containing Triton X-100 as emulsifying agent. After incubation at each optimum temperature for 5 min, the absorbance at 405 nm was measured to detect the released p-nitrophenol. One unit of esterase and lipase activity was defined as the amount of enzyme required to release 1 μmol of p-nitrophenol from p-nitrophenyl butyrate or p-nitrophenyl palmitate per min.
The optimum temperature of the enzymes was determined at various temperatures of 5 to 65°C. The optimum pH was determined over a pH range of 4.0 to 10.0, using the following buffer systems: 50 mM sodium acetate (pH 4.0 to 6.0), 50 mM sodium phosphate (pH 6.0 to 7.5), 50 mM Tris-HCl (pH 7.5 to 8.5), and 50 mM CHES (pH 8.5 to 10.0). The substrate specificity was determined with different aliphatic side chain, C2 (acetate), C4 (butyrate), C6 (hexanoate), C8 (octanoate), C10 (decanoate), C12 (laurate), C14 (myristate), C16 (palmitate), and C18 (stearate) as substrates.
Various metal ions (MnCl2, MgCl2, CaCl2, CuCl2, ZnSO4, FeSO4, CoSO4, and NiSO4) and enzyme inhibitors, Phenylmethylsulfonyl fluoride (PMSF) and Ethylenediaminetetraacetic acid (EDTA), at a final concentration of 1 mM were added to the enzyme in 50 mM Tris-HCl buffer (pH 7.5) then assayed for enzyme activity after preincubation at 35°C for 1 hr. The effects of detergents on enzyme activity were determined by incubating the enzyme in 50 mM Tris-HCl buffer (pH 7.5) containing 1% (w/v) of the detergents SDS, Triton X-100, and Tween-20, -40, -60, -80 at 35°C for 1 hr.
The effect of NaCl concentration on enzyme activity was measured using p-nitrophenyl hexanoate as the substrate. The reaction mixture contained enzyme solution in 50 mM Tris-HCl (pH 7.5) containing different final NaCl concentrations ranging from 0.5 to 4 M. The mixture was preincubated at 35°C for 30 min, and then the enzyme activity was detected by adding p-nitrophenyl hexanoate.
2.5. Nucleotide Sequence Accession Numbers
The obtained nucleotide sequences have been deposited in the GenBank database under the accession numbers EM3L1 (GQ340923), EM3L2 (GQ340924), EM3L3 (GQ340925), EM3L4 (GQ340926), EM3L6 (GQ340927), and EM3L7 (GQ340928).
3. Results
3.1. Screening and Primary Sequence Analysis of Lipolytic Enzyme-Encoding Genes
To explore the untapped esterases/lipases in marine environment, metagenomic approach was applied to the samples from deep-sea sediment, located near a small volcanic cone named the Edison Seamount at a depth of 1440 m where there are extensive clam beds associated with the low-temperature vents [34]. Deep-sea sample was chosen to represent the psychrophilic environment with average temperature below 4°C. A metagenomic library consisting of 81,100 fosmid clones was constructed in a fosmid vector, pCC1FOS. Each clone contained an insert of approximately 15 to 33 kb. Fosmid clones having lipolytic activity were identified by activity screening on agar plates containing 1% tributyrin (TBN). As a result, 6 positive clones (designated as pFosEM3L1, pFosEM3L2, pFosEM3L3, pFosEM3L4, pFosEM3L6, and pFosEM3L7) were obtained by zones of clearance around the colonies.
The 6 fosmid clones were subjected to further subcloning into the pBluescript SK(−) plasmid. The subclones with lipolytic activity were selected on TBN plates again. Sequence analyses of the 2-3 kb DNA inserts in the subclones revealed the presence of ORFs encoding putative α/β hydrolases with G + C contents of 46.5–66.6%. A BLAST search of the amino acid sequences of six ORFs indicated that they yielded identities of less than 60% to proteins in the database. The deduced amino acid sequences of EM3L1, EM3L2, EM3L3, EM3L4, EM3L6, and EM3L7 showed the highest similarity to α/β hydrolase fold protein (YP_003395264) from Conexibacter woesei DSM 14684 (39% identity), hypothetical protein (ZP_01915829) from Limnobacter sp. MED105 (49% identity), triacylglycerol acyl hydrolase (AAK07450) from Moritella marina (49% identity), LpqC (ZP_01463024) from Stigmatella aurantiaca DW4/3-1 (39% identity), α/β hydrolase fold protein (ZP_01697334) from Bacillus coagulans 36D1 (33% identity), and lipase (ACJ13070) from uncultured bacterium (58% identity), respectively.
To see how the ORFs were related to known esterases/lipases, the phylogenetic relationship was analyzed based on the esterase/lipase classification (family I–VIII) by Arpigny and Jaeger [1] including recently reported families such as LipG [35], EstA [36], LipEH166 [20], FLS18 [18], and EstD2 [17] (Figure 1). EM3L1, EM3L3, and EM3L6 were grouped into family V in Figure 1, retaining the G-X-S-M-G pentapeptide motif and the HG sequence for the oxyanion hole (Figures 2(a) and 2(b)). The His residue consisting of the catalytic triad could not be predicted in EM3L1 and EM3L6 by the sequence alignment (Figure 2(a)). EM3L2 was clustered together with EstD2 reported from metagenomic library of plant rhizosphere soil recently [17]. Multiple alignment analysis of EM3L2 revealed that the active site serine was encompassed by the conserved pentapeptide G-H-S-Q-G in EstD2 family (Figure 2(c)).
Figure 1.
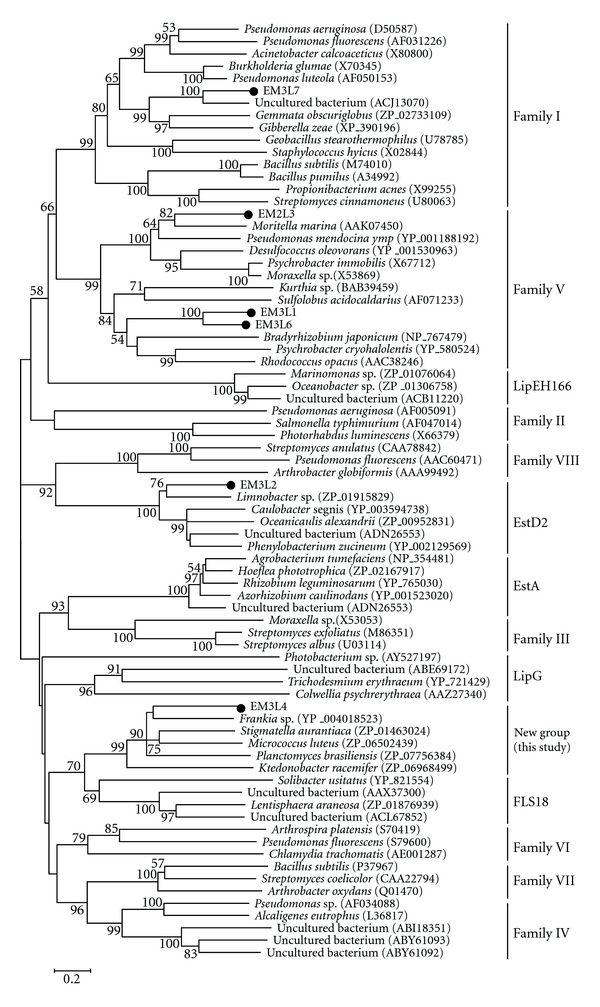
Phylogenetic tree of the lipolytic enzymes. The tree was constructed using the MEGA 4.1 program with the neighbor-joining algorithm. Only bootstrap values greater than 50% are shown. Bar: 0.2 substitutions per amino acid site.
Figure 2.
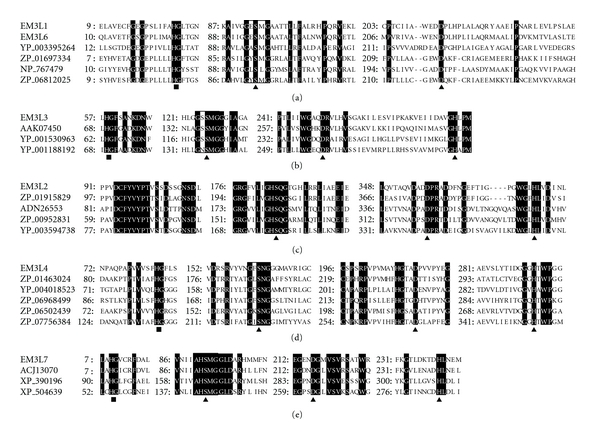
Multiple alignments of the conserved motifs of the ORFs isolated from metagenomic library with the homologs of each family or group. (a) Family V including EM3L1 and EM3L6: YP_003395264, an α/β hydrolase fold protein from Conexibacter woesei DSM 14684; ZP_01697334, an α/β hydrolase fold from Bacillus coagulans 36D1; NP_767479, a hypothetical protein bll0839 from Bradyrhizobium japonicum USDA 110; ZP_06812025, an α/β hydrolase fold protein from Geobacillus thermoglucosidasius C56-YS93. (b) Family V including EM3L3: AAK07450, a triacylglycerol acyl hydrolase from Moritella marina; YP_001530963, an α/β hydrolase fold from Desulfococcus oleovorans Hxd3; YP_001188192, an α/β hydrolase fold from Pseudomonas mendocina ymp (YP_001188192). (c) EstD2 family including EM3L2: ZP_01915829, a hypothetical protein from Limnobacter sp. MED105; ADN26553, an EstD2 from uncultured bacterium; ZP_00952831, a hypothetical protein from Oceanicaulis alexandrii HTCC2633; YP_003594738, a hypothetical protein from Caulobacter segnis ATCC 21756. (d) New group including EM3L4: ZP_01463024, an LpqC from Stigmatella aurantiaca DW4/3-1; YP_004018523, a lipoprotein from Frankia sp. EuI1c; ZP_06968499, a putative lipoprotein from Ktedonobacter racemifer DSM 44963; ZP_06502439, a conserved hypothetical protein from Micrococcus luteus SK58. (e) Family I including EM3L7: ACJ13070, a lipase from uncultured bacterium; (ACJ13070); XP_390196, a hypothetical protein from Gibberella zeae PH-1; XP_504639, a YALI0E31515p from Yarrowia lipolytica. Triangles and squares represent the residues involved in the formation of the catalytic triad and the oxyanion hole, respectively, and the conserved pentapeptide motifs are boxed.
On the other hand, it was found that EM3L4 did not show significant similarity to any member in the eight families and branched out to a group together with close homologous genes. It is noteworthy that the comparative analysis of the deduced amino acid sequence revealed that EM3L4 displayed low similarity to poly(3-hydroxyalkanoate) depolymerase (ZP_07602911) from Streptomyces violaceusniger (34% identity), and both the active site serines in the two proteins were encompassed by G-X-S-N-G pentapeptide in common (Figure 2(d)). From the sequence analysis, it seems likely that EM3L4 could be a novel enzyme (Figure 1). Lastly, EM3L7 was clustered together with family I bacterial lipolytic enzymes, and the active site serine was encompassed by the characteristic A-X-S-X-G motif (Figure 2(e)). The motif was usually found in Bacillus lipases belonging to Subfamily I.4 even if no significant homology (less than 14% identity) was found between them. Taken together, the metagenomic study applied to marine sediment sample of deep sea has the potential to yield novel molecular entities unrelated to the known sequences.
3.2. Expression and Characterization of the Lipolytic Enzyme-Encoding Genes
To express six genes, EM3L1, EM3L2, EM3L3, EM3L4, EM3L6, and EM3L7, we investigated the presence of signal sequences using the SignalP 3.0 program and found that EM3L2, EM3L3, and EM3L4 retained a putative signal sequence 18–25 amino acids long. All the genes with signal sequence removed were amplified, and the resulting expression constructs were expressed in E. coli BL21 strain. Total cell lysates, soluble fractions, and insoluble fractions were analyzed by 12% SDS-PAGE, and we found that some portion of the proteins encoded by three of the genes (EM3L1, EM3L2, and EM3L3) were detected in the soluble fraction (Figure 3(a)), whereas the proteins encoded by the three genes (EM3L4, EM3L6, and EM3L7) were expressed as insoluble forms.
Figure 3.
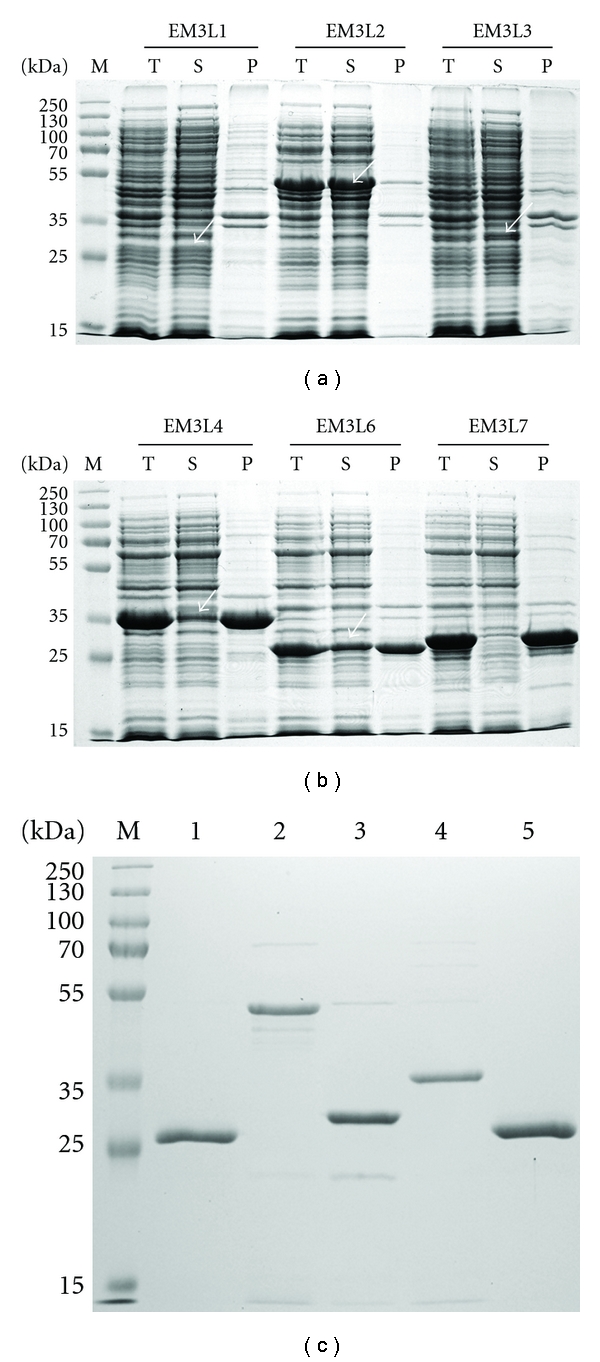
Expression of esterase/lipase genes. (a) Expression analysis of EM3L1, EM3L2, and EM3L3; (b) expression analysis of EM3L4, EM3L6, and EM3L7 with co-expression of the GroEL-GroES chaperone genes at 16°C; (c) purified enzymes. The bands corresponding to the proteins are indicated by arrows. Lane M: molecular mass standards, lane T: total cell lysate, lane S: soluble fraction, lane P: insoluble fraction, lane 1: purified EM3L1, lane 2: purified EM3L2, lane3: purified EM3L3, lane 4: purified EM3L4, and lane 5: purified EM3L6.
Based on the fact that expression at low temperature increases the stability and proper folding of proteins, possibly due to the fact that the hydrophobic interactions that determine inclusion body formation are temperature dependent [37–39], we made three proteins (EM3L4, EM3L6, and EM3L7) being induced at the low temperature of 16°C. However, the induction of expression at low temperature alone was not successful in enhancing the solubility of the expressed proteins (data not shown). Since it has been reported that GroEL-GroES is effective in facilitating their correct folding by minimizing aggregation and misfolding [40], we employed GroEL-GroES co-expression in solubilizing the proteins. It turns out that EM3L4 and EM3L6 proteins were solubly expressed by coexpressing GroEL-GroES at 16°C. In contrast to the other proteins, the majority of the expressed EM3L7 remained in the insoluble fraction regardless of the experimental conditions we tried in this study (Figure 3(b)). The expressed proteins were purified as described in Section 2, and SDS-PAGE analysis of purified EM3L1, EM3L2, EM3L3, EM3L4, and EM3L6 showed a protein band which correlated well to the predicted molecular weight (Figure 3(c)). This result demonstrated that the combination of co-expression of chaperones and induction at low temperature may be effective in achieving soluble expression of lipolytic enzyme-encoding genes.
The purified enzymes showed an optimum activity in the temperature range of 30–35°C and at a neural or slightly alkaline pH (pH 7.5–8.5) (Table 2 and see Figures S1 and S2 in Supplementry Material available on line at doi:10.1155/2011/271419). It is noteworthy that they could be classified as cold-active enzymes based on the temperature profiles and activation energy values. As the source of the genes was a deep-sea sediment sample, we predicted that they are cold-adapted enzymes. A similar phenomenon has been reported in other studies; for example, two cold-adapted esterase from Arctic sediment metagenome showed an optimum temperature at 30°C [23] and thermostable esterase from hot spring metagenome showed enzyme activity above 30°C up to 95°C [41]. This result suggested that the property of lipolytic enzymes derived from metagenomic library might be reflected by their environmental characters.
Table 2.
Characteristics of the purified lipolytic enzymes.
| Enzyme | Temperaturea
(optimum) (°C) |
Activation energy (kcal/mol) |
pHb
(optimum) |
Substratec
(optimum) |
Specific activity (U/mg) |
|---|---|---|---|---|---|
| EM3L1 | 15–50 (35) | 3.6 | 7.5–9.5 (8.5) | C2–C10 (C6) | 558.2 |
| EM3L2 | 15–40 (30) | 8.6 | 7.0–10.0 (8.5) | C6 (C6) | 1.7 |
| EM3L3 | 15–40 (35) | 3.5 | 7.0–10.0 (8.5) | C2–C10 (C6) | 29.7 |
| EM3L4 | 20–50 (35) | 10.6 | 7.0–10.0 (7.5) | C16–C18 (C16) | 5.3 |
| EM3L6 | 15–40 (35) | 4.1 | 7.5–9.0 (8.5) | C2–C10 (C6) | 23.3 |
aThe temperature range is the temperatures at which the activities are greater than 50% of the highest value (Figure S1). bpH range is the pHs at which the activities are greater than 50% of the highest value (Figure S2). cSubstrates are the p-nitrophenyl esters for which the enzyme shows an activity greater than 20% of the highest value (Figure S3).
EM3L1, EM3L2, EM3L3, and EM3L6 could hydrolyze short chain substrates (C2 to C12) and showed the highest activity toward p-Nitrophenyl hexanoate (C6) (Table 2 and Figure S3). On the other hand, EM3L4 showed enzymatic activity toward long-chain substrates (C16–C18), which can be classified as a lipase. The specific activities of the enzymes were determined to be in the range of 1.7–558.2 U/mg at the optimal conditions (Table 2).
Since EM3L4 belonged to a new group and the homologous proteins have never been characterized, the biochemical property of EM3L4 was further analyzed. To determine the resistance to various chemical agents, the purified EM3L4 was incubated with chemical agents, and the remaining activity was measured with p-nitrophenyl palmitate or p-nitrophenyl hexanoate as substrate at 35°C. The activity of EM3L4 was increased by the presence of Mn2+, Mg2+, Ca2+, Cu2+, and Ni2+(Table 3). It was inhibited by Zn2+, Fe2+, and Co2+ and was completely inhibited by PMSF (Table 3). Besides, it was also inhibited by various nonionic detergents such as Tween-20, -40, -60, -80 and Triton X-100 and ionic detergent, SDS (Table 3). The enzyme activity of EM3L4 was significantly affected by salinity. As shown in Figure 4, EM3L4 displayed the highest enzyme activity in the presence of 1 M NaCl. Moreover, the activity was maintained with up to 4 M NaCl (Figure 4).
Table 3.
Effect of metal ions and detergents on EM3L4.
| Metal ions (1 mM) | Relative activity (%) | Detergent (1%) | Relative activity (%) |
|---|---|---|---|
| None | 100 | None | 100 |
| MnCl2 | 147 | Tween 20 | 74 |
| MgCl2 | 175 | Tween 40 | 70 |
| CaCl2 | 175 | Tween 60 | 54 |
| CuCl2 | 165 | Tween 80 | 69 |
| ZnSO4 | 52 | Triton X-100 | 92 |
| FeSO4 | 0 | SDS | 0 |
| CoSO4 | 35 | ||
| NiSO4 | 112 | ||
| PMSF | 42 | ||
| EDTA | 107 |
Figure 4.
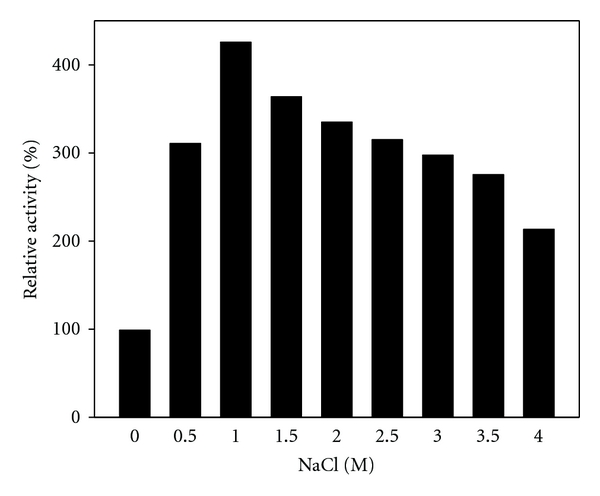
Effect of NaCl on p-nitrophenyl hexanoate hydrolysis activity of EM3L4. The activity of the enzyme preparation in the absence of NaCl before incubation was defined as the 100% level.
4. Discussion
Our approach to search for novel lipolytic enzymes started with the construction of metagenomic library from deep sea sediment. As a result, we could discover six lipolytic enzyme-encoding genes with low sequence similarity (33–58% identity) to the known proteins. By removing signal sequence in the N-terminus and co-expression of chaperone gene at low temperature, we could obtain soluble recombinant proteins encoded by 5 genes from E. coli. The biochemical characterization of them revealed being cold-adapted, reflecting the environmental feature of the origin of metagenome.
Particularly, EM3L4 branched out to a new group in the phylogenetic tree, and any of homologous proteins to EM3L4 has never been characterized. The purified EM3L4 preferred to hydrolyze longer fatty acids, and highly active at high NaCl concentration. Recently, some esterases whose activities were increased in the presence of NaCl have been identified using metagenomic libraries of various marine environments such as deep-sea water [42], sea water [36], and sea sediment [18]. We suggest that EM3L4 is a novel lipase retaining salt-resistance property. Then, a question arises whether homologous proteins belonging to the same group with EM3L4 show similar properties, and it will need further investigation.
Conclusively, this study demonstrated that novel lipolytic enzymes in terms of primary sequence, activity profile, and substrate specificity could be obtained by metagenomic approach using deep-sea sediment sample. They could be potentially used as a biocatalyst in pharmaceutical and fine chemical industries.
Acknowledgments
This work was supported by the KORDI in-house program (PE98513) and the Marine and Extreme Genome Research Center Program and the Development of Biohydrogen Production Technology Using Hyperthermophilic Archaea Program of the Ministry of Land, Transport, and Maritime Affairs, Republic of Korea.
References
- 1.Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochemical Journal. 1999;343(1):177–183. [PMC free article] [PubMed] [Google Scholar]
- 2.Nardini M, Dijkstra BW. α/β hydrolase fold enzymes: the family keeps growing. Current Opinion in Structural Biology. 1999;9(6):732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 3.Jaeger KE, Dijkstra BW, Reetz MT. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annual Review of Microbiology. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- 4.Jaeger KE, Eggert T. Lipases for biotechnology. Current Opinion in Biotechnology. 2002;13(4):390–397. doi: 10.1016/s0958-1669(02)00341-5. [DOI] [PubMed] [Google Scholar]
- 5.Pandey A, Benjamin S, Soccol CR, Nigam P, Krieger N, Soccol VT. The realm of microbial lipases in biotechnology. Biotechnology and Applied Biochemistry. 1999;29(2):119–131. [PubMed] [Google Scholar]
- 6.Sharma R, Chisti Y, Banerjee UC. Production, purification, characterization, and applications of lipases. Biotechnology Advances. 2001;19(8):627–662. doi: 10.1016/s0734-9750(01)00086-6. [DOI] [PubMed] [Google Scholar]
- 7.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & Biology. 1998;5(10):R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 8.Schloss PD, Handelsman J. Biotechnological prospects from metagenomics. Current Opinion in Biotechnology. 2003;14(3):303–310. doi: 10.1016/s0958-1669(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 9.Steele HL, Streit WR. Metagenomics: advances in ecology and biotechnology. FEMS Microbiology Letters. 2005;247(2):105–111. doi: 10.1016/j.femsle.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 11.Rondon MR, August PR, Bettermann AD, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Applied and Environmental Microbiology. 2000;66(6):2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Entcheva P, Liebl W, Johann A, Hartsch T, Streit WR. Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia. Applied and Environmental Microbiology. 2001;67(1):89–99. doi: 10.1128/AEM.67.1.89-99.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Béjà O, Koonin EV, Aravind L, et al. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Applied and Environmental Microbiology. 2002;68(1):335–345. doi: 10.1128/AEM.68.1.335-345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiology and Molecular Biology Reviews. 2004;68(4):669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streit WR, Daniel R, Jaeger KE. Prospecting for biocatalysts and drugs in the genomes of non-cultured microorganisms. Current Opinion in Biotechnology. 2004;15(4):285–290. doi: 10.1016/j.copbio.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Streit WR, Schmitz RA. Metagenomics—the key to the uncultured microbes. Current Opinion in Microbiology. 2004;7(5):492–498. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Lee MH, Hong KS, Malhotra S, et al. A new esterase EstD2 isolated from plant rhizosphere soil metagenome. Applied Microbiology and Biotechnology. 2010;88(5):1125–1134. doi: 10.1007/s00253-010-2729-6. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Fu C, Huang Y, et al. Novel lipolytic genes from the microbial metagenomic library of the South China Sea marine sediment. FEMS Microbiology Ecology. 2010;72(2):228–237. doi: 10.1111/j.1574-6941.2010.00851.x. [DOI] [PubMed] [Google Scholar]
- 19.Wei P, Bai L, Song W, Hao G. Characterization of two soil metagenome-derived lipases with high specificity for p-nitrophenyl palmitate. Archives of Microbiology. 2009;191(3):233–240. doi: 10.1007/s00203-008-0448-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim EY, Oh KH, Lee MH, Kang CH, Oh TK, Yoon JH. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Applied and Environmental Microbiology. 2009;75(1):257–260. doi: 10.1128/AEM.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tirawongsaroj P, Sriprang R, Harnpicharnchai P, et al. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. Journal of Biotechnology. 2008;133(1):42–49. doi: 10.1016/j.jbiotec.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 22.Jeon JH, Kim JT, Kim YJ, et al. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Applied Microbiology and Biotechnology. 2009;81(5):865–874. doi: 10.1007/s00253-008-1656-2. [DOI] [PubMed] [Google Scholar]
- 23.Jeon JH, Kim JT, Kang SG, Lee JH, Kim SJ. Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Marine Biotechnology. 2009;11(3):307–316. doi: 10.1007/s10126-008-9145-2. [DOI] [PubMed] [Google Scholar]
- 24.Park HJ, Jeon JH, Kang SG, Lee JH, Lee SA, Kim HK. Functional expression and refolding of new alkaline esterase, EM2L8 from deep-sea sediment metagenome. Protein Expression and Purification. 2007;52(2):340–347. doi: 10.1016/j.pep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Hurt RA, Qiu X, Wu L, et al. Simultaneous recovery of RNA and DNA from soils and sediments. Applied and Environmental Microbiology. 2001;67(10):4495–4503. doi: 10.1128/AEM.67.10.4495-4503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bimboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research. 1979;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 32.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Herzig P, Hannington M, McInnes B, et al. Submarine volcanism and hydrothermal venting Studied in Papua, New Guinea. EOS. 1994;75(44):513–520. [Google Scholar]
- 35.Lee MH, Lee CH, Oh TK, Song JK, Yoon JH. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacterial lipases. Applied and Environmental Microbiology. 2006;72(11):7406–7409. doi: 10.1128/AEM.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu X, He H, Guo C, Sun B. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Applied Microbiology and Biotechnology. 2008;80(4):615–625. doi: 10.1007/s00253-008-1566-3. [DOI] [PubMed] [Google Scholar]
- 37.Niiranen L, Espelid S, Karlsen CR, et al. Comparative expression study to increase the solubility of cold adapted Vibrio proteins in Escherichia coli . Protein Expression and Purification. 2007;52(1):210–218. doi: 10.1016/j.pep.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Vera A, González-Montalbán N, Arís A, Villaverde A. The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnology and Bioengineering. 2007;96(6):1101–1106. doi: 10.1002/bit.21218. [DOI] [PubMed] [Google Scholar]
- 39.Chesshyre JA, Hipkiss AR. Low temperatures stabilize interferon α-2 against proteolysis in Methylophilus methylotrophus and Escherichia coli . Applied Microbiology and Biotechnology. 1989;31(2):158–162. [Google Scholar]
- 40.Braig K. Chaperonins. Current Opinion in Structural Biology. 1998;8(2):159–165. doi: 10.1016/s0959-440x(98)80033-x. [DOI] [PubMed] [Google Scholar]
- 41.Rhee JK, Ahn DG, Kim YG, Oh JW. New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Applied and Environmental Microbiology. 2005;71(2):817–825. doi: 10.1128/AEM.71.2.817-825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrer M, Golyshina OV, Chernikova TN, et al. Microbial enzymes mined from the Urania deep-sea hypersaline anoxic basin. Chemistry & Biology. 2005;12(8):895–904. doi: 10.1016/j.chembiol.2005.05.020. [DOI] [PubMed] [Google Scholar]


