Abstract
The etiologic paradigm of complex human disorders such as autism is that genetic and environmental risk factors are independent and additive, but the interactive effects at the epigenetic interface are largely ignored. Genomic technologies have radically changed perspective on the human genome and how the epigenetic interface may impact complex human disorders. Here, I review recent genomic, environmental and epigenetic findings that suggest a new paradigm of “integrative genomics” in which genetic variation in genomic size may be impacted by dietary and environmental factors that influence the genomic saturation of DNA methylation. Human genomes are highly repetitive, but the interface of large-scale genomic differences with environmental factors that alter the DNA methylome such as dietary folate is under-explored. In addition to obvious direct effects of some environmental toxins on the genome by causing chromosomal breaks, non-mutagenic toxin exposures correlate with DNA hypomethylation that can lead to rearrangements between repeats or increased retrotransposition. Since human neurodevelopment appears to be particularly sensitive to alterations in epigenetic pathways, a further focus will be on how developing neurons may be particularly impacted by even subtle alterations to DNA methylation and proposing new directions towards understanding the quixotic etiology of autism by integrative genomic approaches.
Key words: DNA methylation, copy number variation, autism, neurodevelopment, genomics, epigenomics, epigenetics, folate, folic acid, environmental exposures, Alu, MeCP2, LINE-1
Environmental Toxins Negatively Impact Global DNA Methylation
The epigenetic modification of DNA methylation acts at the interface of genetic and environmental factors. As part of the emerging field of “environmental epigenetics,” a variety of environmental toxins with known adverse impacts on human health or neurodevelopment have been investigated for their potential effects on DNA methylation and other epigenetic effects (reviewed in ref. 1). Arsenic, cadmium, benzene and air pollution are exposures associated with reduction of DNA methylation levels at LINE1 and/or Alu repeats in human tissues.2–6 In mouse models of human exposures, methylmercury resulted in hypermethylation of brain derived neurotropic factor (Bdnf) in hippocampus7 and diethylstilbestrol exposure reduced global methylation in the uterus.8 In a human Greenlandic Inuit population with high persistent organic pollutant (POP) levels, reduced global DNA methylation (LINE1) was observed with increased POP levels.9 Furthermore, prenatal exposure of a rat model with organochloride pesticides, methylmercury, POPs or a mixture of all three chemical classes showed that POPs in the mixture correlated with reduced DNA methylation levels in liver.10
Within the class of POPs, polychlorinated biphenyls (PCBs) are a widely distributed class of environmental pollutants previously used in industrial products until adverse health effects were recognized in the 1970s resulting in discontinued use. The developmental neurotoxicity of PCBs became devastatingly apparent after the large-scale consumption of PCB-contaminated rice oil that occurred in 1968 in Japan and in 1979 in Taiwan.11,12 PCBs are known to disrupt neurotransmitter systems, endocrine systems and intracellular signaling pathways.13,14 While PCB levels are gradually declining in the environment following the discontinued use, a related class of POPs, the polybrominated diphenyl ethers (PBDEs), are currently used as commercial flame-retardants and are a growing concern for human exposures. Our recent analyses of perinatal PBDE exposure in a genetic mouse model susceptible to social behavioral deficits showed global hypomethylation in brain associated with adverse social and cognitive behavioral outcomes (Woods et al., in preparation).
The major future challenge that will be explored in this review is understanding how DNA methylation of human repetitive elements interacts with environmental risk and protective factors in the etiology of a complex genetic disorder such as autism.
The Human Genome Is Highly Repetitive and Repeats Are Highly Polymorphic
Current estimates are that approximately half of the human genome is made up of repeats. Common repeats include transposon-derived repeats, simple sequence repeats, processed pseudogenes and tandemly repeated sequences, together making up almost half of repeats in the human genome.15 In addition to the common repeat categories observed in other mammalian species, the primate lineage has evolved a unique group of “low copy” repeats (called segmental dups), whose breakpoints are enriched for a CpG-dense class of repeats called Alu.16 Segmental dups predispose certain chromosomal loci to copy-number variation (CNV), which are gains or losses of DNA segments ranging from several kb to several Mb.17–19
Interestingly, segmental dups and the CNVs that arise from these low copy repeats appear to have played an important role in primate evolution and species differences in levels of brain expressed transcripts. Comparisons of human gene expression patterns with other primates have shown brain-specific transcript level differences that are unique to human.20,21 Interestingly, the human chromosomal regions that are significantly enriched for segmental dups22 also frequently overlap with regions of human/chimpanzee differences in gene neighborhood and brain expression.23
In addition to their contribution to primate species differences, repeats are also quite polymorphic between individual human genomes. The largest duplication blocks are largely invariant between individual humans, although 3% of assignable duplications are predicted to be unique to an individual.24 In addition, 2.7% of microsatellite repeats were recently estimated to be polymorphic between individuals, with long microsatellites showing the most variability.25 Additional variation in human genomes comes from the large blocks of ribosomal DNA repeats26,27 and interstitial telomeric repeats28 that are estimated to vary greatly between individuals. LINE-1 transposable elements also continue to undergo retrotransposition, particularly in the neuronal lineage, suggesting that repeats may contribute to somatic mosaicism and individual variation.29,30
Human Neurodevelopmental Disorders Show Increased CNV Burden
While some common CNVs are polymorphic and often inherited in humans, a higher frequency of de novo rare CNVs are found in patients with autism and schizophrenia compared to unaffected controls.31–33 In addition, individuals affected by neurodevelopmental disorders in general appear to have a greater overall burden of common polymorphic CNVs than unaffected controls. Chromosomal regions that are hotspots for primate-specific segmental dups and chimpanzee/human differences frequently coincide with the breakpoints of CNVs found in autism and schizophrenia, including 1q21.1, 15q11.2 and 15q13.3.31–34
While progress in CNV detection and their frequent occurrences in autism and other neurodevelopmental disorders is an exciting development for understanding genetic bases, challenges to understanding causality of specific genes within specific CNVs remain.35 Gains and losses of the same locus often leads to overlapping phenotypes, as is observed in 22q11.2, 7q11.23, 17p11.2 and 15q11-q13 deletion and duplication syndromes with co-morbid autism.35,36 Furthermore, although the gains and losses of genes within CNVs are expected to result in predictable changes to corresponding transcript levels, some genes within an autism 15q11-q13 duplication syndrome brain sample were actually expressed significantly lower than expected, likely due to the known epigenetic complexity of this locus.37,38
The other major challenge to interpreting the genetic relevance of CNVs is understanding how the loss or gain of specific chromosomal loci may cause disease. Since the loss of a gene copy is generally expected to be more pathogenic than a gain, the goal of human CNV disease association studies is often to find genes within small rare CNV deletions. However, this worthy approach may be unjustifiably ignoring the “elephant in the room” of rare CNV duplications associated with autism that are much larger than deletions (>500 kb) and contain more genes implicated in autism than the CNV deletions that are more intensely investigated.34
Complicating CNV genotype-phenotype studies even further are issues of variable penetrance or variable expressivity of the phenotype. For instance, while maternal duplication of 15q11-q13 (15q dup or idic15) is associated with autism in 85% of cases,38 paternal 15q11-q13 duplication has been observed in healthy unaffected individuals39 as well as cases with autism or language and social defects.40–44 Therefore, a simple genotype-phenotype correlation with CNVs and neurodevelopmental disorders is not apparent.
DNA Methylation Suppresses Genome Rearrangements and Hypomethylation Leads to Instability
DNA methylation is the first layer of epigenetic information, resulting from a covalent modification of cytosine at CpG sites. In the mammalian genome, CpG sites are de-enriched because of frequent C to T mutations that occur at methylated CpG sites.45 Therefore, large clusters of CpG sites called CpG islands are frequently assayed for DNA methylation because they are also often associated with gene promoters or other regulatory elements and allow the analysis of multiple CpG sites in a single assay. However, this CpG island bias in methylation analyses may be a case of “looking under a streetlight for lost keys,” as gene promoters are vastly de-enriched for DNA methylation, compared to gene bodies, intergenic and repetitive regions, where DNA methylation is extensive.46–49 However, DNA methylation even at repetitive regions is not saturating, as globally human genomes have ∼79% total methylation and the most methylated sequences are not higher than ∼90% methylation of available CpG sites.46–48
Collectively, 25% of the total DNA methylation in human cells is at Alu sequences and the most recent Alu subtypes are the most methylated.50 Interestingly, a correlation between the methylation state of Alu and structural genome variation in evolution has been suggested by a recent study of white-cheeked gibbons, a hybrid primate species. White-cheeked gibbons show multiple chromosomal rearrangements at Alu breakpoints and these breakpoints correspond to lower Alu methylation levels compared to humans.51 In human cell culture models and in cancer cell lines, DNA hypomethylation leads to genome instability.52 Furthermore, treatment with the DNA methylation inhibitor 5-aza-cytidine resulted in global hypomethylation, genomic instability and structural rearrangements,53 while Dnmt1 deficiency led to microsatellite instability in mouse embryonic stem cells.54
Diet Influences DNA Methylation and Folic Acid Supplementation is Protective for Autism
Nutritional modification of DNA methylation can have profound effects on phenotypic outcome of social animals such as queen determination in honeybees55 as well as agouti coat color and obesity in the genetic mutant Avy allele in mouse.56 Folate is the major methyl donor to S-adenosylmethionine (SAM), a key enzyme for DNA methylation in the one carbon metabolism pathway, and thus has been investigated in DNA methylation pathways, as well as protection or reversal of DNA hypomethylation caused by environmental pollutants.57 The synthetic form of folate, folic acid, is protective for human colon cancer through regulation of DNA repair and DNA methylation.58 Folic acid has well characterized protective effects on genome instability in cell culture of primary and transformed cell lines, likely mediated by DNA hypomethylation.
Folic acid supplementation recommendations for women of childbearing age in 1991 and widespread cereal fortification in 1998 are widely regarded as major advances in modern preventative medicine in the US, as these measures have been credited in preventing up to 70% of neural tube defects. In addition, folic acid appears to protect against a wide range of human cancers and neurodevelopmental and neurodegenerative disorders. Recent data from a large case-control epidemiological study demonstrates a significant protective association for prenatal vitamin use and high periconceptional folate intake in relation to risk for autism102. The strongest protective effect was observed with prenatal vitamins taken in the month before and first two months of pregnancy and for mothers with the MTHFR T/T risk allele, similar to that observed with neural tube defects.
This new finding revealing a protective effect of prenatal vitamin usage for autism risk is somewhat paradoxical to multiple hypotheses suggesting that folic acid supplementation may be responsible for the recent decade's apparent increase in autism diagnoses.59–61 Because the timing and level of folic acid supplementation in the form of prenatal vitamins is critical for the protective effect, these findings and hypotheses may not be in absolute conflict and there is middle ground to be reached with future studies. Perhaps the low-level of folic acid fortification in cereal grains in the diet may select for the survival of fetuses with increased number of repeats and structural duplications compared to previous generations. In this case, additional folic acid and B12 in prenatal vitamins taken prior to conception may buffer the genetic risk of increased repeat burden by providing additional methyl donors to methylate the additional DNA.
Interactions of Genetics and Folic Acid Protection from an Epigenomic Perspective
Previous research on genetic susceptibilities for folic acid protection have been limited to common single nucleotide polymorphisms (SNPs) in several genes in the one-carbon folate metabolism pathway, such as the common polymorphism in MTHFR. The protective nature of prenatal vitamin use for autism risk suggests that there could be an additional genomic susceptibility at the level of increased genomic repeat burden. This effect could be counteracted by dietary methyl donors in the critical first month of pregnancy when most women are not supplementing because they are unaware that they are pregnant. In support of this hypothesis, the incidence of Down syndrome, a common genetic disorder resulting from trisomy of chromosome 21 (adding ∼1.5% to total genome size), is reduced by the use of nutritional supplements (mainly folic acid and antioxidant vitamins C and E) during the first month of pregnancy.62 Furthermore, the MTHFR genotype of mothers has been implicated in the risk of Down syndrome by numerous studies (reviewed in ref. 63). Interestingly, a recent genomic analysis of Down syndrome lymphocytes showed altered DNA methylation of multiple genes on autosomes other than chromosome 21, suggesting genome-wide defects in DNA methylation for this genetic disorder.64
Why are the earliest stages of pregnancy the most critical for dietary methyl donors from an epigenomic point-of-view? Oocytes, early pre-implantation embryos and embryonic stem cells are characterized by having a greater need for DNA methylation, as they utilize non-CpG methylation in addition to higher CpG methylation as compared to differentiated fibroblasts.49 Recent next-generation sequencing efforts have now provided access to the whole human DNA methylome at single base resolution, revealing striking differences in the epigenomic landscapes of pluripotent and lineage-committed human cells.49,65 These studies have confirmed that CpG island promoters are generally unmethylated, while rare promoter methylation is inversely correlated with gene expression, often for a small subset of genes that regulate pluripotency and early lineage commitment. For the majority of genes, the “shores” of their CpG islands, located up to 2 kb distant from the promoter itself appear to be more relevant for tissue-type discrimination.46 Interestingly, gene bodies (spanning exons and introns) and intergenic regions show high (>75%) methylation in many human tissues, including embryonic stem cells and cerebral cortex.46–49 Gene body methylation levels and patterns vary considerably between cell types and developmental stages and are positively associated with gene expression.46,66,67 Emerging evidence is therefore in favor of distinct DNA methylation patterns that dynamically regulate cellular differentiation. Perhaps the reason that the pre- and periconception stages are the critical window for folic acid protection of neural tube defects and autism is because the early embryo has a critical high-need for methyl donors for dynamic DNA methylation changes during cellular differentiation while also quite vulnerable to environmental chemical exposures.
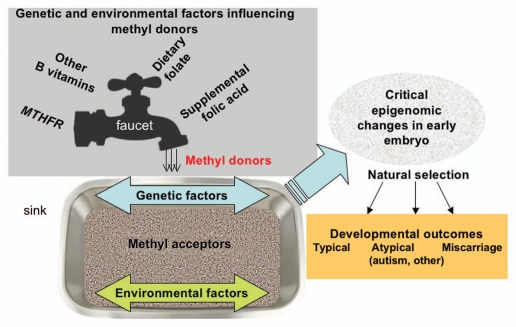
Figure 1 shows a model using the analogy of the whole human genome as a large “sink” of methyl acceptors, coming not only from DNA, but also RNA and protein molecules that become methylated. The size of the genomic sink varies between individuals and is influenced by both genetic and environmental factors that predispose to genomic changes. The genetic environmental interactions that have been investigated to date have focused on the genes and dietary factors that regulate the supply of methyl donors, shown here as a “faucet.” Since DNA methylation reactions are not saturated, especially in the critical peri-implantation stage, the size of the genomic cytosine “sink” may be modified by increased genomic duplications, requiring more methyl donors for DNA methylation. Genomic repeats and duplications that influence the quantity of methyl acceptors in the sink may self-perpetuate through genome instability when segmental dups are hypomethylated. An additional environmental factor could be increased maternal age, which conveys higher risk for genomic instability as well as for autism.68,69 The total burden of environmental chemical exposures may also impact DNA stability as well as methylation levels. Developmental outcomes such as autism may be impacted by the size of the genomic sink, but supplemental folic acid may influence the selection of implanted embryos and the severity of outcome for genetically high-risk individuals.
Figure 1.
The epigenomic “sink” analogy for the interaction of genomic structural changes and factors affecting methylation. Prior studies of genetic and environmental factors influencing DNA methylation have focused on those factors influencing the supply of methyl donors, such as MTHFR, dietary folate, prenatal vitamins containing folic acid and other B vitamins. In this model, an analogy is made of the “faucet” for the supply of methyl donors, while the genome is represented as a large “sink.” The genomic sink varies in size with common repeats (Alu and LINE-1), segmental dups and the structural variants and CNVs that arise from repeat rearrangements. Trisomy, such as in Down syndrome, also changes the size of the genomic sink. Environmental factors such as increased maternal age and a variety of chemical exposures may also influence the genomic sink size by increased risk of genomic instability. The implanted embryo is perhaps at the most vulnerable stage for the epigenomic interface of genetic and environmental risk factors because DNA methylation levels are high and methylomic patterns are dynamically changing. Since most human fertilizations do not implant or make it to successful live births, there is a is a strong natural selection for embryo implantation and survival that may be directly influenced by dietary folate or other environmental factors. For fetuses that do implant, subtle alterations in DNA methylation patterns may impact the developmental outcome, as either typical development, atypical (resulting in neurodevelopmental disorders such as autism) or miscarriage.
A Unifying Model for Environmental Impacts on the Human Methylomic “Sink”
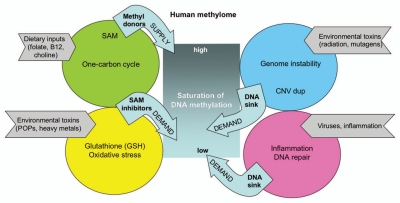
What appears to be a unifying theme from the emerging literature of epigenetic impacts of environmental exposures is that a wide variety of environmental toxins with known adverse effects on human neurodevelopment also appear to result in reduced global DNA methylation.1 On the protection side, dietary methyl donors and antioxidants appear to counteract the detrimental epigenetic effects of environmental toxins. A unifying mechanism for the biochemical connection of pathways between diet and environmental exposures that interface DNA methylation levels has been previously proposed.70 This important unifying hypothesis is expanded to include an integrative genomic point-of-view and the methylomic sink hypothesis in the model presented in Figure 2.
Figure 2.
An integrative genomic model of the major genetic and environmental pathways influencing the human methylome. The total human methylome is represented in the middle grey box that may vary in its overall level of saturation of DNA methylation. Four different categories of pathways that appear to influence saturation of DNA methylation are shown by the colored circles that are either creating supply or demand for methyl donors. Dietary inputs are required by SAM for DNA methylation in the one carbon cycle, while the biochemically linked pathway of GSH synthesis acts as inhibitors of SAM, creating a demand for more dietary inputs when chemicals are present and oxidative stress pathways are activated. Environmental toxins may also act directly as mutagens on the genome that may create more genome instability and potentially more CNV duplication events, creating more demand by increasing the size of the sink and could be self-perpetuating through hypomethylation. Lastly, viral infections or general inflammatory pathways may also increase demand on the need for methylation by promoting increased cell division, repression of viral DNA and DNA repair pathways.
Figure 2 outlines an oversimplified model of the pathways that act on both the “supply” and the “demand” sides affecting DNA methylation saturation levels in the total genomic sink. The one-carbon metabolic pathway is the major supplier of methyl groups for saturation of DNA methylation. The glutathione (GSH) synthesis pathway is upregulated in response to environmental chemicals as GSH becomes conjugated to diverse chemicals.70 Because the one-carbon methylation and the glutathione (GSH) synthesis pathways are biochemically linked, enhanced need for GSH to conjugate environmental toxins impairs SAM synthesis, acting as an added “demand” or drain on the availability of methyl donors for DNA methylation. In addition, increased DNA content resulting from genome instability between segmental dups leading to CNV duplications (that may also be increased due to DNA hypomethylation) increase demand for methyl donors by increasing the size of the sink. Furthermore, since new methyl groups need to be added with every cell division in an immune response, following DNA repair and following viral infections, inflammatory pathways are expected to increase demand on the methylome.
DNA Methylation Pathways Are of Critical Importance for Maturing Neurons
The developing mammalian brain is particularly sensitive to epigenetic alterations, as observed by the fact that mutations in epigenetic effectors can result in human neurodevelopmental disorders.71,72 DNA methylation is highly dynamic in mammalian postnatal neurons and these cells express DNA methyltransferases (DNMT) at high levels.73 Importantly, deficiency in both DNMT1 and DNMT3A in forebrain excitatory neurons showed deficits in learning and memory,74 similar to deficits in fear memory observed with DNMT1 chemical inhibition.75 Likewise, DNMT3A regulates emotional behavior and spine plasticity76 and ensures the expression of key neurogenic genes by targeting non-promoter DNA methylation around active genes.77
In mammals, DNA methylation at CpG sites has traditionally been considered a repressive mark in the genome, as high DNA methylation levels in intergenic sequences and repetitive elements prevents spurious transcription while lack of DNA methylation in promoters and CpG islands promotes gene transcription.78,79 Surprisingly, recent genome-wide DNA methylation studies have shown that high CpG methylation is common in gene bodies (genomic loci spanning exons and introns) where it positively correlates with transcription.47,80 Furthermore, gene bodies contained in partially methylated domains (PMDs, continuous domains of <70% methylated CpGs) in human fibroblasts showed decreased expression compared to those contained in highly methylated domains.49 We have recently demonstrated that human neurons contain differential PMDs and that neuronal-specific large-scale methylation domains contain a functional subset of neuronally expressed genes that are highly enriched for autism candidate genes (Schroeder et al. in revision). Collectively, these studies suggest that neurons may require high levels of DNA methylation particularly around gene bodies for genes involved in calcium signaling, synaptic transmission and neuronal differentiation.
In addition to neuronal DNA methylation, the DNA methyl binding protein MeCP2 is highly expressed in mature neurons in the brain. MeCP2 is an essential “reader” of DNA methylation marks in brain and has diverse roles in transcriptional modulation,81,82 chromatin structural organization,83,84 RNA splicing85 and DNA repair.86 Mutations in MECP2 cause the autism spectrum disorder Rett syndrome,87 duplication of MECP2 causes intellectual disability with autism,88,89 and significantly lower MeCP2 levels are observed in 79% of autism postmortem brain samples, correlating with increased MECP2 promoter methylation.90 These combined studies suggest that both DNA methylation and MeCP2 are part of a pathway critical for neuronal maturation in human development.
Interestingly, Rett syndrome brain and embryonic stem cell derived neural progenitor cells show increased levels of LINE-1 retrotransposition limited to neuronal tissues.91 These results suggest that neurons may be somewhat unique among human cell types in their controlled use of increased retrotransposon for genetic repeat diversity, and therefore may be more susceptible to changes in the genomic sink size. These results further implicate the role of DNA methylation and MeCP2 in the controlled regulation of LINE-1 retrotransposition and suggest that reduced DNA methylation and MeCP2 levels observed in autism90 may be contributing to genomic alterations of individual neurons in brain.
A New Perspective for Autism in “Integrative Genomics”
Autism includes a complex mixture of human disorders with the shared diagnostic features of social defects, language impairments and stereotyped or repetitive behaviors and interests.94–96 Based on family studies and twin studies, autism is considered to have a high heritability, but the genetic basis is decidedly complex (reviewed in ref. 97). Both “common disease-common variant” strategies based on genome-wide association and “common disease-rare variant” strategies are widely utilized to study the genetics etiology of autism.98 While rare genetic variants account for 10–20% of autism cases, epigenetic variation is predicted to be a potentially more common cause of dysregulation to synaptic pathways.36,97,99 Epigenetic alterations have been observed in several candidate genes associated with decreased expression in autism brain.90,100,101 Furthermore, deficiencies in methylation, GSH and oxidative stress pathways have been implicated in autism.92
The hypotheses in this review are proposed to stimulate thought and future investigation from an interdisciplinary perspective and are not intended to replace traditional genetic approaches, but simply to broaden their scope to include the whole genome and its interactions with environmental factors. The integrative genome perspective is also not intended to supplant the continued investigation of specific genes or chromosomal loci implicated in autism, although it may help in understanding how multiple genes relevant to neurodevelopment may be connected and impacted through global epigenetic changes to the whole genome. Future investigations under the heading of “integrative genomics” could include understanding how human structural variation may be impacted by environmental toxins and dietary factors. Of particular importance would be to understand how folic acid supplementation at levels found in cereal grains and in prenatal vitamins may be globally impacting the human genome and the developing brain methylome. Understanding how large-scale DNA methylation patterns change during neuronal differentiation and maturation and how both genetic and environmental factors may result in alterations to these pathways is also likely to be critical. All of these types of studies are expected to be of importance for treatment and prevention of autism and other complex genetic disorders.
Acknowledgments
I thank Dr. Dag Yasui for critical reading of the manuscript, as well as Dr. Rebecca Schmidt, Dr. Irva Hertz-Picciotto and Dr. Isaac Pessah for important interdisciplinary discussions. Thanks to NIH awards 1R01ES015171, 1R01HD048799 and 2R01HD041462 for support of research.
Abbreviations
- CNV
copy-number variant
- DNMT
DNA methyltransferase
- GSH
glutathione
- MeCP2
methyl CpG binding protein 2
- PBDE
polybrominated diphenyl ether
- PCB
polychlorinated biphenyl
- POP
persistent organic pollutants
- SAM
S-adenosylmethionine
References
- 1.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 4.Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 118:790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onishchenko N, Karpova N, Sabri F, Castren E, Ceccatelli S. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem. 2008;106:1378–1387. doi: 10.1111/j.1471-4159.2008.05484.x. [DOI] [PubMed] [Google Scholar]
- 8.Sato K, Fukata H, Kogo Y, Ohgane J, Shiota K, Mori C. Neonatal exposure to diethylstilbestrol alters expression of DNA methyltransferases and methylation of genomic DNA in the mouse uterus. Endocr J. 2009;56:131–139. doi: 10.1507/endocrj.k08e-239. [DOI] [PubMed] [Google Scholar]
- 9.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desaulniers D, Xiao GH, Lian H, Feng YL, Zhu J, Nakai J, et al. Effects of mixtures of polychlorinated biphenyls, methylmercury and organochlorine pesticides on hepatic DNA methylation in prepubertal female Sprague-Dawley rats. Int J Toxicol. 2009;28:294–307. doi: 10.1177/1091581809337918. [DOI] [PubMed] [Google Scholar]
- 11.Kuratsune M, Yoshimura T, Matsuzaka J, Yamaguchi A. Yusho, a poisoning caused by rice oil contaminated with polychlorinated biphenyls. HSMHA Health Rep. 1971;86:1083–1091. [PMC free article] [PubMed] [Google Scholar]
- 12.Rogan WJ, Gladen BC, Hung KL, Koong SL, Shih LY, Taylor JS, et al. Congenital poisoning by polychlorinated biphenyls and their contaminants in Taiwan. Science. 1988;241:334–336. doi: 10.1126/science.3133768. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson P, Jakobsson E, Fredriksson A. Brominated flame retardants: a novel class of developmental neurotoxicants in our environment? Environ Health Perspect. 2001;109:903–908. doi: 10.1289/ehp.01109903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guvenius DM, Aronsson A, Ekman-Ordeberg G, Bergman A, Noren K. Human prenatal and postnatal exposure to polybrominated diphenyl ethers, polychlorinated biphenyls, polychlorobiphenylols and pentachlorophenol. Environ Health Perspect. 2003;111:1235–1241. doi: 10.1289/ehp.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zepeda-Mendoza CJ, Lemus T, Yanez O, Garcia D, Valle-Garcia D, Meza-Sosa KF, et al. Identical repeated backbone of the human genome. BMC Genomics. 11:60. doi: 10.1186/1471-2164-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and functional impact of copy number variation in the human genome. Nature. 464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 18.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 19.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 20.Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Z, Tang H, Ventura M, Cardone MF, Marques-Bonet T, She X, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 23.De S, Teichmann SA, Babu MM. The impact of genomic neighborhood on the evolution of human and chimpanzee transcriptome. Genome Res. 2009;19:785–794. doi: 10.1101/gr.086165.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payseur BA, Jing P, Haasl RJ. A genomic portrait of human microsatellite variation. Mol Biol Evol. 28:303–312. doi: 10.1093/molbev/msq198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stults DM, Killen MW, Pierce HH, Pierce AJ. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18:13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA. 2009;106:17829–17834. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin KW, Yan J. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutat Res. 2008;658:95–110. doi: 10.1016/j.mrrev.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 30.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 36.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogart A, Leung KN, Wang NJ, Wu DJ, Driscoll J, Vallero RO, et al. Chromosome 15q11-13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J Med Genet. 2009;46:86–93. doi: 10.1136/jmg.2008.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hogart A, Wu D, Lasalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38:181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cook EH, Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- 40.Mohandas TK, Park JP, Spellman RA, Filiano JJ, Mamourian AC, Hawk AB, et al. Paternally derived de novo interstitial duplication of proximal 15q in a patient with developmental delay. Am J Med Genet. 1999;82:294–300. [PubMed] [Google Scholar]
- 41.Mao R, Jalal SM, Snow K, Michels VV, Szabo SM, Babovic-Vuksanovic D. Characteristics of two cases with dup(15)(q11.2-q12): one of maternal and one of paternal origin. Genet Med. 2000;2:131–135. doi: 10.1097/00125817-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 42.Veltman MW, Craig EE, Bolton PF. Autism spectrum disorders in Prader-Willi and Angelman syndromes: a systematic review. Psychiatr Genet. 2005;15:243–254. doi: 10.1097/00041444-200512000-00006. [DOI] [PubMed] [Google Scholar]
- 43.Veltman MW, Thompson RJ, Craig EE, Dennis NR, Roberts SE, Moore V, et al. A paternally inherited duplication in the Prader-Willi/Angelman syndrome critical region: a case and family study. J Autism Dev Disord. 2005;35:117–127. doi: 10.1007/s10803-004-1039-1. [DOI] [PubMed] [Google Scholar]
- 44.Depienne C, Moreno-De-Luca D, Heron D, Bouteiller D, Gennetier A, Delorme R, et al. Screening for genomic rearrangements and methylation abnormalities of the 15q11-q13 region in autism spectrum disorders. Biol Psychiatry. 2009;66:349–359. doi: 10.1016/j.biopsych.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 45.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 46.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc Natl Acad Sci USA. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie H, Wang M, Bonaldo Mde F, Smith C, Rajaram V, Goldman S, et al. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res. 2009;37:4331–4340. doi: 10.1093/nar/gkp393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbone L, Harris RA, Vessere GM, Mootnick AR, Humphray S, Rogers J, et al. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 2009;5:1000538. doi: 10.1371/journal.pgen.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 53.Shvachko LP. DNA hypomethylation as Achilles' heel of tumorigenesis: a working hypothesis. Cell Biol Int. 2009;33:904–910. doi: 10.1016/j.cellbi.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Kim M, Trinh BN, Long TI, Oghamian S, Laird PW. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 2004;32:5742–5749. doi: 10.1093/nar/gkh912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 56.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duthie SJ. Folate and cancer: how DNA damage, repair and methylation impact on colon carcinogenesis. J Inherit Metab Dis. 2011;34:101–109. doi: 10.1007/s10545-010-9128-0. [DOI] [PubMed] [Google Scholar]
- 59.Beard CM, Panser LA, Katusic SK. Is excess folic acid supplementation a risk factor for autism? Med Hypotheses. 2011;77:15–17. doi: 10.1016/j.mehy.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 60.Leeming RJ, Lucock M. Autism: Is there a folate connection? J Inherit Metab Dis. 2009;32:400–402. doi: 10.1007/s10545-009-1093-0. [DOI] [PubMed] [Google Scholar]
- 61.Rogers EJ. Has enhanced folate status during pregnancy altered natural selection and possibly autism prevalence? A closer look at a possible link. Med Hypotheses. 2008;71:406–410. doi: 10.1016/j.mehy.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 62.Czeizel AE, Puho E. Maternal use of nutritional supplements during the first month of pregnancy and decreased risk of Down's syndrome: case-control study. Nutrition. 2005;21:698–704. doi: 10.1016/j.nut.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 63.Patterson D. Folate metabolism and the risk of Down syndrome. Downs Syndr Res Pract. 2008;12:93–97. doi: 10.3104/updates.2051. [DOI] [PubMed] [Google Scholar]
- 64.Kerkel K, Schupf N, Hatta K, Pang D, Salas M, Kratz A, et al. Altered DNA methylation in leukocytes with trisomy 21. PLoS Genet. 6:1001212. doi: 10.1371/journal.pgen.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyall K, Pauls DL, Santangelo S, Spiegelman D, Ascherio A. Maternal early life factors associated with hormone levels and the risk of having a child with an autism spectrum disorder in the nurses health study II. J Autism Dev Disord. 2011;41:618–627. doi: 10.1007/s10803-010-1079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shelton JF, Tancredi DJ, Hertz-Picciotto I. Independent and dependent contributions of advanced maternal and paternal ages to autism risk. Autism Res. 2010;3:30–39. doi: 10.1002/aur.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee DH, Jacobs DR, Jr, Porta M. Hypothesis: a unifying mechanism for nutrition and chemicals as lifelong modulators of DNA hypomethylation. Environ Health Perspect. 2009;117:1799–1802. doi: 10.1289/ehp.0900741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lasalle JM, Yasui DH. Evolving role of MeCP2 in Rett syndrome and autism. Epigenomics. 2009;1:119–130. doi: 10.2217/epi.09.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;15:138–150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 73.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 74.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 76.LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bird AP, Wolffe AP. Methylation-induced repression—belts, braces and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 79.Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 80.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 81.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 84.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 85.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Squillaro T, Alessio N, Cipollaro M, Renieri A, Giordano A, Galderisi U. Partial silencing of methyl cytosine protein binding 2 (MECP2) in mesenchymal stem cells induces senescence with an increase in damaged DNA. Faseb J. 24:1593–1603. doi: 10.1096/fj.09-143057. [DOI] [PubMed] [Google Scholar]
- 87.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 88.Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, Hollanders K, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melissa BR, Sarika UP, Tavyev YJ, Feng Z, Claudia MBC, Christian PS, et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MECP2 duplication syndrome. Annals of Neurology. 2009;66:771–782. doi: 10.1002/ana.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, Lasalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:172–182. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, et al. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80:1611–1617. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- 93.Nagarajan RP, Patzel KA, Martin M, Yasui DH, Swanberg SE, Hertz-Picciotto I, et al. MECP2 promoter methylation and X chromosome inactivation in autism. Autism Res. 2008;1:169–178. doi: 10.1002/aur.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Volkmar FR, Pauls D. Autism. Lancet. 2003;362:1133–1141. doi: 10.1016/S0140-6736(03)14471-6. [DOI] [PubMed] [Google Scholar]
- 95.Lawler CP, Croen LA, Grether JK, Van de Water J. Identifying environmental contributions to autism: provocative clues and false leads. Ment Retard Dev Disabil Res Rev. 2004;10:292–302. doi: 10.1002/mrdd.20043. [DOI] [PubMed] [Google Scholar]
- 96.Veenstra-Vanderweele J, Christian SL, Cook EH., Jr Autism as a paradigmatic complex genetic disorder. Annu Rev Genomics Hum Genet. 2004;5:379–405. doi: 10.1146/annurev.genom.5.061903.180050. [DOI] [PubMed] [Google Scholar]
- 97.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32:402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Currenti SA. Understanding and determining the etiology of Autism. Cell Mol Neurobiol. 2010;30:161–171. doi: 10.1007/s10571-009-9453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med. 2009;7:62. doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. Faseb J. 24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmidt RJ, Hansen RL, Hartiala J, Allayee H, Schmidt LC, Tancredi DJ, et al. Prenatal vitamins, one-carbon metabolism gene variants, and risk for Autism. Epidemiology. 2011;22:476–485. doi: 10.1097/EDE.0b013e31821d0e30. [DOI] [PMC free article] [PubMed] [Google Scholar]