Abstract
A proper balance between synthesis, maturation and degradation of cellular proteins is crucial for cells to maintain physiological functions. The costly process of protein synthesis is tightly coupled to energy status and nutrient levels by the mammalian target of rapamycin (mTOR), whereas the quality of newly synthesized polypeptides is largely maintained by molecular chaperones and the ubiquitin-proteasome system. There is a wealth of evidence indicating close ties between the nutrient signaling pathway and the intracellular stress response. Dysregulation of both systems has been implicated in aging and age-associated pathologies. In this review, we describe molecular mechanisms underlying the connection between mTOR and the chaperone network and discuss the importance of their functional interaction in growth and aging.
Key words: target of rapamycin, stress response, ribosome, chaperone, translation, folding, degradation, aging
Introduction
Protein homeostasis—a delicate equilibrium between synthesizing proteins, maintaining protein conformations, refolding misfolded proteins and removing damaged proteins—is normally maintained by cellular networks involving ribosomes, chaperones and the ubiquitin-proteasome system. Ribosomes decode genetic information and convert it into the amino acid sequences of proteins.1,2 Molecular chaperones govern the integrity of the proteome as “cellular lifeguards.”3,4 On the other hand, the ubiquitin-proteasome system removes damaged proteins from cells.5,6 Protein synthesis has a defining role in forming the proteome and promotes cell growth, whereas failure to eliminate misfolded proteins can lead to the formation of toxic aggregates, inactivation of functional proteins and, ultimately, cell death. Growing evidence indicates that loss of defense against proteotoxic stress often leads to age-related diseases, such as type-2 diabetes, cardiovascular diseases, cancer and neurodegenerative disorders.7,8 Cells also experience stress as a result of nutrient excess. As a result, excessive nutrients have caused an obesity epidemic while promoting carcinogenesis and shortening life span.9,10
During the past decade, it has become clear that the rate of aging, like many other processes in biology, is subject to regulation.11 In many animals, longevity is regulated by a conserved insulin and insulin-like growth factor-1 (IGF-1) signaling pathway. Most recent studies have weaved the mammalian target of rapamycin (mTOR) into this intricate tapestry of the insulin/IGF-1 signaling network.12–14 mTOR is a well-conserved serine/threonine protein kinase that controls protein synthesis, cell growth and metabolism by integrating both extracellular cues and intracellular energy status. In many organisms, decreased TOR signaling is associated with the extension of life span.15 Interestingly, these longevity benefits are accompanied by increased stress resistance.16 These findings suggest an intimate connection between the nutrient signaling and the stress response. Much of the current knowledge about the chaperone network and mTOR signaling remains limited to their own context. In this review, we attempt to explore the functional connection between mTOR and stress response pathways and discuss the physiological implications in maintaining protein homeostasis.
Chaperone Network and Stress Response
Molecular chaperones are ubiquitous, highly conserved proteins, and are key elements of the maintenance of protein homeostasis in cells.4 The main chaperone families are HSP70, HSP90, HSP104, HSP40 (DnaJ) and small HSPs (HSP27, α-crystallins). Chaperones have diverse roles in regulating protein conformation and are essential in protecting nascent polypeptide chains from misfolding by facilitating co- and post-translational folding, assisting in assembly and disassembly of macromolecular complexes and regulating translocation.17 A defining characteristic of chaperones is to hold substrates by binding hydrophobic residues, a necessary step to suppress misfolding and aggregation of the proteins, allowing them to remain competent for subsequent folding. Molecular chaperones also facilitate degradation of misfolded polypeptides by the ubiquitin-proteasome system when the polypeptides cannot be refolded.18
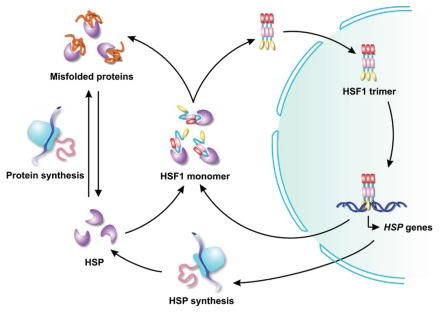
Despite the abundance and apparent capacity of chaperones to restore folding equilibrium in cells, the concentration of chaperones is titrated closely to the folding requirements within a specific cell type.19 Under conditions that cause widespread damage to cell proteins (e.g., heat shock), the cell's need for these chaperones increases further. The stress response or heat shock response enables the cell to elevate the expression of chaperone and to protect against the accumulation of misfolded proteins. In mammalian cells, the heat shock transcription factor 1 (HSF1) is the major transcriptional regulator of stress response.20 Under normal conditions, HSF1 is sequestered in the cytosol in a complex with molecular chaperones. During conditions that perturb cytosolic protein-folding homeostasis, the chaperones are diverted elsewhere. The released HSF1 then trimerizes and enters the nucleus, where it rapidly drives transcription of numerous genes involved in protein folding, refolding, degradation of misfolded proteins and other proteins that regulate stress tolerance (Fig. 1). Thus, the activation status of HSF1 is governed in large part by the balance between the amount of misfolded proteins and the chaperone availability in cells.21 As chaperones are growth-regulated and stress-responsive, they afford the cell with a stress sensor to link stress signaling processes with protein homeostasis.
Figure 1.
HSF1-mediated stress response. The conversion between inactive HSF1 monomer and active HSF1 trimer is controlled by chaperone availability in cells. Chaperone availability, on the other hand, is governed by the balance between protein synthesis and folding requirement. Stress conditions, such as heat shock, lead to the accumulation of misfolded proteins, which subsequently triggers the stress response in order to achieve protein homeostasis.
The protein components of eukaryotic cells face acute and chronic challenges to their integrity. Transient cellular adaptation of protein homeostasis is essential because of an everchanging proteome during development and the presence of new proteins and the accumulation of misfolded proteins upon aging.22 Aging challenges proteome homeostasis because of decreasing chaperone capacity and increasing protein damage. It has been reported that protein oxidation, damage, misfolding and aggregation together with the simultaneously impaired function and induction of chaperones in aged organisms disturb the balance between chaperone requirements and availability.23 The HSF1-mediated stress signaling provides a compelling example for a stress-resistance mechanism that regulates lifespan. HSF1 overexpression induces longevity, while HSF1 knockout shortens life span in C. elegans.24 Moreover, HSF1 is indispensible to yield lifespan extension in classical long-lived insulin-signaling mutants.25,26 Thus, a robust stress response is required for lifespan extension in these organisms.
mTOR Signaling Pathway
Coupling of the availability of nutrients and growth factors to cellular growth is essential for all organisms. mTOR, a highly conserved serine/threonine kinase, is a central regulator of cell growth and metabolism in eukaryotes.12 mTOR activates cell growth in response to nutrients (amino acids), growth factors (insulin and insulin-like growth factor) and cellular energy status (ATP) (Fig. 2). The core kinase mTOR is present in two functionally and structurally distinct multiprotein complexes termed TOR complex 1 (TORC1) and TOR complex 2 (TORC2).13,14 In mammals, the rapamycin-sensitive mTORC1 consists of mTOR, raptor and mLST8. Recent work reveals that PRAS40 and Deptor are components as well as substrates of mTORC1.27,28 mTORC2 contains mTOR, Rictor, mSIN1, mLST8 as well as Deptor.29–32 In contrast to mTORC1, mTORC2 is not directly inhibited by rapamycin. In the past decade, mTORC1 has been acknowledged for controlling many cellular processes that ultimately determine cell growth including protein synthesis, ribosome biogenesis, nutrient transport and autophagy. The two best-characterized substrates of mTORC1 are S6 kinase (S6K) and 4E-BP1, via which mTORC1 controls protein synthesis.29,30,32 Phosphorylation of 4E-BP1 by mTORC1 results in its dissociation from eIF4E, promoting assembly of the eIF4F complex to allow proper initiation. S6K1 phosphorylation promotes protein synthesis and cell growth, presumably by phosphorylating multiple substrates (e.g., ribosomal protein S6, translational regulators eIF4B and PDCD4). In this way, S6K enhances the overall translation capacity of cells.33
Figure 2.
mTOR signaling pathway. mTORC1 and mTORC2 receive distinct upstream signals from both extracellular and intracellular stimuli and have different downstream targets. Notably, interconnection between mTORC1 and mTORC2 occurs at several levels. For example, one of the mTORC2 downstream targets, Akt, serves as an upstream regulator of mTORC1, whereas one important mTORC1 target ribosome acts as an activator of mTORC2. Most of the signaling components are involved in growth and aging, and lots of mutations have been found in human diseases such as cancer. For simplicity, only typical mTOR components are shown. See the main text for details.
Growth factors regulate mTORC1 activity via either the PI3K/Akt pathway or the Ras/MAPK pathway, which converge on the Tuberous Sclerosis heterodimeric complex (TSC1–TSC2) to inhibit the GTPase activating (GAP activity) toward the small GTPase Rheb.14 Cells lacking a functional TSC-Rheb-GAP exhibit constitutive activation of mTORC1 signaling, and this is not increased further by insulin. The mechanism by which Rheb-GTP controls the catalytic competence of the mTORC1 is not fully understood. However, it is generally believed that the activation of mTORC1 involves two indistinguishable steps: first, a Rheb-GTP-induced activation of the mTOR catalytic function and second, recruitment of the raptor-associated substrate.34 This feature implies that a precise regulation of mTORC1 assembly is crucial for signaling transmission.
Many conditions that shift cells from states of nutrient utilization and growth to states of cell maintenance extend lifespan. The TOR pathway has warranted increased attention from the aging-research community due to its highly conserved influence on life span in a number of organisms. Decreased TOR signaling (using RNAi or a hypomorphic TOR mutant) has been shown to extend life span in the nematode C. elegans.35 Likewise, overexpression of a dominant-negative allele of TOR or inhibitors of TOR (Tsc1 and Tsc2) extends life span in Drosophila.36 Deletion of the S. cerevisiae TOR1 gene was shown to increase replicative life span.37 A recent high-throughput screen for gene deletions that extend chronological life span yielded a number of genes involved in nutrient sensing and that are influenced in part by the TOR pathway.38 In addition, it was shown that treatment of stationary phase yeast culture with rapamycin, a specific inhibitor of TOR, also extends chronological life span.38 Most recently, mice fed with a diet containing rapamycin also demonstrated longevity benefit.39 In fact, cellular growth and aging share a common molecular mechanism.40 It was demonstrated that mTOR drives cellular aging and rapamycin prevented conversion of cell cycle arrest into senescence.41,42 Thus mTOR overactivation is involved in both cellular senescence, organismal aging and age-related diseases.43
mTOR-Mediated Translational Regulation of Hsp70
It has been well established that hyperactive mTORC1 signaling enhances global protein synthesis. Uncontrolled protein synthesis and dysfunctional nutrient sensing challenge the integrity of protein homeostasis. A recent study reported that mouse embryonic fibroblasts (MEFs) lacking TSC induced unfolded protein response (UPR) in the endoplasmic reticulum (ER).44 It has been suggested that hyperactive mTOR activity triggers the stress response, because higher levels of protein synthesis increased the cellular load of erroneously synthesized polypeptides. To our surprise, we observed a defective cytosolic stress response in these cells.45 Despite the upregulated HSF1 transcriptional activity, there is a clear deficiency in heat shock-induced Hsp70 expression in MEFs lacking TSC2. It was not due to the lack of Hsp70 mRNA. Rather, the Hsp70 mRNA failed to undergo selective translation under stress conditions. In addition, Hsp70 expression is also significantly reduced in cells overexpressing Rheb, the upstream positive regulator of mTORC1.46 Importantly, decreasing mTORC1 signaling by raptor knockdown or PI3K inhibition augments the heat shock-induced Hsp70 expression. These findings provide an explanation for why unrestrained mTORC1 signaling is always accompanied by reduced stress resistance. Conversely, decreasing PI3K-mTOR signaling potentially enhances the stress response by promoting Hsp70 expression, thereby increasing the availability of proteolytic and chaperone functions that may contribute to the observed increase in organism stress resistance and lifespan.
What's the molecular mechanism then? It has long been known that some cellular proteins continue to be synthesized under stress conditions where global translation is severely compromised.47,48 One prominent example is the selective translation of heat shock proteins (HSPs) under stress conditions.49 An important mode of translational regulation during stress is the selective recruitment of mRNAs through the internal ribosome entry site (IRES).50,51 Accumulating evidence has supported the notion that mTORC1 signaling, while promoting cap-dependent mRNA translation, suppresses IRES-mediated translation.52 However, no IRES activity has been validated in the Hsp70 mRNA 5′ untranslated region (5′UTR).53 Despite the lack of IRES feature for Hsp70 5′ UTR, we confirmed that the selective translation of Hsp70 mRNA occurs via the cap-independent mechanism.45 It remains obscure how the 5′ UTR of Hsp70 mRNA drives the cap-independent translation without acting as an IRES. Another interesting question is how Hsp70 mRNA adopts the cap-independent translation, when all the eukaryotic mRNAs are synthesized in a capped form. Most recently, it has been reported that the expression of several decapping enzymes was enhanced during heat stress.54 This phenomenon could lead to the selective translation of Hsp70 mRNA due to the unique features of the Hsp70 5′UTR in mediating capindependent translation. In summary, the stress-induced switch between cap-dependent and cap-independent translation plays an important role in cellular adaptation under adverse conditions. Dysregulated mTORC1 signaling could cripple the stress response necessary for cell survival.
Hsp90-Mediated Regulation of mTOR
It is not surprising that the interaction between stress response and nutrient signaling pathways could be a two-way communication. Adverse environmental and metabolic conditions (including nutrient limitation, hypoxia and DNA damage) causes a decrease in mTORC1 signaling.55 In addition to conserving cellular energy, the reduction in translation that accompanies a decrease in mTORC1 activity also prevents the synthesis of unwanted proteins that could interfere with the stress response. However, the effect of stress conditions on mTORC1 signaling appears to be ambiguous, varying from exposure length, dose/concentration and time between stimulus and assay. Each stress condition may have distinct effects on mTOR-dependent cellular events.56 Several stress stimuli, such as UV light exposure, H2O2 addition, heat shock and fluid sheer stress, have been shown to cause an initial increase of mTOR-dependent S6K phosphorylation, followed by decrease in mTORC1 activity occurring after prolonged or severe exposure.57–59 The initial increase in mTORC1 signaling upon imposition of stress should not be viewed as the cell misinterpreting a life-threatening insult. Rather, this feature enables the cell to distinguish physiological fluctuations in the quality of translational products from the devastating accumulation of misfolded proteins. It is generally accepted that global translation is suppressed in response to severe stress conditions, but little is known about how cells respond to fluctuations in the quality of protein products. Like the variation of nutrients availability in the environment, there is a wide fluctuation in the quality of synthesized proteins during cell growth and an organism's development.
mTOR forms a huge complex (up to ∼2 MDa) with other subunits in order to achieve signal transmission. These multicomponent complexes present special challenges for adjusting signaling pathways, which must sense both increasing and decreasing signal intensities, in order to terminate and reinitiate signaling efficiently.60 A dynamic mode of mTORC1 complexes would fulfill this regulatory requirement. According to this scheme, termination of mTORC1 signaling would not require a separate mechanism, but rather would be an intrinsic consequence of continuous disassembly of mTORC1. Hsp90 is an optimal candidate to mediate the dynamic remodeling.61 In general, Hsp90 is highly abundant and has evolved to act promiscuously and associate with client proteins.62 Indeed, mTOR and other regulator components like raptor are chaperone “clients,” as evidenced by their selective binding to Hsp90 when not forming a complex.63 We propose that chaperone-mediated mTORC1 disassembly coordinates with nutrients signaling-triggered assembly. Supporting this notion, we observed a complete resistance to insulin stimulation in cells lacking HSF1 or in the presence of an Hsp90 inhibitor such as geldanamycin.63 This mechanism enables mTORC1 to rapidly detect and respond to environmental cues while also sensing intracellular protein misfolding. The tight linkage between protein quality and quantity control provides a plausible mechanism coupling protein misfolding with metabolic dyshomeostasis.
mTOR-Controlled Ribosome Biogenesis
An important prerequisite for the regulation of global protein synthesis is the continuous supply of translation machinery. Therefore, ribosome biogenesis has to meet the demand for protein synthesis. In eukaryotic cells, the ribosome biogenesis is tightly controlled based on the availability of growth factors and nutrients.33,47,64 As growth conditions change, cells must accurately and rapidly rebalance ribosome production with the availability of resources, because ribosome biogenesis accounts for a large segment of total energy consumption. In particular, cells must limit the production of ribosomes when nutrients are limited. The fact that mTORC1 signaling controls ribosome biogenesis at many levels underscores the involvement of mTOR in linking nutrient availability to the biosynthesis of ribosomes. In yeast, research has shown that TORC1 regulates transcription factors for RP genes, mostly by affecting sub-cellular localization in response to environmental cues.65 Also, rapamycin treatment promotes rDNA chromatin remodeling to reduce transcription.66 Studies have further identified mTORC1-mediated regulation of Pol III transcription.67 Another recent study revealed that endogenous mTORC1 directly associates with a Pol I and Pol III transcription factor, TFIIIC.68 This direct interaction allows for a rapid response to changes in cell homeostasis at the transcriptional level to affect global protein synthesis.
Ribosome biogenesis is most notably controlled at the level of translation. A common feature of RP transcripts is the presence of a characteristic 5′UTR: an uninterrupted sequence of 6–12 pyrimidines at the 5′ end called the 5′-terminal oligopyrimidine (5′TOP) sequence.69,70 The 5′TOP motif is necessary for a growth-associated translational regulation of RP mRNAs. As a consequence, the translation of RP is poor in quiescent cells but is strongly and rapidly activated on serum refeeding, by insulin and by amino acid availability.70 Despite years of study, how the 5′TOP sequence regulates the translation initiation remains unclear. It was initially suggested that the translational activation of TOP mRNAs is controlled by S6Ks. However, the role of S6Ks in ribosome biogenesis has been challenged in a series of recent studies.71,72 Thus, the mechanism of translational regulation of TOP mRNAs remains a mystery. It has been reported that the 5′TOP is recognized by specific transacting factors.69 Yet, the functional significance of some TOP-interacting proteins remains to be confirmed.73,74 More surprisingly, rapamycin treatment does not seem to affect the polysome formation of TOP mRNAs.75 These observations raise the question of how mTORC1 signaling mediates the translational control of ribosome biogenesis. Current opinion is that regulation of TOP mRNAs is unlikely to be by a straightforward repressor mechanism. A recent study reported that the TSC-mTOR pathway mediates translational activation of TOP mRNAs largely in a raptor- and rictor-independent manner, suggesting an intriguing possibility for mTORC3.75 Given the pivotal role of mTOR in cellular and organismal homeostasis, one of the pressing needs is to better understand the mechanisms of mRNA translation at multiple stages using a systemic approach.
Ribosome-Mediated Regulation of mTOR
In contrast to mTORC1, the knowledge concerning mTORC2 is significantly lagging. mTORC2 controls cell survival and proliferation in addition to cell morphology.29 Although compelling evidence has placed mTORC2 downstream of PI3K and upstream of the serine/threonine kinase AKT,76 it has been unknown how mTORC2 is regulated. Recently a stimulating new story emerged after identifying that mTORC2 activation relies on association with ribosome.77 Active mTORC2 was physically associated with the ribosome, and insulin-stimulated PI3K signaling promoted mTORC2-ribosome binding. It appears that the PI3K-dependent mTORC2-ribosome interaction plays a major role in mTORC2 activation. Thus, the ribosome serves as a missing link between PI3K and mTORC2. Intriguingly, this function of ribosomes is independent of translation. It remains intangible how exactly ribosomes regulate the kinase activities of mTORC2.
Identification of the ribosome as one of the crucial players in regulating mTORC2 activity not only reveals that the ribosome can act as a kinase platform, but also raises new questions concerning the functional interaction between mTORC1 and mTORC2 in cellular processes. Given that mTORC1 regulates ribosome biogenesis, it is conceivable that mTORC2 modulates mTORC1 by activating AKT, and mTORC1 affects mTORC2 by increasing ribosome biogenesis. Furthermore mTORC1 regulates mTORC2 via S6K-mediated feedback inhibition.78 It is worth noting that some ribosomal proteins are involved in extraribosomal functions.79 For example, free RPS7, RPL5, RPL11 and RPL27 interact with the p53 system, which leads to cell cycle arrest and apoptosis.80 Thus, the assembly and disassembly of ribosomes may constitute a cellular surveillance network by connecting nutrient signaling-mediated cell growth and p53-controlled cell proliferation. A great deal of investigation is needed to unfold this exciting chapter of the TOR story.
Ribosome Dynamics and Co-Translational Folding
Polypeptides emerging from the ribosome must fold into stable 3-dimensional structures and maintain that structure throughout their functional lifetimes. While molecular chaperones promote proper protein folding, the successful folding process of newly synthesized polypeptides primarily depends on the fidelity of transcription and translation. Mistranslated proteins should not be subject to chaperone rescue and are efficiently identified by degradation systems.81 This translational challenge is illustrated by observations that, even under optimal conditions, nearly 10% of newly synthesized proteins are mistranslated,82 and 20–30% of all nascent polypeptides are rapidly degraded owing to folding errors.83 Previous studies have concentrated on how changes in mTORC1 signaling lead to alteration of ribosome biogenesis and mRNA translation initiation.84 Less attention has focused on how alterations of translation speed (elongation rate) may influence the translation fidelity. As discussed above, our recent studies have begun to reveal the interconnection between the mTORC1 signaling pathway and the chaperone network, which affects the co-translational folding of nascent polypeptides. However, it remains possible that mTORC1-controlled elongation may directly influence the accuracy of amino acid incorporation.
mTORC1 regulates protein translation at multiple stages, including initiation and elongation. Although the regulatory mechanisms impinging on the initiation steps have received considerable attention, accumulating information points to the elongation phase as a target for control under defined circumstances. Most of the recent advances relate to the regulation of eEF2 and its cognate kinase eEF2 kinase. eEF2 mediates the translocation step of elongation, where the ribosome moves relative to the mRNA by one codon, and the peptidyl-tRNA shifts from the A site to the P site. eEF2 undergoes phosphorylation at Thr56 within the GTP-binding domain, and this modification interferes with its ability to bind the ribosome, thus inhibiting its function.85 mTORC1 negatively regulates eEF2 kinase and thereby activates eEF2. The multiplicity of regulatory inputs into the enzyme that phosphorylates eEF2 suggests that it has a key role in cellular regulation, in particular in the overall control of protein synthesis.86
Increasing evidence has supported the notion that local discontinuous translation (ribosome pausing) temporally separates the translation of segments of the peptide chain and actively coordinates their co-translational folding.87 An interesting recent report indicated that slowing bacterial translation speed enhances eukaryotic protein folding efficiency.88 We have observed that, in cells with hyperactive mTORC1 signaling, increased elongation speed is accompanied by the deficiency of luciferase folding. Importantly, slowing down translation elongation by rapamycin treatment increases the folding efficiency (unpublished results). Several possibilities could contribute to the inverse correlation between elongation speed and folding quality of nascent polypeptides. First, the higher protein production could exceed the chaperone availability, thereby reducing the overall co-translational folding capacity. Second, the faster translation speed may eliminate the ribosome pausing necessary for co-translational folding. Third, the increased elongation rate potentially compromises the translational fidelity by promoting mis-incorporation of amino acids. mRNA translation is the most error-prone step in gene expression, as approximately three codons in 10,000 are mistranslated.82 Therefore, it is conceivable that the increased translation speed under hyperactive mTOR signaling generates more error proteins. Given the fact that the most common molecular signal of aging is an accumulation of altered proteins, derived from both erroneous biosynthesis and post-synthetic modification, one of the future challenges will be to elucidate how mTOR-controlled translational regulation influences the translation fidelity as well as the folding process of translational products.
Conclusions and Outlook
The interface between chaperone-mediated stress response and the mTOR-mediated nutrient-sensing system can be viewed as a central homeostatic mechanism (Fig. 3). The concept of “less is more” was originally derived from the relationship between mTOR signaling and aging based on the observation that, in a wide range of organisms, reduced TOR signaling extends lifespan.15 It is clear that the same concept also applies to protein homeostasis. Although accumulating evidence has begun to divulge an important cellular surveillance mechanism linking protein quantity and quality control, more questions than answers arise. For example, how does the cell differentiate the physiological fluctuation from severe stresses? What is the physiological switch between cell growth signaling and cell survival pathways? What is the common mechanism for the ribosome acting as a central platform for multiple signaling pathways? These mutual connections may help formulate the cellular decision between life and death under different conditions. This framework not only offers an excellent opportunity for exploring the biological significance of the link between nutrient signaling and chaperone network, but also provides an array of putative drug targets for slowing down aging and age-associated pathologies, including cancer, diabetes and neurodegenerative disorders.
Figure 3.
The interface between chaperone-mediated stress response and mTOR-mediated nutrient-sensing system serves as a central homeostatic mechanism. Dysregulation of both pathways has been implicated in aging and age-associated diseases.
Acknowledgments
We would like to thank members of Qian lab for many discussions and ideas. Work in Qian lab is supported by grants from NIH Director's New Innovator Award and Ellison Medical Foundation Young Scholar Award (to S.B.Q.).
References
- 1.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 4.Feldman DE, Frydman J. Protein folding in vivo: the importance of molecular chaperones. Curr Opin Struct Biol. 2000;10:26–33. doi: 10.1016/S0959-440X(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 6.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 7.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 8.Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci. 2009;64:167–170. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Wellen KE, Thompson CB. Cellular metabolic stress: considering how cells respond to nutrient excess. Mol Cell. 2010;40:323–332. doi: 10.1016/j.molcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov D, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Inoki K, Guan KL. Complexity of the TOR signaling network. Trends Cell Biol. 2006;16:206–212. doi: 10.1016/j.tcb.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/S0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 19.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 21.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 22.Soti C, Csermely P. Aging and molecular chaperones. Exp Gerontol. 2003;38:1037–1040. doi: 10.1016/S0531-5565(03)00185-2. [DOI] [PubMed] [Google Scholar]
- 23.Sampayo JN, Gill MS, Lithgow GJ. Oxidative stress and aging-the use of superoxide dismutase/catalase mimetics to extend lifespan. Biochem Soc Trans. 2003;31:1305–1307. doi: 10.1042/BST0311305. [DOI] [PubMed] [Google Scholar]
- 24.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 26.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 31.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 32.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 33.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 34.Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:592–602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 36.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaeberlein M, Powers RW, III, Steffen KK, Westman EA, Hu D, Dang N, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 38.Powers RW, III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blagosklonny MV, Campisi J, Sinclair DA. Aging: past, present and future. Aging (Albany, NY) 2009;1:1–5. doi: 10.18632/aging.100009. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 42.Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- 43.Blagosklonny MV. Increasing healthy lifespan by suppressing aging in our lifetime: preliminary proposal. Cell Cycle. 2010;9:4788–4794. doi: 10.4161/cc.9.24.14360. [DOI] [PubMed] [Google Scholar]
- 44.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3KmTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J Biol Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/S1097-2765(03)00220-X. [DOI] [PubMed] [Google Scholar]
- 47.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 48.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 49.McGarry TJ, Lindquist S. The preferential translation of Drosophila hsp70 mRNA requires sequences in the untranslated leader. Cell. 1985;42:903–911. doi: 10.1016/0092-8674(85)90286-7. [DOI] [PubMed] [Google Scholar]
- 50.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 51.Sarnow P, Cevallos RC, Jan E. Takeover of host ribosomes by divergent IRES elements. Biochem Soc Trans. 2005;33:1479–1482. doi: 10.1042/BST20051479. [DOI] [PubMed] [Google Scholar]
- 52.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neef DW, Thiele DJ. Enhancer of decapping proteins 1 and 2 are important for translation during heat stress in Saccharomyces cerevisiae. Mol Microbiol. 2009;73:1032–1042. doi: 10.1111/j.1365-2958.2009.06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- 57.Ding M, Li J, Leonard SS, Shi X, Costa M, Castranova V, et al. Differential role of hydrogen peroxide in UV-induced signal transduction. Mol Cell Biochem. 2002;235:81–90. doi: 10.1023/A:1015901232124. [DOI] [PubMed] [Google Scholar]
- 58.Jurivich DA, Chung J, Blenis J. Heat shock induces two distinct S6 protein kinase activities in quiescent mammalian fibroblasts. J Cell Physiol. 1991;148:252–259. doi: 10.1002/jcp.1041480210. [DOI] [PubMed] [Google Scholar]
- 59.Kraiss LW, Ennis TM, Alto NM. Flow-induced DNA synthesis requires signaling to a translational control pathway. J Surg Res. 2001;97:20–26. doi: 10.1006/jsre.2001.6091. [DOI] [PubMed] [Google Scholar]
- 60.Morimoto RI. Dynamic remodeling of transcription complexes by molecular chaperones. Cell. 2002;110:281–284. doi: 10.1016/S0092-8674(02)00860-7. [DOI] [PubMed] [Google Scholar]
- 61.Dezwaan DC, Freeman BC. HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- 62.Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian SB, Zhang X, Sun J, Bennink JR, Yewdell JW, Patterson C. mTORC1 links protein quality and quantity control by sensing chaperone availability. J Biol Chem. 2010;285:27385–27395. doi: 10.1074/jbc.M110.120295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- 65.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 66.Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XF. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsang CK, Liu H, Zheng XF. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010;9:953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci USA. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 70.Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPs and their regulation. Biochem Soc Trans. 2006;34:12–16. doi: 10.1042/BST0340012. [DOI] [PubMed] [Google Scholar]
- 71.Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, et al. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol-3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol. 2001;21:8671–8683. doi: 10.1128/MCB.21.24.8671-83.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, et al. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-24.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cardinali B, Carissimi C, Gravina P, Pierandrei-Amaldi P. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J Biol Chem. 2003;278:35145–35151. doi: 10.1074/jbc.M300722200. [DOI] [PubMed] [Google Scholar]
- 74.Intine RV, Dundr M, Vassilev A, Schwartz E, Zhao Y, Zhao Y, et al. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol. 2004;24:10894–10904. doi: 10.1128/MCB.24.24.10894-904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J, et al. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol. 2009;29:640–649. doi: 10.1128/MCB.00980-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 77.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by Association with the Ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 78.Xie X, Guan KL. The Ribosome and TORC2: Collaborators for Cell Growth. Cell. 2011;144:640–642. doi: 10.1016/j.cell.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 82.Kirkwood TB, Holliday R, Rosenberger RF. Stability of the cellular translation process. Int Rev Cytol. 1984;92:93–132. doi: 10.1016/S0074-7696(08)61325-X. [DOI] [PubMed] [Google Scholar]
- 83.Qian SB, Princiotta MF, Bennink JR, Yewdell JW. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J Biol Chem. 2006;281:392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- 84.Raught B, Gingras AC. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. pp. 369–400. [Google Scholar]
- 85.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 86.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 88.Siller E, Dezwaan DC, Anderson JF, Freeman BC, Barral JM. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol. 2010;396:1310–1318. doi: 10.1016/j.jmb.2009.12.042. [DOI] [PubMed] [Google Scholar]