Abstract
XPG is a structure-specific endonuclease required for nucleotide excision repair (NER). XPG incision defects result in the cancer-prone syndrome xeroderma pigmentosum, whereas truncating mutations of XPG cause the severe postnatal progeroid developmental disorder Cockayne syndrome. We show that XPG interacts directly with WRN protein, which is defective in the premature aging disorder Werner syndrome, and that the two proteins undergo similar subnuclear redistribution in S phase and colocalize in nuclear foci. The co-localization was observed in mid- to late S phase, when WRN moves from nucleoli to nuclear foci that have been shown to contain both protein markers of stalled replication forks and telomeric proteins. We mapped the interaction between XPG and WRN to the C-terminal domains of each, and show that interaction with the C-terminal domain of XPG strongly stimulates WRN helicase activity. WRN also possesses a competing DNA single-strand annealing activity that, combined with unwinding, has been shown to coordinate regression of model replication forks to form Holliday junction/chicken foot intermediate structures. We tested whether XPG stimulated WRN annealing activity, and found that XPG itself has intrinsic strand annealing activity that requires the unstructured R- and C-terminal domains but not the conserved catalytic core or endonuclease activity. Annealing by XPG is cooperative, rather than additive, with WRN annealing. Taken together, our results suggest a novel function for XPG in S phase that is, at least in part, performed coordinately with WRN, and which may contribute to the severity of the phenotypes that occur upon loss of XPG.
Key words: Cockayne syndrome, progeria, DNA annealing, DNA replication, DNA damage response
Introduction
The maintenance of genomic integrity is persistently threatened both by endogenous damage from the cellular metabolism and exogenous environmental sources, such as UV and ionizing radiation and mutagenic chemicals. To counter this damage, organisms across the evolutionary scale are equipped with complex and interconnected DNA repair pathways that detect and remove lesions in DNA. XPG is a structure-specific endonuclease that cleaves DNA bubbles and flaps near the junctions of single-stranded and double-stranded DNA with specific polarity.1,2 It also binds strongly to various structured DNAs that it does not cleave, implying separate biological functions for its binding and incision activities.2,3 The incision activity of XPG is essential for removing bulky DNA adducts by the nucleotide excision repair (NER) pathway,1 and point mutations that inactivate the XPG endonuclease function cause the cancer-prone, sun-sensitive disorder xeroderma pigmentosum (XP) in XP-G patients.4,5 XPG also has a nonenzymatic scaffolding role in several steps of NER, including coordination of incision with the resynthesis step.6,7 Regions of XPG share extensive sequence homology with flap endonuclease 1 (FEN1), a much smaller structure-specific endonuclease that removes RNA primers from Okazaki fragments.8In addition, XPG has unique regions with no homology to other known proteins, including an R-(recognition) domain (also called the spacer region) that is larger than the entire FEN1 plus a C-terminal domain that is significantly longer than the corresponding region in FEN1 (Fig. 2A).3,9 The C terminus contains a PCNA binding motif.10
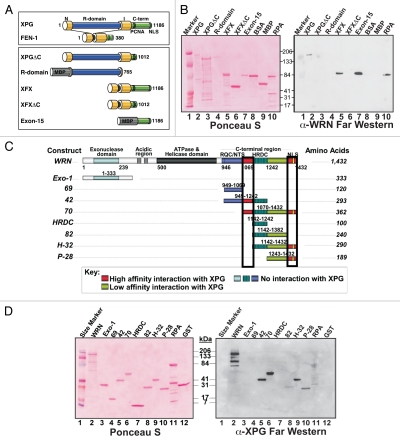
Figure 2.
Mapping the interacting regions in XPG and WRN by far western analysis. (A) XPG domain organization and domain constructs. XPG catalytic domains (N, I), R domain, C-terminal domain encoded by exon 15 (C-term), PCNA binding motif and nuclear localization signal (NLS) are shown. The C-terminal domain is deleted in XPGΔC. The R domain (aa 86–765) and Exon 15 (aa 1,007–1,186) domains were expressed as fusions to maltose binding protein (MBP). In XFX, the R domain was replaced by the corresponding FEN1-loop domain. In XFXΔC, R- and C-terminal domains were removed. (B) WRN interacts with the C-terminal domain of XPG. Purified proteins were separated by SDS-PAGE, blotted onto nitrocellulose, stained with Ponceau-S (left), incubated with purified WRN protein, and bound WRN was detected by mouse anti-WRN antibody (right). BSA and MBP served as negative controls; RPA, which is known to interact with WRN, is a positive control. (C) WRN domain organization and domain constructs. (D) XPG interacts with the C-terminal domain of WRN. Purified proteins were separated by SDS-PAGE, blotted onto nitrocellulose, stained with Ponceau-S (left), incubated with purified XPG protein, and bound XPG was detected by mouse anti-XPG antibody (right).
Separate from its required function in NER, XPG is essential for normal postnatal development in mammals. Patients with rare truncating mutations in XPG have the combined diseases of XP with Cockayne syndrome (XP-G/CS).5,11–13 XP-G/CS presents as severe, primarily postnatal, neurological and developmental dysfunction, with mental retardation, wasting, greatly accelerated symptoms of segmental aging and death in early childhood. XPG-knockout mice recapitulate this patient phenotype, exhibiting severe postnatal wasting and death before 3 weeks of age.14In contrast, mice that lack NER, owing to point mutations that inactivate XPG enzymatic activity, are normal except for UV sensitivity.15,16 Thus, loss of NER is evidently not responsible, per se, for the CS phenotypes that develop in mice and humans with severely truncating or null mutations of XPG.15,16 In particular, loss of the extended C terminus of XPG has been implicated in the CS phenotype by knock-in mouse mutations.16,17
We and others have shown that XPG has multiple nonenzymatic functions that might contribute to the fatal postnatal phenotype associated with its loss. It has a role in the early steps of base excision repair (BER) of oxidative DNA damage through direct interaction with and stimulation of NTH118–20 and other DNA glycosylases (A.H. Sarker et al., in preparation). Moreover, XPG interacts directly3 with both RNA polymerase II and the CSB protein that is essential for initiation of transcription-coupled repair (TCR), a process that preferentially removes transcription-blocking lesions through recognition of stalled RNA polymerase.21 Since oxidative damage to DNA is a principal source of endogenously generated lesions, and since inability to carry out TCR is the molecular hallmark of CS, loss of BER and/or TCR functions of XPG could contribute to the XP-G/CS phenotype. In addition, XPG forms a complex with the transcription and repair factor TFIIH and is important for stable association of the CAK kinase subunit with TFIIH, with the consequence that XP-G/CS cells are deficient in phosphorylating and activating nuclear receptors such as ERα.22 Although these activities of XPG are important, and therefore likely contribute to its postnatal requirement, the exact nature of the defect leading to XP-G/CS and very early death is still not clear.
Werner syndrome (WS) is an adult-onset premature aging disorder caused by deficiency in the WRN protein. Individuals with WS develop cancer, cardiovascular disease, osteoporosis and cataracts, among other pathologies, approximately 2–3 decades early.23 WRN is a 3′–5′ RECQ-like DNA helicase with a marked preference for substrates with extensive secondary structure.24 It also possesses an opposing DNA annealing activity that, when coordinated with its helicase activity, is capable of performing strand exchange.25 Alone among the RECQ helicases, WRN has an exonuclease domain26 and 3′–5′ exonuclease activity.27 Additionally, WRN has been reported to stimulate BER,28–31 and it clearly plays a role in non-homologous endjoining (NHEJ) of DNA double-strand breaks (DSBs).32–34 WRN localizes primarily to nucleoli, but in S phase, it moves from the nucleoli to stalled replication forks, evident as distinct nuclear foci that also contain RPA and RAD51.35,36 In addition, a fraction of WRN associates with telomeres during S phase, where it is important for lagging strand synthesis in the replication of telomeric DNA.37 Loss of WRN leads to increased chromosome aberrations, defective resolution of Holliday junctions, abnormal DNA replication intermediates, large deletions and increased incidence of telomere sister chromatid exchange.37–41 Direct measurement of replication fork kinetics showed that, after DNA damage, replication fork progression was significantly slower in the absence of WRN.42 Thus WRN clearly has an important, though poorly understood, role during replication, and it, like XPG, participates in multiple DNA transactions.
We provide evidence here that XPG interacts directly with WRN and colocalizes with it in nuclear foci in mid- to late S phase. We mapped the interaction to the C terminus of both proteins, and show that the C terminus of XPG strongly stimulates WRN helicase activity, while possessing a previously unrecognized single-strand DNA annealing activity that functions cooperatively with WRN. These results suggest a novel role for XPG in S phase, either separately or together with WRN, and provide additional insight into the severe nature of the defects observed in XP-G/CS patients.
Results
XPG directly interacts with WRN, undergoes similar subnuclear redistribution in S phase and colocalizes with WRN in nuclear foci.
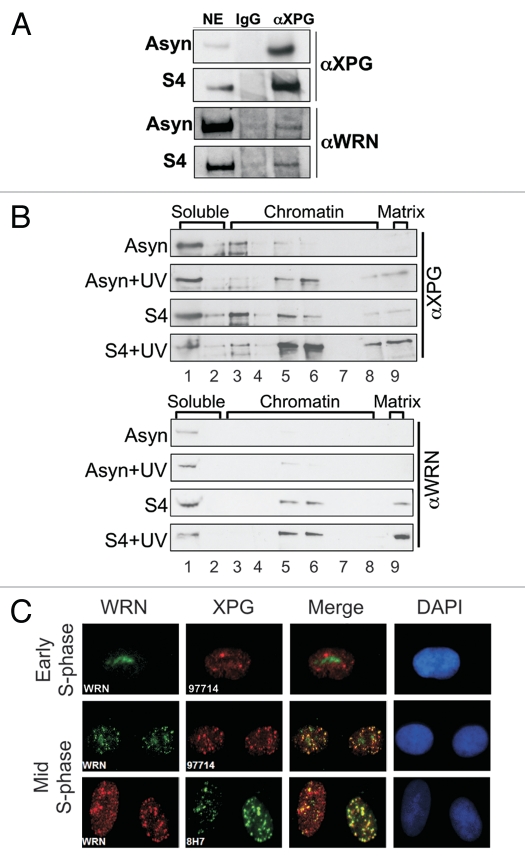
To better understand the nonenzymatic role(s) of XPG and the basis for its postnatal requirement, we performed a search to identify new XPG-interacting partner proteins in cells. We immunoprecipitated nuclear extracts of hTERT-immortalized normal human HCA2 fibroblasts with anti-XPG or control antibody, and found that WRN co-immunoprecipitated with XPG. This interaction was evident in both asynchronous cells and cells in mid-S phase (Fig. 1A). To expand upon this observation, we sought other lines of evidence for an interaction of XPG and WRN, using both biochemical approaches and cellular immunofluorescence.
Figure 1.
XPG interacts with WRN. (A) Western analysis showing co-immunoprecipitation of WRN with XPG from undamaged hTERTimmortalized HCA2 cells, either asynchronous (Asyn) or in mid-S phase 4 h after release from the G1/S boundary (S4). Nuclear extracts were immunoprecipitated with mouse anti-XPG antibody 8H7 and analyzed for WRN and XPG by protein gel blotting. (B) Association of XPG and WRN with chromatin and the nuclear matrix during S phase. The cells, asynchronous or in mid-S phase (S4), were mock treated or UV irradiated (30 J/m2) and, 30 min after damage, were separated into nine fractions as described in Materials and Methods. The lane numbers correspond to the fraction numbers. (C) XPG colocalizes with WRN foci in mid-S phase. Cells were synchronized in early (2 h after release; top part) or in mid-S phase (6 h after release; middle/bottom parts), permeabilized, fixed and immunostained. The top and middle parts show immunostaining using mouse anti-WRN (green), rabbit anti-XPG (red), the merged image (co-localization, yellow) and DAPI (blue) stained nuclei. The bottom part shows immunostaining using rabbit anti-WRN (red), mouse anti-XPG (green), the merged image (co-localization, yellow) and DAPI (blue) stained nuclei.
To understand how XPG and WRN respond to cell cycle position and DNA damage, we employed a serial biochemical fractionation protocol that extracts proteins in nine steps, separating them into soluble, chromatin and nuclear matrix fractions.43 The nuclear matrix is a structural scaffold that anchors arrays of chromatin loops and plays an important role in replication, transcription and most likely also DNA repair.44 The fractions were analyzed by protein gel blotting (Fig. 1B), and successful fractionation was confirmed by the presence of histone H1 in the soluble fraction and vimentin in the nuclear matrix fraction (Sup. Fig. 1).
In undamaged asynchronous cells, both WRN and XPG were largely in the soluble fraction (Fig. 1B, lane 1). After UV damage, a fraction of XPG moved to the chromatin (top, lanes 5–6). By analogy with other proteins, this presumably reflects its function in global NER.45 Strikingly, XPG was also detected in the insoluble nuclear matrix fraction (top, lane 9), likely reflecting XPG function in TCR.43 As expected for a protein not thought to be involved in NER or TCR, WRN did not leave the soluble fraction when asynchronous cells were damaged by UV (Fig. 1B, bottom). However, in cells synchronized in S phase, a substantial fraction of WRN became bound to the chromatin and nuclear matrix at 4 h after release from the G1/S boundary (S4), consistent with its known relocalization to replication forks. Unexpectedly, a significant amount of XPG protein also relocalized to chromatin during mid-S phase in undamaged cells, and a minor fraction was also detectable at the nuclear matrix. When cells in S phase were UV irradiated, causing collapsed or stalled replication forks, even greater amounts of both XPG and WRN associated with the chromatin and nuclear matrix fractions. The similar behavior of the two proteins with respect to subnuclear redistribution in S phase, and particularly in response to replication-blocking damage in S, is consistent with their observed interaction.
We next used indirect immunofluorescence microscopy to investigate the spatial localization of XPG relative to WRN in S-phase cells. To visualize only proteins bound to the chromatin or nuclear matrix, we permeabilized and gently washed the cells prior to fixation.46 This step was critical to our ability to observe discrete foci containing XPG (Sup. Fig. 2, top and middle rows). That the foci observed in this procedure do indeed represent XPG was confirmed by their absence in cells from an XP-G/CS patient with severely truncating xpg mutations5 and no XPG protein by western analysis (Sup. Fig. 2, bottom row and data not shown). In G1 and early S-phase cells, WRN localized primarily to the nucleoli, as evident for cells 2 h after release from the G1/S boundary (Fig. 1C, top row). XPG was not nucleolar in early S phase, but did form foci that were devoid of WRN. In contrast, when we examined cells in mid- to late S phase, WRN had moved out of the nucleoli, and XPG and WRN foci extensively colocalized in a fraction of cells (∼10–15%). The same result was obtained by immunostaining using either of two different antibodies against XPG (Fig. 1C, middle and bottom rows), providing strong support for the conclusion of co-localization.
WRN and XPG interact through the C-terminal regions of each.
We used recombinant proteins containing known XPG domains to map the region in XPG that mediates interaction with WRN. Full-length XPG consists of conserved N- and I-nuclease subdomains, which together form the catalytic domain and non-conserved R- and C-terminal (Exon 15) domains. The latter contains a PCNA binding motif and nuclear localization signal (NLS). We tested full-length XPG and a series of XPG domain constructs purified from baculovirus-infected insect cells or bacteria (Fig. 2A),3 for ability to interact with purified WRN protein by far western analysis (Fig. 2B), using the single-strand DNA binding Replication Protein A (RPA) as a positive control.47 Proteins containing the C-terminal domain [XPG, XFX and Exon-15 (a maltose binding protein (MBP)-Exon15 fusion construct)] interacted with WRN. By contrast, proteins lacking this domain (XPGΔC, R-domain, XFXΔC) and control proteins [MBP; bovine serum albumin (BSA)] did not. Thus, a region within the C-terminal 180 amino acids of XPG (exon 15) is both necessary and sufficient for interaction with WRN. Furthermore, these far western results with purified proteins establish that the interaction between XPG and WRN is direct, not requiring either DNA or other proteins.
Similarly, we tested a variety of WRN protein subdomains (Fig. 2C) by far western analysis for interaction with full-length XPG (Fig. 2D). XPG failed to interact with the exonuclease domain of WRN (1–333). However, it strongly interacted with the WRN C-terminal region [construct 70; amino acids (aa) 1,070–1,432]. The lack of interaction with the HRDC domain (aa 1,142–1,242), construct 69 (aa 949–1,069) or construct 82 (aa 1,142–1,382), together with the strong interaction with the H-32 construct (aa 1,142–1,432) and construct 42 (aa 949–1,242), mapped the interaction to two distinct domains, denoted in red and boxed in Figure 2C. Confirming the mapping studies with full-length XPG, the XPG C-terminal domain (exon 15) also directly interacted with WRN construct 70 (aa 1,070–1,432) when used as a probe in additional far western analyses (data not shown). These results, coupled with our finding that a partially purified WRN construct spanning aa 2–993 does not interact with XPG (not shown), isolates the XPG interaction to two small regions within the WRN C terminus. These regions are aa 1,070–1,142 and aa 1,382–1,432, which contains the NLS of WRN (Fig. 2C).
XPG stimulates WRN helicase activity.
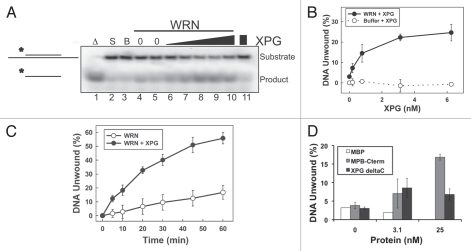
To investigate whether the novel interaction between XPG and WRN has functional significance, we examined the effect of XPG on WRN helicase activity. We first titrated WRN helicase activity (Sup. Fig. 3) to determine a limiting amount of purified WRN protein, which we then tested in the absence or presence of varying concentrations of XPG. Reactions were initiated by addition of WRN to a mixture of XPG and a partial duplex DNA substrate (Fig. 3A). Unwound DNA products were separated from duplex DNA by gel electrophoresis. XPG substantially stimulated WRN unwinding activity in a concentration-dependent manner (Fig. 3A, lanes 6–10). As expected, there was no unwinding by XPG alone (Fig. 3A, lane 11 and 3B). Under these experimental conditions, WRN alone unwound 3% of the substrate, and XPG increased the unwinding 6.5-fold (24.6% of substrate) (Fig. 3B). Next, we examined the effect of XPG on the kinetics of WRN unwinding. Notably, the presence of full-length XPG increased the rate of WRN unwinding > 5-fold (Fig. 3C).
Figure 3.
XPG increases WRN helicase activity. (A) XPG stimulates WRN helicase activity. Labeled DNA substrate (0.5 nM) was incubated with buffer (lane 3), WRN alone (0.1 nM; lanes 4, 5), WRN (0.1 nM) and increasing concentrations of XPG protein (lanes 6–10; 0.2, 0.8, 3.1, 6.3, 25 nM) or XPG (25 nM) without WRN (lane 11) for 30 min. As controls, the substrate alone was boiled for 5 min to mark the position of fully unwound product (lane 1; Δ) or retained on ice (lane 2; S = Substrate). (B) Quantification of XPG stimulation of WRN helicase activity. WRN (0.1 nM) was assayed for helicase activity in the presence of increasing concentration of XPG (solid circles) and compared with XPG without WRN (open circles). DNA unwinding (%) from three independent experiments was plotted (error bars are the SD). (C) XPG increases the rate of WRN unwinding. Helicase reactions containing 0.1 nM WRN protein with or without 3.1 nM XPG were stopped at the indicated times, and the percentage of unwound DNA was determined. Error bars are the SD from three independent experiments. (D) The C-terminal domain of XPG is required to significantly stimulate WRN helicase activity. WRN (0.1 nM) was assayed for helicase activity in the presence of increasing concentration of XPG, MBP-C-terminally, XPGΔC or MBP protein as a control. DNA unwinding (%) from three independent experiments was plotted (error bars are the SD).
Having shown that interaction of XPG with WRN is through the C terminus of XPG, we used XPG constructs that lacked or contained the C terminus (XPGΔC and MBP-C-term) to determine whether direct interaction between WRN and XPG is required for the stimulation of WRN unwinding activity. XPGΔC did not appreciably alter WRN helicase activity, whereas the XPG C terminus alone stimulated unwinding, although only at a much higher concentration than full-length XPG (Fig. 3D). Thus, the C-terminal domain of XPG is both necessary and sufficient for stimulation of WRN helicase activity, as it is for direct interaction of the two proteins.
XPG anneals ssDNA alone and cooperatively with WRN.
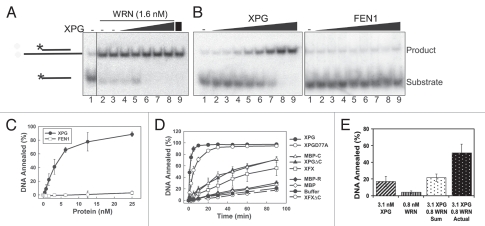
RECQ helicases, including WRN, not only unwind duplex DNA but also catalyze the opposing activity of ssDNA annealing. Both properties may be important for replication fork regression and/or the re-start of stalled replication forks. To determine whether XPG also affects the annealing activity of WRN, we assayed for DNA strand annealing using a labeled ssDNA substrate and unlabeled complementary DNA oligonucleotide. Assays were performed in the absence of ATP to prevent helicase activity. We first used an amount of WRN that efficiently anneals DNA (Fig. 4A, lanes 2 and 3), and tested whether XPG inhibits annealing by WRN. However, addition of increasing concentrations of XPG to the reaction not only failed to inhibit annealing, but appeared to stimulate it (Fig. 4A, lanes 4–8). Very surprisingly, the XPG-only control reaction revealed that XPG itself possesses intrinsic ssDNA annealing (Fig. 4A, lane 9).
Figure 4.
XPG has DNA strand annealing activity that is cooperative with WRN. (A) XPG does not inhibit WRN annealing activity. WRN (1.6 nM) was incubated with labeled 20-mer DNA substrate and unlabeled, partially complementary 46-mer DNA (lane 1, no added protein) in the absence (lanes 2 and 3) or presence of increasing concentrations of XPG (lanes 4 to 8: 0.2, 0.8, 3.1, 6.3, 25 nM). Lane 9 shows DNA annealing by 25 nM XPG alone (no WRN). (B) DNA annealing activity of XPG but not FEN1. DNA substrate was incubated with increasing concentration of XPG or FEN1 protein (0, 0.2, 0.4, 0.8, 1.6, 3.1, 6.3, 12.5, 25 nM) for 10 min, and the products were separated by native PAGE. (C) Quantification of XPG and FEN1 annealing activity in (B). Error bars are the SD from four independent experiments. (D) DNA annealing kinetics of XPG, catalytically inactive XPG and XPG domain constructs. Annealing reactions contained 25 nM of each protein. Samples were removed at the indicated times and the products separated by native PAGE. The percent DNA annealed was determined. Error bars are the SD from three independent experiments. (E) XPG and WRN anneal DNA cooperatively. DNA annealing was measured for XPG (3.1 1nM) and WRN (0.8 nM) separately or together. The sum of annealed products from each independent reaction was compared with the actual amount of annealing when WRN and XPG were both present. Error bars are the SD from three independent experiments.
To examine this new function of XPG, we titrated either XPG or its smaller homolog FEN1 into the ssDNA annealing assay. XPG efficiently annealed DNA in a concentration dependent manner, whereas FEN1 completely lacked this activity (Fig. 4B, quantified in 4C). Testing various XPG domain constructs (Fig. 2A) showed that proteins lacking either the R- or C-terminal domains performed some annealing but at very low levels compared with the full-length protein (Fig. 4D). The XFXΔC construct, which retains the endonuclease domain but lacks the R- and C-terminal domains (thus resembling FEN1), lacked annealing activity (indistinguishable from buffer alone) (Fig. 4D). A catalytically inactive mutant XPG, XPGD77A,48was almost as competent as wt protein for annealing (Fig. 4D). Thus both the R- and C-terminal domains, but not endonuclease activity, are important for this unexpected activity of XPG.
Having found that XPG interacts directly with WRN and possesses similar DNA annealing activity, we asked whether XPG and WRN annealing activities are additive or cooperative. We measured the DNA annealing activities of sub-stoichiometric concentrations of XPG and WRN separately and in combination at the same concentrations. Annealing by both proteins together was 140% greater than the sum of the individual reactions (Fig. 4E). Thus, ssDNA annealing by XPG and WRN is cooperative. These findings, together with the ability of XPG to stimulate WRN helicase activity, indicate functional significance for the direct interaction of XPG with WRN.
Discussion
We have identified a direct functional interaction between XPG and WRN proteins, revealed an unexpected strand annealing activity of XPG, and demonstrated that WRN and XPG both undergo similar subnuclear redistributions to foci in S phase. Further, we found that XPG and WRN colocalize in nuclear foci specifically during mid-S phase. Although XPG foci formed throughout S phase, the co-localization was not observed until mid- to late S phase, when WRN moves from nucleoli to nuclear foci that contain markers of stalled replication forks such as RAD51 and RPA as well as telomeric proteins and telomeric DNA.37
The hypothesized function of WRN in S phase nuclear foci is informed by the phenotype of cells lacking WRN and by the known biochemical activities of purified WRN protein. Cells from Werner syndrome patients display abnormal DNA replication intermediates, chromosomal aberrations, defective resolution of Holliday junctions, reduced replication fork rates after DNA damage and large deletions dependent on DNA replication.38–40,42 Together, these data indicate that WRN likely functions in the protection of stalled replication forks after DNA damage. Of relevance to this role, the combined unwinding and annealing activities of WRN have been shown to coordinate regression of model replication forks to form Holliday junction/chicken foot intermediate structures, considered to be an early event in the processing of a blocked replication fork.49,50 Because XPG both stimulates WRN helicase activity and anneals DNA cooperatively with WRN, it is possible that XPG may act to promote fork regression with WRN (Fig. 5). Recently, it was shown that WRN efficiently catalyzed regression even through RPA-bound ssDNA, resulting in RPA displacement.51 Because XPG also physically interacts with RPA,52 WRN, XPG and RPA may function together to mediate the process of replication fork regression and/or fork restart (Fig. 5). The absence of this pathway could lead to an increased incidence of replication fork breaks and might explain the increase in recombination events34 and genomic instability observed in WS cells.
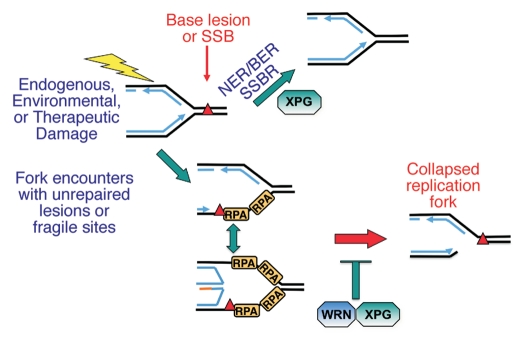
Figure 5.
Model for possible roles of XPG and WRN during S phase. XPG contributes to removal of DNA damage before a replication fork encounters it, thus facilitating replication by providing a clean template. XPG and WRN together may mediate either fork reversal or restart by coupled action in unwinding DNA and in cooperative DNA strand annealing. Absence of this latter pathway in WS or XP-G/CS cells may lead to increased breaks at stalled replication forks and fork collapse.
Multiple lines of evidence additionally support a role for WRN at telomeres. WRN foci not only colocalize with telomeric proteins, but WRN directly binds telomeric chromatin37 and interacts directly with the shelterin proteins TRF2 and POT1, both of which stimulate its helicase activity.53,54 Further, WRN-deficient cells undergo rapid replicative senescence without accelerated telomere shortening but with aberrant telomeric structures, including increased telomere associations and telomere sister chromatid exchanges and loss of the G-rich lagging strand DNA.37,41 These data suggest that telomere dysfunction is one cause of the observed genomic instability and premature aging observed in WS. It is possible that the interaction of XPG with WRN is functionally important in telomere maintenance, and so it will be of interest to learn whether XPG has a role at telomeres in S phase. This novel possibility may partially explain the severe progeroid developmental disorder seen in XP-G/CS patients, although the early postnatal appearance of progeria is in marked contrast to the relatively late onset of WS symptoms.
WRN additionally has a role in the processing of DSBs, particularly in the NHEJ pathway. WS fibroblasts display a mild but distinct sensitivity to ionizing radiation, and WRN additionally interacts physically and functionally with DNA-PK, which phosphorylates WRN and stimulates WRN exonuclease activity.32 Notably, WRN accumulates at sites of laser-induced DSBs but not at single-strand breaks or oxidative base damage.55 Whereas the helicase and exonuclease domains were shown to be dispensable for WRN recruitment to DSBs, the annealing domain, which has been mapped to WRN residues 1,072–1,150,56 was absolutely required for recruitment.55 In this context, it is of particular significance that our mapping results show that one of the two XPG interaction domains in WRN is coincident with the annealing domain. Thus, it is possible that the interaction between XPG and WRN promotes WRN recruitment to this type of damage and regulates its activity at the break site.
The regulation and coordination of the multiple biochemical functions of WRN—DNA binding, unwinding, annealing and exonuclease activities—and its multiple cellular functions are poorly understood. It is likely that protein partner interactions serve to both localize WRN to specific cellular sites and to modulate its enzymatic activities. Several domains of WRN have been structurally characterized, including the unique N-terminal exonuclease domain, which is homologous to the E. coli DNA polymerase I proofreading domain,26 the C-terminal HRDC domain,57 which is important for substrate specificity, the RECQ C-terminal (RQC) domain (949–1,079)58 and a novel heptad-repeat, coiled-coil region between the nuclease and helicase domains that facilitates multimerization of WRN.59 The RQC domain is particularly interesting, because it contains a novel winged-helix motif that contributes to base unpairing,58 and one of the two XPG interaction domains with WRN maps to WRN amino acids 1,070–1,142, immediately adjacent to the core RQC domain (Fig. 2C). This region, which was recently shown to be necessary and sufficient for the intrinsic ssDNA annealing activity of WRN, is currently structurally uncharacterized.56 The PONDR secondary structure prediction program60 suggests that the domain is largely disordered. It is possible that the interaction between XPG and WRN may confer a disorder-to-order transition that serves to modulate WRN protein activity, as illustrated by their cooperative DNA strand annealing. Whether the interaction confers structure to an otherwise unstructured region will be an interesting hypothesis to test in future studies.
The direct, functional interaction of XPG with WRN suggests that XPG may participate in any one of the aforementioned critical pathways with WRN. However, it is important to note that the progeroid developmental phenotype seen in XP-G/CS patients is far more severe than the phenotypes observed with WS patients. This discrepancy implies that XPG functions in important cellular pathways, independently from WRN. While the severity of the XP-G/CS phenotype likely results in part from the requirement for XPG in TCR and/or BER (Fig. 5), it is additionally possible that XPG plays a role during S phase independently of WRN. Notably, the XPG foci that form in S phase do not entirely colocalize with WRN. We observed XPG foci both in early S phase, at a time when WRN is still predominantly nucleolar, and in mid/late S phase when a majority of cells contain WRN and/or XPG foci. At this time, however, only 10–15% of the cells showed substantial co-localization between XPG and WRN, although in those cells, the degree of co-localization was nearly complete. These data strongly suggest that XPG functions with WRN at a particular stage in S phase, and that it also has other replication-associated functions and likely other novel protein partners. Discerning these additional new functions of XPG should provide further clues to the extreme phenotype of patients lacking functional XPG. In addition, the demonstrated interaction between XPG and WRN in mid- to late S phase opens the way for understanding the mechanisms by which genomic integrity is maintained during replication by interactions of the DNA repair pathways mediated by these two multifunctional proteins.
Materials and Methods
Cells and cell culture.
HCA2 foreskin fibroblasts were from J. Smith (University of Texas) and were immortalized by infection with an hTERT-expressing retrovirus as previously described in reference 61. XPCS2LV fibroblasts (GM13370) were from the Coriell NIGMS Human Genetic Cell Repository. Cells were cultured under ambient oxygen levels and 10% CO2 in DMEM supplemented with 10% fetal calf serum. Synchronization into S phase was performed as previously described in reference 62.
Antibodies.
The XPG antibodies used were mouse monoclonal anti-XPG 8H7 (Abcam or Neomarkers), which recognizes an epitope in the XPG C-terminal region and polyclonal rabbit anti-XPG 97714, which we raised against a conserved R domain peptide and purified as described in reference 3. Other antibodies used were rabbit anti-WRN (ab200, Abcam, for western analysis and immunofluorescence), mouse anti-WRN (ab66606, Abcam, for far western analysis), mouse anti-WRN (195C, Sigma, for immunofluorescence), rabbit anti-Histone H3 (ab1791, Abcam) and mouse anti-Vimentin (Ab-2, Neomarkers). We used secondary goat antibodies conjugated to Alexa Fluors 488 or 594 (Molecular Probes) and used DAPI to stain nuclear DNA.
Cellular fractionation into soluble, chromatin-bound and nuclear matrix-associated proteins.
Cell lysates were fractionated by sequential extraction with salt and detergent, essentially as previously described in references 43 and 63. Briefly, soluble proteins were extracted by a low concentration of non-ionic detergent (fractions 1, 2), then loosely bound chromatin proteins were released by DNase I (fraction 3). More tightly bound chromatin-associated proteins were progressively released by increasing salt concentrations (fractions 4–7), ending with high salt containing 1% Triton X-100 (fraction 8). Proteins associated with the insoluble nuclear matrix were released by 2–4% SDS (fraction 9).
Indirect immunofluorescence.
For XPG immunostaining, cells in 4- or 8-well chamber slides (Invitrogen) were permeabilized with 0.5% Triton in CSK buffer63 and then fixed with 4% paraformaldehyde. Samples were blocked with 10% goat serum prior to incubating overnight at 4°C with primary antibody in 10% serum. Following washes with PBS, we incubated samples with conjugated secondary antibodies for 30 min at room temperature, followed by three washes with PBS. The final wash contained DAPI (0.1 mg/ml). We mounted slides in Vectashield and viewed by epifluorescence. Images of cells were acquired on a microscope (BX60; Olympus) using a 40x UPlanFl 0.5 NA (Olympus) lens without oil and captured with a charge-coupled device camera (Diagnostic Instruments, Inc.,) into SPOT imaging software (Diagnostic Instruments, Inc.). All modifications were applied to the whole image using Photoshop CS2 (Adobe).
Expression and purification of proteins.
Full-length WRN, XPG and XPG constructs XPGΔC, XFX, XFXΔC and XPG D77A were expressed in insect cells and purified essentially as described in references 3 and 27. For creation of fusion constructs with Maltose Binding Protein (MBP), R domain (XPG residues 86–765) and C-terminal domain (XPG residues 1,007 to 1,186) segments were cloned into pGAZ/MBP1 (kind gift of Gareth Williams, LBNL) containing 6-His and MBP tags upstream of the XPG sequence. The domain fusion constructs and 6-His-MBP without XPG sequences were expressed and purified from E. coli by a combination of nickel affinity, ion exchange and gel-filtration chromatography. The WRN exonuclease construct (1–333) was expressed and purified as previously described in reference 26. The WRN C-terminus constructs (WRN residues 949–1,069, 949– 1,242, 1,070–1,432, 1,142–1,242, 1,142–1,382, 1,142–1,432, 1,243–1,432) were designed based on results of limited proteolysis studies of the WRN C terminus, with proteolytic fragments identified by MALDI-TOF mass spectroscopy, N-terminal sequencing and SDS-PAGE. Each construct was expressed with a 6x-His tag and purified from E. coli by a combination of nickel affinity, ion exchange and gel-filtration chromatography. Bovine serum albumin (BSA) was obtained from New England Biolabs. FEN1 protein was a kind gift from Yoshihiro Matsumoto (Fox Chase Cancer Center), RPA protein was a gift from Susan Tsutakawa (Lawrence Berkeley National Laboratory), and GST was a gift from Jill Fuss (Lawrence Berkeley National Laboratory).
Helicase and annealing assays.
Helicase reactions (20 µl) were performed at 37°C for the times indicated in buffer containing 25 mM HEPES pH 7.5, 50 mM NaCl, 5 mM ATP, 2 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA and 0.5 nM labeled DNA substrate. The substrate was formed by annealing a 5′ end labeled 20-mer (5′-CGC TAG CAA TAT TCT GCA GC) to an unlabeled 46-mer (5′-GCG CGC AAG CTT GGC TGC AGA ATA TTG CTA GCG GGA ATT CGG CGC G) to create a partial duplex substrate with 5′ and 3′ unpaired regions. XPG was pre-incubated with substrate for 5 min at room temperature prior to addition of WRN to initiate the reaction. Reactions were terminated by addition of 15 mM EDTA, 1% sodium dodecyl sulfate, 5% glycerol, 0.04% bromo-phenyl blue and 0.04% xylene cyanol (final concentrations), quenched on ice and immediately loaded onto native 12% polyacrylamide gels and separated by electrophoresis. Annealing reactions were performed as described for helicase assays but without addition of ATP and contained labeled 20-mer (0.5 nM). Reactions were initiated by addition of protein and unlabeled 46-mer substrate (1.0 nM).
Nuclear extraction and immunoprecipitation.
Cells were washed in 5 cell pellet volumes (CPV) with ice-cold hypotonic buffer (Buffer A) containing 10 mM Tris, pH 7.9, 1.5 mM MgCl2, 5 mM KCl, 0.5 mM DTT, 0.1 mM PMSF and protease inhibitors, followed by centrifugation at 250 g. Cell pellets were resuspended in 3 CPV Buffer A and dounce homogenized. Nuclei were collected by centrifugation at 250 g and resuspended in 50 mM Tris, pH 7.9, 2 mM EDTA, 10% sucrose, 10% glycerol, 0.1 mM PMSF and protease inhibitors. NaCl was added to a final concentration of 300 mM, followed by incubation on ice for 30 min and centrifugation at 1,000 g. The nuclear extracts (supernatant) were diluted to 150 mM NaCl prior to immunoprecipitation. Extracts were precleared with mouse IgG prior to addition of XPG antibody or control mouse IgG.
Far western analysis.
Far westerns were performed as previously described in reference 3.
Acknowledgments
We thank Yoshihiro Matsumoto for the generous gift of FEN1 protein, Altaf H. Sarker for advice on nuclear matrix fractionation protocols, Chiharu Hitomi for substantial contributions toward the cloning and purification of selected WRN protein constructs, and Kiran Rangaraj for performing far western analyses and for generating the diagram of WRN protein constructs. This work was supported by NIH/National Cancer Institute grants R01 CA063503 (P.K.C.), P01 CA092584 (P.K.C., J.A.T.) and R01 CA104660 (J.A.T., S.M.Y.) and by NIH/National Institute on Aging grant P01 AG017242 (J.C.). K.S.T. was supported by a Ruth Kirschstein National Research Service Award, F32GM083590 and National Institute on Aging Training grant T32 AG00266. Work at LBNL was performed under US Department of Energy Contract Number DE-AC02-05CH11231.
Supplementary Material
References
- 1.O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 2.Hohl M, Thorel F, Clarkson SG, Scharer OD. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J Biol Chem. 2003;278:19500–19508. doi: 10.1074/jbc.M213155200. [DOI] [PubMed] [Google Scholar]
- 3.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, et al. Recognition of RNA polymerase II and transcription bubbles by XPG CSB and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 4.O'Donovan A, Wood RD. Identical defects in DNA repair in xeroderma pigmentosum group G and rodent ERCC group 5. Nature. 1993;363:185–188. doi: 10.1038/363185a0. [see comments] [DOI] [PubMed] [Google Scholar]
- 5.Nouspikel T, Lalle P, Leadon SA, Cooper PK, Clarkson SG. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc Natl Acad Sci USA. 1997;94:3116–3121. doi: 10.1073/pnas.94.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsutakawa SE, Classen S, Chapados BR, Arvai A, Finger LD, Guenther G, et al. Human Flap Endonuclease Structures, DNA Double Base Flipping and a Unified Understanding of the FEN1 Superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohl M, Dunand-Sauthier I, Staresincic L, Jaquier-Gubler P, Thorel F, Modesti M, et al. Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res. 2007;35:3053–3063. doi: 10.1093/nar/gkm092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 11.Emmert S, Slor H, Busch DB, Batko S, Albert RB, Coleman D, et al. Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J Invest Dermatol. 2002;118:972–982. doi: 10.1046/j.1523-747.2002.01782.x. [DOI] [PubMed] [Google Scholar]
- 12.Lindenbaum Y, Dickson D, Rosenbaum P, Kraemer K, Robbins I, Rapin I. Xeroderma pigmentosum/cockayne syndrome complex: first neuropathological study and review of eight other cases. Eur J Paediatr Neurol. 2001;5:225–242. doi: 10.1053/ejpn.2001.0523. [DOI] [PubMed] [Google Scholar]
- 13.Schärer OD. XPG: its products and biological roles. Adv Exp Med Biol. 2008;637:83–92. doi: 10.1007/978-0-387-09599-8_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harada YN, Shiomi N, Koike M, Ikawa M, Okabe M, Hirota S, et al. Postnatal growth failure, short life span and early onset of cellular senescence and subsequent immortalization in mice lacking the xeroderma pigmentosum group G gene. Mol Cell Biol. 1999;19:2366–2372; PubMed. doi: 10.1128/mcb.19.3.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian M, Jones DA, Smith M, Shinkura R, Alt FW. Deficiency in the nuclease activity of xeroderma pigmentosum G in mice leads to hypersensitivity to UV irradiation. Mol Cell Biol. 2004;24:2237–2242. doi: 10.1128/MCB.24.6.2237-42.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiomi N, Kito S, Oyama M, Matsunaga T, Harada YN, Ikawa M, et al. Identification of the XPG region that causes the onset of Cockayne syndrome by using Xpg mutant mice generated by the cDNA-mediated knock-in method. Mol Cell Biol. 2004;24:3712–3719. doi: 10.1128/MCB.24.9.3712-9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiomi N, Mori M, Kito S, Harada YN, Tanaka K, Shiomi T. Severe growth retardation and short life span of double-mutant mice lacking Xpa and exon 15 of Xpg. DNA Repair (Amst) 2005;4:351–357. doi: 10.1016/j.dnarep.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bessho T. Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repair enzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 1999;27:979–983. doi: 10.1093/nar/27.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinfeld M, Xing JZ, Lee J, Leadon SA, Cooper PK, Le XC. Factors influencing the removal of thymine glycol from DNA in gamma-irradiated human cells. Prog Nucleic Acid Res Mol Biol. 2001;68:139–149. doi: 10.1016/S0079-6603(01)68096-6. [DOI] [PubMed] [Google Scholar]
- 21.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 22.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, et al. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Oshima J. The Werner syndrome protein: an update. Bioessays. 2000;22:894–901. doi: 10.1002/1521-1878(200010)22:10<894::AIDBIES4> 3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 24.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–2407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 26.Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, et al. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 27.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brosh RM, Jr, Driscoll HC, Dianov GL, Sommers JA. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- 29.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J Biol Chem. 2007;282:26591–2602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 30.Harrigan JA, Opresko PL, Von Kobbe C, Kedar PS, Prasad R, Wilson SH, et al. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J Biol Chem. 2003;278:22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 31.Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 32.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, et al. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. PubMed. [DOI] [PubMed] [Google Scholar]
- 33.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahn JJ, Lowery MP, Della-Coletta L, Adair GM, Nairn RS. Depletion of Werner helicase results in mitotic hyperrecombination and pleiotropic homologous and nonhomologous recombination phenotypes. Mech Ageing Dev. 2010;131:562–573. doi: 10.1016/j.mad.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, et al. Werner's syndrome protein (WRN) migrates Holliday junctions and colocalizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakamoto S, Nishikawa K, Heo SJ, Goto M, Furuichi Y, Shimamoto A. Werner helicase relocates into nuclear foci in response to DNA damaging agents and colocalizes with RPA and Rad51. Genes Cells. 2001;6:421–430. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- 37.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 38.Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:70920. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 41.Hagelstrom RT, Blagoev KB, Niedernhofer LJ, Goodwin EH, Bailey SM. Hyper telomere recombination accelerates replicative senescence and may promote premature aging. Proc Natl Acad Sci USA. 2010;107:15768–15773. doi: 10.1073/pnas.1006338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamiuchi S, Saijo M, Citterio E, de Jager M, Hoeijmakers JH, Tanaka K. Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc Natl Acad Sci USA. 2002;99:201–206. doi: 10.1073/pnas.012473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anachkova B, Djeliova V, Russev G. Nuclear matrix support of DNA replication. J Cell Biochem. 2005;96:951–961. doi: 10.1002/jcb.20610. [DOI] [PubMed] [Google Scholar]
- 45.Otrin VR, McLenigan M, Takao M, Levine AS, Protic M. Translocation of a UV-damaged DNA binding protein into a tight association with chromatin after treatment of mammalian cells with UV light. J Cell Sci. 1997;110:1159–1168. doi: 10.1242/jcs.110.10.1159. PubMed. [DOI] [PubMed] [Google Scholar]
- 46.Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.2818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, et al. Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 48.Constantinou A, Gunz D, Evans E, Lalle P, Bates PA, Wood RD, et al. Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J Biol Chem. 1999;274:5637–5648. doi: 10.1074/jbc.274.9.5637. [DOI] [PubMed] [Google Scholar]
- 49.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:1393946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 50.Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machwe A, Lozada E, Wold MS, Li GM, Orren DK. Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. J Biol Chem. 2011;286:3497–3508. doi: 10.1074/jbc.M110.105411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Z, Henricksen LA, Wold MS, Ingles CJ. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 53.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 54.Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, et al. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 55.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, et al. Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 56.Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008;47:10247–10254. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitano K, Yoshihara N, Hakoshima T. Crystal structure of the HRDC domain of human Werner syndrome protein, WRN. J Biol Chem. 2007;282:2717–2728. doi: 10.1074/jbc.M610142200. [DOI] [PubMed] [Google Scholar]
- 58.Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18:177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 59.Perry JJ, Asaithamby A, Barnebey A, Kiamanesch F, Chen DJ, Han S, et al. Identification of a coiled coil in werner syndrome protein that facilitates multimerization and promotes exonuclease processivity. J Biol Chem. 2010;285:25699–25707. doi: 10.1074/jbc. M110.124941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AIDPROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 61.Rubio MA, Kim SH, Campisi J. Reversible manipulation of telomerase expression and telomere length. Implications for the ionizing radiation response and replicative senescence of human cells. J Biol Chem. 2002;277:28609–28617. doi: 10.1074/jbc.M203747200. [DOI] [PubMed] [Google Scholar]
- 62.Davalos AR, Campisi J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J Cell Biol. 2003;162:1197–1209. doi: 10.1083/jcb.200304016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He DC, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.