Abstract
Productive virus infection requires evasion, inhibition, or subversion of innate immune responses. West Nile virus (WNV), a human pathogen that can cause symptomatic infections associated with meningitis and encephalitis, inhibits the interferon (IFN) signal transduction pathway by preventing phosphorylation of Janus kinases and STAT transcription factors. Inhibition of the IFN signal cascade abrogates activation of IFN-induced genes, thus attenuating an antiviral response. We investigated the mechanism responsible for this inhibition and found that WNV infection prevents accumulation of the IFN-α receptor subunit 1 (IFNAR1). The WNV-induced depletion of IFNAR1 was conserved across multiple cell types. Our results indicated that expression of WNV nonstructural proteins resulted in activated lysosomal and proteasomal protein degradation pathways independent of the unfolded protein response (UPR). Furthermore, WNV infection did not induce serine phosphorylation, a modification on IFNAR1 that precedes its natural turnover. These data demonstrate that WNV infection results in a reduction of IFNAR1 protein through a non-canonical protein degradation pathway, and may participate in the inhibition of the IFN response.
Introduction
West Nile virus and other arboviruses have the remarkable ability to replicate and assemble virus particles despite activating innate immune responses. While animals have evolved highly complex and powerful immune signals, viruses have adopted mechanisms to evade, subvert, disrupt, or inhibit them. Hence the relationship between cellular agonists and viral antagonists of the innate immune response is a driving force in viral pathogenesis.
WNV has an approximately 11 kb-long positive-sense RNA genome encoding a single polyprotein, which is then proteolytically processed into 10 individual proteins, including three structural proteins (the capsid C, membrane M, and envelope E), and seven nonstructural proteins (glycoprotein NS1, NS2A, protease cofactor NS2B, protease and helicase NS3, NS4A, NS4B, and the polymerase NS5) (reviewed in 5,33,34). The virus is maintained in an enzootic cycle between mosquitoes and birds, but can infect mammals, including horses and humans. In humans, WNV infection typically presents as a febrile illness that can generally be resolved in healthy individuals (49,50). However, in some cases WNV infection can progress to more serious CNS-associated sequelae, including lethal encephalitis (46,53–55). Since its introduction into North America in 1999, WNV has spread rapidly across the continental United States, and subsequently into Mexico and South America, and has emerged as the major cause of viral encephalitis in the Western hemisphere (18,20). So far, antiviral therapies or vaccines are not available to treat or prevent WNV infections.
WNV has evolved the ability to block the interferon (IFN) signal transduction pathway (19,38). In WNV-infected cells, IFN exposure fails to induce phosphorylation of the Janus kinases JAK1 and Tyk2, and as a consequence, the STAT transcription factors remain latent, preventing transcriptional activation of interferon-stimulated genes (ISGs) (19). Mutagenesis experiments revealed that NS4B is a determinant for blocking IFN signaling in cells harboring a replicating genome. However, homologous mutations in infectious virus did not phenocopy these results, indicating that additional viral factors contribute to IFN antagonism during virus infections (13). Results derived from transfection studies with individual proteins remained inconclusive, invoking roles for NS4B and several other NS proteins in inhibition of the IFN response (31,38,44,45). Recently, various studies with WNV and dengue virus (DENV), a closely related flavivirus, have suggested a number of potential mechanisms employed to block IFN-stimulated signals and antiviral responses (1,22,29,41).
The IFN-α receptor (IFNAR) has two major subunits, IFNAR1 and IFNAR2c, which tightly dimerize upon IFN-α binding (58). IFNAR1 is a highly glycosylated protein with an apparent molecular weight of ∼110 kDa (10,35). Tyk2 is associated with IFNAR1, while JAK1, STAT1, and STAT2 are associated with IFNAR2 (8,9,11,30). Exposure to IFN induces dimerization of the receptor components and leads to the activation of the associated kinases (8,9,16). Following activation of Jak1 and Tyk2 kinases, the STAT proteins are phosphorylated, which leads to their translocation to the nucleus, resulting in transcription of ISGs. In response to IFN, IFNAR1 is phosphorylated on serines 535 (S535) and 539 (S539), leading to ubiquitination on several lysines (K501, K525, and K526) by the E3 ligase complex SCF-Trcp (27). Ubiquitinated IFNAR1, together with IFNAR2, translocate into endosomes. While IFNAR2 is recycled to the plasma membrane, IFNAR1 is proteolytically degraded in lysosomes (25,36,40,51). IFN-induced downregulation of IFNARs represents one of several negative feedback loops used by cells to counter potential cytotoxic effects associated with the expression of ISGs.
Since WNV infection inhibits IFN responses at a receptor-proximal point in the pathway, we envisioned a scenario in which WNV infection alters the biochemical composition of the IFN receptor complex. To this end, we predicted that WNV activates or deregulates natural mechanisms for control of IFNAR complex components that play a role in negative feedback regulation of the IFN response. The results presented in this study demonstrate that WNV infections lead to a reduction in IFNAR1 expression.
Materials and Methods
Plasmids and transfections
Plasmids encoding IFNAR1 (pUNO-IFNAR1) and IL-12Rβ1 (pORF9-IL12Rβ1) were purchased from Invivogen (San Diego, CA). Point mutants of IFNAR1 were generated with the QuikChange™ mutagenesis kit (Stratagene, Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. The overlapping primers used for mutagenesis were 5′-CAA ACT AGC CAA GAT GCA GGA AAT TAT TCT AAT G-3′ and 5′-CAT TAG AAT AAT TTC CTG CAT CTT GGC TAG TTT G-3′. The mutation was verified by nucleotide sequence analysis. HeLa cells were transfected with lipofectamine 2000 (Invitrogen, Carlsbad, CA) with 1 μg plasmid per 106 cells.
Cell culture and virus infections
HeLa, 293, A549, Vero, and BHK-21 cells were maintained at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco's modified Eagle medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal calf serum and 100 μg/mL penicillin, 100 μg/mL streptomycin, 1% non-essential amino acids (Gibco), and 50 μg/mL glutamine. Prior to infection, transfected or untransfected cells were seeded at 1 × 106 cells in 60-mm plates. Cells were infected with virus in PBS containing 2% FBS for 1 h. Unadsorbed virus was removed by washing, and DMEM supplemented with 2% FBS was added to the cells. After 24 h, the cells were treated with IFN-α2a or left untreated as indicated in the text.
To produce infectious virus, West Nile virus strain NY99 was produced from the plasmid construct pFL-WNV-NY99 or mutants as previously described (56). BHK-21 cells were electroporated with 2 μg in vitro transcribed RNA. Media were collected 3 d post-transfection, cleared of cell debris, and stored at −70°C. Titers were determined by plaque assay on BHK21 cells as previously described (60).
Antibodies
Mouse monoclonal antibodies against tubulin (T5168; Sigma-Aldrich, St. Louis, MO), IFNAR1 (sc7391; Santa Cruz Biotechnology, Santa Cruz, CA), IFNAR2 (sc30014; Santa Cruz Biotechnology), IL-12Rβ1 (sc25479; Santa Cruz Biotechnology), STAT1α p91 (sc417; Santa Cruz Biotechnology), rabbit monoclonal antibodies against phosphorylated IFNAR1 (PhosphoDetect, PS1003; Calbiochem, San Diego, CA), EGFR (15F8; Cell Signaling Technology, Inc., Beverly, MA), rabbit polyclonal antibodies against phosphotyrosine 701-STAT1 (9171; Cell Signaling Technology), and goat polyclonal antibodies against WNV NS3 (AF2907; R&D Systems, Inc., Minneapolis, MN) were purchased. Rabbit polyclonal antibodies to WNV NS5 were a gift from R. Padmanabhan (Georgetown University).
Western blots and immunoprecipitations
Cells were lysed in 1% Triton lysis buffer (1% Triton X-100, 150 mM sodium chloride, and 50 mM Tris, pH 8.0) and protein amounts were quantified by Bradford assay (Bio-Rad Laboratories, Inc., Hercules, CA). Proteins (15 μg) were separated by SDS-PAGE and transferred to Immobilon-P (Millipore, Billerica, MA). Membranes were cut according to molecular weight markers and membrane strips were probed with the indicated antibodies. Horseradish peroxidase-conjugated secondary antibodies against goat, mouse, and rabbit IgG (Amersham Biosciences, Piscataway, NJ) were used. The bands were visualized using SuperSignal West Pico solutions (Pierce Protein Research Products, Rockford, IL).
Biotin labeling
Cell surface proteins were labeled with sulfosuccinimidyl-2-(biotinamido)ethyl-1-1,3-dithiopropinate (sulfo-NHS-SS-biotin) provided in a cell surface protein isolation kit (Pierce Protein Research Products) according to the manufacturer's instructions. Briefly, cells were washed with PBS and incubated with sulfo-NHS-SS-biotin for 30 min at 4°C. The reaction was stopped with quenching buffer and the cell monolayer was scraped and lysed in a buffer containing 1% Brij and 0.1% SDS for 30 min on ice. One-seventh of extract was set aside for total protein control and the remainder of the biotin-labeled protein was affinity purified with immobilized agarose NeutrAvidin beads by incubation overnight at 4°C.
RNA analysis
Total RNA was extracted with TRIzol reagent (Invitrogen) following the manufacturer's instructions. For PCR, total RNA (1 μg) was used for cDNA synthesis with primer 5′-GGGGCTTGGTATATATGTGG-3′. XBP-1 cDNA was amplified with a forward primer 5′-CCTTGTAGTTGAGAACCAGG-3′, and reverse primer 5′-AGTTGTCCAGAATGCCCAAC-3′. PCR products were analyzed on 4% Nu-Sieve agarose gel. Real-time PCR was performed in triplicate using 200 pg of amplified cDNA and primer/probe sets specific to β-actin, IFNAR1, IFNAR2, and CHOP (Applied Biosystems Inc., Carlsbad, CA) on an ABI 7000 instrument. Results were normalized to β-actin prior to determination of fold induction.
Results
WNV infection induces a decrease in IFNAR1 protein levels
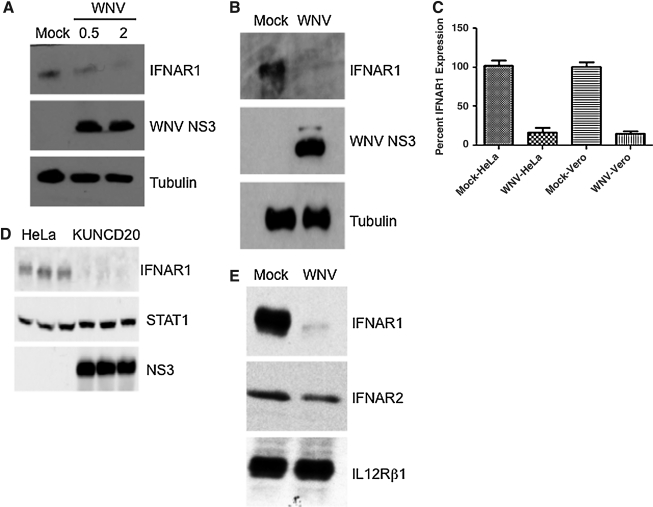
Preliminary efforts to investigate how WNV inhibits the IFN response pointed to defects in phosphorylation of Jak1, and in particular, Tyk2. However, expression levels of both kinases did not seem to be affected by WNV infection (19). While we confirmed those original observations, we conducted a more detailed analysis of expression levels of the two IFNAR subunits, IFNAR1 and IFNAR2. We examined the protein levels of both proteins during wild-type WNV NY99 infections of HeLa and Vero cells. In all cases examined, WNV replication significantly reduced accumulation of IFNAR1 (Fig. 1A and B). In multiple experiments with HeLa and Vero cells, IFNAR1 protein levels were decreased in a range from 75 to almost 99%. Protein levels were determined by densitometry and quantified by ImageJ software, standardized against tubulin (Fig. 1A and B). When HeLa cells were infected with WNV NY99 at MOI 0.5 or 2 for 24 h, IFNAR1 protein levels declined in relation to the MOI (Fig. 1A and Supplemental Fig. 1A; see online supplementary material at http://www.liebertonline.com). At times, it was noted that an additional band was observed migrating with an apparent MW of 60 kDa. This band does not represent IFNAR1, although it would be consistent with the calculated MW of 58 kD for the 557-long receptor subunit. Instead, the band corresponding to a MW of 110 kD represents bona-fide IRNAR1, which is known to be highly glycosylated at over 15 predicted sites (35). Consistent with glycosylation, treatment of cell extracts with the endoglycosylase PNGase F resulted in dissolution of the single higher molecular weight band into several smaller bands (Supplemental Fig. 1B; see online supplementary material at http://www.liebertonline.com) (35). Vero cells are defective in IFN production (12). Therefore, the decrease in IFNAR1 is not due to IFN being produced by infected cells and inducing degradation.
FIG. 1.
WNV inhibits accumulation of IFNAR1. (A) HeLa cells were infected with WNV NY99 at an MOI of 0.5 or 2 for 24 h and lysates were analyzed by Western blotting with anti-IFNAR1, anti-NS3 (WNV), and anti-tubulin antibody. (B) Vero cells were infected with WNV NY99 at an MOI of 2 for 24 h and lysates were analyzed by Western blotting with anti-IFNAR1, anti-NS3 (WNV), and anti-tubulin antibody. (C) Graph representing densitometry of IFNAR1 bands in multiple experiments with HeLa and Vero cells. (D) HeLa and WNV replicon-bearing HeLa cells, KUNCD20, and cell lysates were analyzed by Western blotting with anti-IFNAR1, anti-STAT1, and anti-NS3 (WNV) antibody. Triplicate samples were run for comparison. (E) HeLa cells were transfected with plasmids expressing IFNAR1, IFNAR2, or IL-12Rβ1 and 48 h later were infected with WNV NY99 (MOI of 2) for 24 h. Cell lysates were analyzed by Western blotting with the indicated antibodies. Data from one representative experiment of three are shown.
To determine if viral replication as well as nonstructural proteins were responsible for the IFNAR1 decrease, we examined previously characterized subgenomic replicon-bearing cells, KUNCD20 (13,19). IFNAR1 molecules were immunoprecipitated from KUNCD20 cells and parental HeLa cell lysates. KUNCD20 cells expressed approximately 70% less IFNAR1 protein than HeLa cells, demonstrating a significant depletion of this protein by WNV nonstructural proteins (Fig. 1C). Thus these results showed that infection and replication of two different strains of WNV, NY99 and Kunjin, cause a decrease in IFNAR1 protein levels, suggesting conservation of the mechanism between highly neuropathogenic (NY99) and more attenuated strains (Kunjin).
To determine the specificity of the IFNAR1 decrease, we analyzed the effect of WNV infections on IFNAR2 and IL-12Rβ1 expression levels. We selected IL-12Rβ1 because it associates with and signals through Tyk2, similarly to IFNAR1 (2,23,63). Following WNV infection, IL-12Rβ1 levels remained unaffected (0–5% decrease), and IFNAR2 levels were only modestly diminished compared to uninfected cells (10–40% decrease) (Fig. 1D).
IFNAR1 depletion is not caused by cell stress
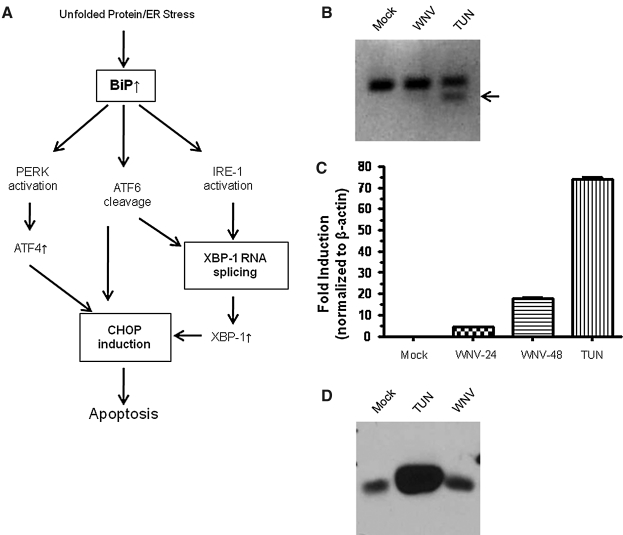
Previous studies have shown that certain flavivirus infections can induce cellular stress response pathways, leading to global inhibition of protein synthesis, enhanced degradation of proteins, and cell death (42,57,59,62). Recently, it has been reported that under selected conditions replication of vesicular stomatitis virus (VSV) and hepatitis C virus (HCV) can induce a cellular stress response leading to a decrease in IFNAR1 expression levels (36). The unfolded protein or ER stress response (UPR/ER stress) pathway consists of a highly defined group of proteins, that when activated, result in expression of proteins that dictate cell survival or apoptosis (Fig. 2A). To determine if the WNV-induced decrease in IFNAR1 protein was the result of a general cell stress response, we measured splicing of X box binding protein 1 (XBP1) mRNA and RNA expression of cyclic AMP response element-binding transcription factor homologous protein (CHOP), both of which increase in response to cellular stress (24,39,47). We found that XBP1 splicing was not induced in WNV-infected HeLa cells under the same conditions used to demonstrate inhibition of IFNAR1 (Fig. 2B). However, activation of XBP1 splicing was observed in A549 cells (data not shown). In HeLa cells, CHOP RNA transcripts were approximately 4.4-fold increased at 24 h post infection, which is low compared to the greater than 60-fold induction observed with tunicamycin (Fig. 2C). BiP is a master regulator of several of the UPR/ER stress pathways. Its upregulation is a hallmark of massive cell stress. WNV infection did not alter BiP expression, especially compared to tunicamycin treatment (Fig. 2D). Hence, based on these results covering multiple possible pathways, we considered it unlikely that IFNAR1 downregulation could be caused solely by a general stress response. Moreover, as shown in the following sections, we did not observe a stress-induced phosphorylation of IFNAR1 that precedes IFNAR1 degradation (36).
FIG. 2.
WNV does not induce cell stress. (A) Schematic representation of the current understanding of ER stress. (B and C) HeLa cells were infected with WNV at an MOI of 2 for 24 and 48 h. Total RNA was analyzed for: (B) XBP1 splicing by reverse transcription PCR (arrow denotes spliced XBP1), and (C) expression of CHOP mRNA by quantitative PCR. (D) HeLa cells were infected with WNV at an MOI of 2 for 24 h or treated with tunicamycin (30 μM) for 3 h. Total protein was analyzed for: BiP induction by Western blot (TUN, tunicamycin). Data from one representative experiment of three are shown.
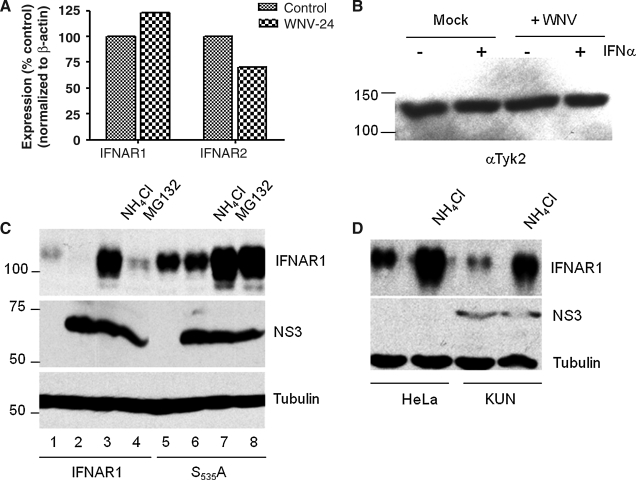
WNV-induced reduction of IFNAR1 occurs at the protein level and depends on both the lysosomal and proteasomal protein degradation pathways. WNV-induced reduction of IFNAR1 levels could be explained by at least three different mechanisms: inhibition of mRNA synthesis, inhibition of protein translation, or increase in protein degradation. Analysis of IFNAR1 and IFNAR2 mRNA levels did not reveal a significant difference between uninfected and WNV-infected HeLa cells (Fig. 3A), indicating that regulation of the receptor occurred at the protein level. IFNAR1 protein stability is dictated by the IFNAR1-associated kinase, Tyk2 (16,40,51,52). To determine if Tyk2 levels were altered by WNV infection, we infected HeLa cells with MOI of 2 and showed that Tyk2 levels were not altered by virus infection (Fig. 3B). These observations agreed with our previous results with HeLa cells expressing subgenomic WNV replicons (19). Thus far, our results indicated that WNV replication leads to a significant reduction in the accumulation of IFNAR1 in the presence of Tyk2 without cell stress, and invoked the possibility that a novel mechanism is responsible for the observed decrease in IFNAR1 accumulation.
FIG. 3.
The WNV-induced decrease of IFNAR1 is mediated by a lysosomal and proteasomal degradation pathway. (A and B) HeLa cells were infected with WNV NY99 (MOI of 2) for 24 h. (A) Total RNA was analyzed for: IFNAR1 and IFNAR2 mRNA by quantitative PCR. (B) WNV-infected cells were treated with 1000 U/mL IFN for 30 min. Total cell lysates were analyzed for Tyk2 expression by Western blot with the appropriate antibodies. (C) HeLa cells transfected with wild-type or mutant IFNAR1S535A were infected with WNV NY99 (MOI of 2) (lanes 2–4 and 6–8) for 8 h. WNV-infected cells were treated with NH4Cl (30 mM) or MG132 (10 μM) for 16 h. Transfected cells that were mock-infected and untreated cells acted as controls (lanes 1 and 5). Cell lysates were analyzed by Western blotting using antibodies against IFNAR1, NS3, and tubulin. (D) Kunjin replicon-bearing cells were treated with NH4Cl (30 mM) for 16 h. Cell lysates were analyzed by Western blotting using antibodies against IFNAR1, NS3, and tubulin. Data from one representative experiment of two are shown.
The current model for IFNAR1 turnover postulates that the receptor is degraded after endocytosis in the lysosomal compartment, which is partially controlled by phosphorylation of serine 535 (S535) (25,27,28,40). To determine the role of S535, we constructed an alanine substitution mutant, IFNAR1-S535A. We analyzed IFNAR1 and IFNAR1-S535A levels in cells treated with ammonium chloride (NH4Cl), which is known to prevent lysosomal acidification, and found that the chemical rescued accumulation of the receptor in both WNV-infected cells and in cells stably expressing subgenomic WNV replicons (Fig. 3C and D). These results indicated that WNV replication increased the rate of receptor turnover and destruction in lysosomes.
To determine if other protein degradation pathways were required to deplete IFNAR1, we treated cells with the proteasome inhibitor MG132. We found that MG132 treatment restored IFNAR1 levels in WNV-infected cells. These data support the previously published reports that IFNAR1 is imported into a lysosomal degradation pathway following IFN treatment (40). However, these data also suggest that WNV infection may also activate proteasomal degradation of IFNAR1, in the absence of functioning lysosomes.
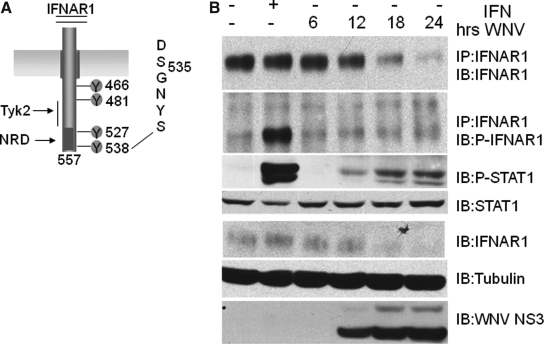
IFN binding induces the phosphorylation of serines S535 and S539 located near the C-terminus of IFNAR1 (Fig. 4A), leading to ubiquitin-dependent internalization and proteolytic digestion in the lysosomal compartment (25,27,28,40). Mutation of S535 to alanine (S535A) attenuates this process (27,40). Consistent with previous reports, the S535A mutation significantly increased accumulation of IFNAR1 in normal, uninfected cells. Importantly, its levels were modestly reduced in WNV-infected HeLa cells (15–25%). As observed with the wild-type receptor, accumulation of the mutant increased in the presence of both ammonium chloride and MG132. It is also possible that the role for the proteasome may increase when the lysosomal pathway is blocked. Regardless, these results were also consistent with a model predicting that WNV caused a reduction in IFNAR1 levels by increasing protein turnover by the lysosomal pathway.
FIG. 4.
The WNV-induced decrease of IFNAR1 occurs through a non-canonical pathway. (A) Schematic representation of IFNAR1 protein. Potential tyrosine phosphorylation sites are noted. The enlarged sequence marks serines 535 and 539 important for IFNAR1 stability. (B) HeLa cells were infected with WNV NY99 at an MOI of 2 for 24 h. Mock-infected cells were left untreated (lane 1) or treated with IFN (1000 U/mL) for 30 min (lane 2). Cell lysates were immunoprecipitated with IFNAR1 antibodies. Immune complexes and lysates were analyzed by Western blotting with antibodies against IFNAR1, phospho-S535/S539-IFNAR1 (P-IFNAR1), phospho-STAT1 (P-STAT1), STAT1, tubulin, and WNV NS3.
IFNAR1 turnover in WNV-infected cells uses a non-canonical pathway. The model for regulation of IFNAR1 expression predicts that S535 phosphorylation triggers ubiquitination and subsequently AP2 and clathrin-dependent endocytosis of the receptor subunit and proteolytic digestion in the lysosomal compartment (25–28,40). We therefore tested whether WNV induces phoshorylation of S535 through activation of a serine kinase. However, the decline of IFNAR1 in HeLa cells infected with WNV did not correlate with a detectable increase in S535 phosphorylation (Fig. 4, second panel, lanes 4–6). These results also showed that the observed reduction of IFNAR1 levels was not due to the expression of IFN-α that could be induced by WNV. If this were the case, S535 phosphorylation should have been detected, as it was after treatment of cells with IFN-α (Fig. 4, second panel, lane 2). Importantly, the results showed that stress-induced phosphorylation of S535 did not occur under our experimental conditions, and hence could not explain the observed reduction in IFNAR1 expression (36). It should be noted that in some experiments we could detect tyrosine phosphorylation of Stat1, indicating that inhibition of IFN signaling was not always complete.
WNV inhibits surface accumulation of IFNAR1
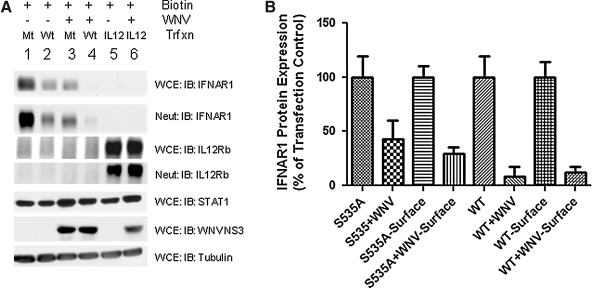
To further validate this model, we determined the surface expression of wild-type and mutant (S535A) IFNAR1 in normal and WNV-infected cells. For this purpose, we biotinylated cell surface proteins of normal and WNV-induced HeLa cells and determined the levels of affinity purified proteins. The results showed that WNV reduced total accumulation of wild-type and mutant IFNAR1 eight- to 10-fold, and two- to threefold, respectively (Fig. 5A and B). The reduction in surface protein expression was similar, reaching levels of eight- to 12-fold for wild-type, and threefold for the mutant receptor.
FIG. 5.
WNV inhibits expression of cell surface IFNAR1 protein. (A) HeLa cells transfected with wild-type (Wt) IFNAR1, IFNAR1-S535A (Mt), or IL-12Rβ1 were infected with WNV NY99 (MOI of 2) for 24 h. Cells were surface biotin-labeled and precipitated with neutravidin beads. Lysates (WCE) and biotin-avidin complexes (Neut) were analyzed by Western blotting with the indicated antibodies. (B) The level of cell surface proteins expressed in transfected and infected cells shown in A were quantified and expressed as a percentage of the protein expressed in uninfected total lysate (WCE) lanes. Data from one representative experiment of two are shown.
These results raised a question about the specificity of the observed reduction in cellular protein expression. To this end we made the following observations. First, among the surface proteins examined, IFNAR2 and IL-12Rβ1 were either not, or only modestly, reduced in WNV-infected cells. Second, a comparison of total surface protein levels between HeLa cells and cells expressing subgenomic replicons did not exhibit any detectable differences (data not shown). Third, expression levels of Tyk2, Stat1, tubulin, and actin were not affected by WNV infections (Figs. 1, 2, 4, and 5). Thus we concluded that WNV reduces IFNAR1 and does not act as a general activator of protein degradation or an inhibitor of protein synthesis.
Discussion
A novel mechanism for innate immune suppression
The current study builds on our previous observations with WNV and HCV, which revealed an unexpected difference between the two viruses in their ability to antagonize the innate immune response. Consistent with clinical results, we and many others found that HCV was very sensitive to the antiviral activity of IFN in tissue culture cells (3,14,19). In contrast to HCV, and in agreement with previous results, we found that WNV was much more resistant to the cytokine, which could be explained by a general inhibition of IFN signaling (19,38). Several other laboratories reported similar observations with other members of the genus Flavivirus, including the Japanese encephalitis and Dengue viruses (4,31,32,44,45). Biochemical studies provided evidence for a mechanism in which one or several viral proteins inhibited the Jak-Stat signal transduction pathway, explaining why WNV-infected cells failed to establish an antiviral state (13,19,38,44). The present study provided a possible explanation for these observations. WNV infections cause a significant reduction in a key component for IFN signaling, IFNAR1. This observation could have been explained by a cellular stress mechanism leading to a general inhibition of protein synthesis (i.e., through activation of PERK and phosphorylation of eIF2α) (36). However, such a scenario was not supported by our results. For example, WNV replication did not significantly alter the stability of the second IFN-α receptor subunit INFAR2, or the Tyk2-binding cytokine receptor subunit IL-12β1, and levels of cytoplasmic or nuclear proteins examined in this study remained normal. Also, Tyk2 expression levels were not altered during infection, so its depletion could not explain the IFNAR1 decrease (Fig. 3). Furthermore, we analyzed Tyk2 association with IFNAR1 during WNV infection. Tyk2 co-precipitated with IFNAR1 as long as IFNAR1 was detectable (data not shown). Furthermore, we did not obtain evidence for activation of a stress response in infected HeLa cells or in HeLa cells stably expressing subgenomic replicons (Fig. 1C). We did not observe XBP-1 splicing or CHOP expression. These data are in contrast to recently published reports suggesting that WNV causes cell death through induction of cell stress (42). The studies differ in multiplicity of infection as well as cell type. We demonstrated that different cell types respond to WNV infection by activating cell stress pathways (Supplementary Fig. 2A and B; see online supplementary material at http://www.liebertonline.com). However, WNV infection did not induce cell stress in our HeLa cells, so this cannot explain the observed loss of IFNAR1 accumulation.
Our model for inhibition of the IFN response differs from others proposed for WNV-related flaviviruses, including Japanese encephalitis virus (JEV) and tick-borne encephalitis virus (TBEV). These viruses appear to inhibit the IFN signaling by a mechanism requiring the viral polymerase encoded by NS5 (4,31,32). Although a physical interaction between the TBEV NS5 protein with IFNAR2 could be demonstrated, it did not lead to an inhibition of expression of either receptor subunit, as we observed with IFNAR1 in WNV-infected cells (4,48). Very recently, other groups also reported the role of flavivirus NS5 in blocking IFN signaling (1,41). Ashour et al. showed that dengue virus NS5 expression resulted in proteasomal degradation of the STAT2 signaling molecule (1). However, Mazzon et al. reported that NS5 inhibits phosphorylation/activation of STAT2, but requires other NS proteins to induce degradation. More importantly, it was shown that NS5 from highly pathogenic WNV could completely inhibit IFN signaling, whereas the NS5 from more attenuated strain could not (41). The exact cause of this discrepancy is unclear. Furthermore, we found that WNV infection causes a decrease in IFNAR1 protein levels, but that NS5 binding to the receptor is not responsible for the observed IFNAR1 decrease (Fig. 1A–D and data not shown). In fact, we found that no WNV NS protein was associated with IFNAR1 during the course of infection. Taken together, these results suggest that WNV and other flaviviruses have evolved several mechanisms to decrease intracellular innate immune signaling pathways.
Comparison with other viruses
Interestingly, a number of DNA and RNA viruses have evolved mechanisms to combat IFN signaling. Human cytomegalovirus initiates the degradation of JAK1, whereas human herpes simplex virus type 1 inhibits the phosphorylation of Jak1, Jak2, and Tyk2 (7,43,61). Conversely, closely related paramyxoviruses target multiple levels of the signaling cascade, including directly inducing the proteasomal degradation of STAT proteins and targeting the scaffolding that connects JAK1 to the receptor complex (21,61). The data presented in our studies suggest that WNV infection initiates the degradation of the IFNAR1 receptor subunit. The former studies and our current results highlight the wide variety of mechanisms employed by viruses to block innate immune responses and emphasize the need for further studies.
Model for WNV-induced inhibition of IFNAR1
The current model for ligand-induced degradation of IFNAR1 predicts phosphorylation of S535 and S539, creating a target site for ubiquitination (27,28). Ubiquitination of three lysine residues (K501, K525, and K526), activates an endocytic motif YVFF (26). Our results showed that WNV replication did not induce the initial phosphorylation of S535 in IFNAR1, thus preventing downstream events (Fig. 4A). Furthermore, the observation that the S535A mutant inhibited IFNAR1 degradation could be interpreted to mean that WNV activates a natural pathway at the level of S535 phosphorylation that functions in the absence of the natural ligands IFN-α or IFN-β, such as has recently been described for VSV and HCV (36). Therefore, we propose a model suggesting that WNV infections increase the rate of turnover of a subset of IFNAR1 proteins during their migration from the trans-Golgi to the plasma membrane. Such a model would make the following predictions. First, increased degradation is restricted to cell surface proteins, which is supported by our results, although we cannot provide an explanation at present for the mechanism selecting only a subset of these proteins. Second, IFNAR1 degradation occurs without S535 phosphorylation, as we observed. Third, IFNAR1 degradation does not appear to depend on a direct interaction with a viral protein, which is consistent with our results. Future studies are required to dissect the exact nature of the IFNAR1 turnover seen during WNV infection.
Implications for WNV biology
The current study provides a possible explanation for the previously described inhibition of IFN signaling. Because WNV infections begin in Langerhans and dermal dendritic cells (DCs) and then propagate in lymph nodes, locales known for efficient IFN production, it is likely that the observed attenuation of the IFN response is necessary for viremia to occur (6,17). Due to IFN's importance in antiviral responses, it is quite possible that WNV employs multiple mechanisms to inhibit IFN signaling, including NS2A inhibition of the IFN-β promoter (37); NS4B and NS5 blocking STAT activation (13,29,44); infection delaying IRF3 dimerization and activation (15); and now IFNAR1 protein depletion. Based on our results, we speculate that this suppression is caused by a novel mechanism involving degradation of IFNAR1 through protein destruction pathways. Hence, the unknown proteins responsible for this downregulation might become an important target for the development of therapeutics that prevent WNV-mediated inhibition of the IFN response, and consequently dissemination of the virus into the bloodstream and CNS disease, thus reducing the most serious neuropathologic sequelae of WNV infections in humans.
Supplementary Material
Acknowledgments
We thank Sid Balachandran (Fox Chase Cancer Center [FCCC]), Ted Ross (University of Pittsburgh), and Sean P. McBurney (University of Pittsburgh) for helpful comments and critical reading of the manuscript. We acknowledge the services provided by the FCCC tissue culture, DNA sequencing, and FACS sorting facilities. We thank Drs. R. Padmanabhan (Georgetown University) and Jon Chernoff (FCCC) for reagents. This work was supported by National Institutes of Health (NIH) grant R01 AI48046, and an appropriation from the Commonwealth of Pennsylvania. J.D.E. was supported by NIH grant T32 CA-09035-30, The Fine Foundation, and University of Pittsburgh CVR funds.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Ashour J. Laurent-Rolle M. Shi OY. Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon C. McVicar DW. Ortaldo JR. Rees RC. O'Shea JJ. Johnston JA. Interleukin 12 (IL-12) induces tyrosine phosphorylation of JAK2 and Tyk2: differential use of Janus family tyrosine kinases by IL-2 and IL-12. J Exp Med. 1995;181:399–404. doi: 10.1084/jem.181.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailly F. Si N. Si A. Trepo C. Treatment of HCV liver disease by recombinant interferon alpha. Nephrol Dial Transplant. 1996;11(Suppl 4):56–57. doi: 10.1093/ndt/11.supp4.56. [DOI] [PubMed] [Google Scholar]
- 4.Best SM. Morris KL. Shannon JG, et al. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005;79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinton MA. The molecular biology of West Nile Virus: a new invader of the Western hemisphere. Annu Rev Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- 6.Byrne S. Halliday GM. Johnston LJ. King NJ. Interleukin-1 beta but not tumor necrosis factor is involved in West Nile virus-induced Langerhans cell migration from the skin in C57BL/6 mice. J Invest Dermatol. 2001;117:702–709. doi: 10.1046/j.0022-202x.2001.01454.x. [DOI] [PubMed] [Google Scholar]
- 7.Chee AV. Roizman B. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J Virol. 2004;78:4185–4196. doi: 10.1128/JVI.78.8.4185-4196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colamonici O. Yan H. Domanski P, et al. Direct binding to and tyrosine phosphorylation of the alpha subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol Cell Biol. 1994;14:8133–8142. doi: 10.1128/mcb.14.12.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colamonici OR. Uyttendaele H. Domanski P. Yan H. Krolewski JJ. p135tyk2, an interferon-alpha-activated tyrosine kinase, is physically associated with an interferon-alpha receptor. J Biol Chem. 1994;269:3518–3522. [PubMed] [Google Scholar]
- 10.Constantinescu SN. Croze E. Murti A, et al. Expression and signaling specificity of the IFNAR chain of the type I interferon receptor complex. Proc Natl Acad Sci USA. 1995;92:10487–10491. doi: 10.1073/pnas.92.23.10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domanski P. Fish E. Nadeau OW. Witte M. Platanias LC. Yan H. Krolewski J. Pitha P. Colamonici OR. A region of the beta subunit of the interferon alpha receptor different from box 1 interacts with Jak1 and is sufficient to activate the Jak-Stat pathway and induce an antiviral state. J Biol Chem. 1997;272:26388–26393. doi: 10.1074/jbc.272.42.26388. [DOI] [PubMed] [Google Scholar]
- 12.Emeny JM. Morgan MJ. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J Gen Virol. 1979;43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- 13.Evans JD. Seeger C. Differential effects of mutations in NS4B on West Nile virus replication and inhibition of interferon signaling. J Virol. 2007;81:11809–11816. doi: 10.1128/JVI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Festi D. Sandri L. Mazzella G, et al. Safety of interferon beta treatment for chronic HCV hepatitis. World J Gastroenterol. 2004;10:12–16. doi: 10.3748/wjg.v10.i1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredericksen BL. Gale M., Jr West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauzzi MC. Velazquez L. McKendry R. Mogensen KE. Fellous M. Pellegrini S. Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J Biol Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- 17.Granwehr BP. Lillibridge KM. Higgs S. Mason PW. Aronson JF. Campbell GA. Barrett AD. West Nile virus: where are we now? Lancet Infect Dis. 2004;4:547–556. doi: 10.1016/S1473-3099(04)01128-4. [DOI] [PubMed] [Google Scholar]
- 18.Gubler DJ. The continuing spread of West Nile virus in the Western hemisphere. Clin Infect Dis. 2007;45:1039–1046. doi: 10.1086/521911. [DOI] [PubMed] [Google Scholar]
- 19.Guo JT. Hayashi J. Seeger C. West Nile virus inhibits the signal transduction pathway of alpha interferon. J Virol. 2005;79:1343–1350. doi: 10.1128/JVI.79.3.1343-1350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes EB. Sejvar JJ. Zaki SR. Lanciotti RS. Bode AV. Campbell GL. Virology, pathology, and clinical manifestations of West Nile virus disease. Emerg Infect Dis. 2005;11:1174–1179. doi: 10.3201/eid1108.050289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath CM. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur J Biochem. 2004;271:4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- 22.Jiang D. Weidner JM. Qing M, et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaleem A. Ahmad I. Hoessli DC. Walker-Nasir E. Saleem M. Shakoori AR. Nasir Ud D. Epidermal growth factor receptors: function modulation by phosphorylation and glycosylation interplay. Mol Biol Rep. 2009;36:631–639. doi: 10.1007/s11033-008-9223-6. [DOI] [PubMed] [Google Scholar]
- 24.Kim R. Emi M. Tanabe K. Murakami S. Role of the unfolded protein response in cell death. Apoptosis. 2006p;11:5–13. doi: 10.1007/s10495-005-3088-0. [DOI] [PubMed] [Google Scholar]
- 25.Kumar K. Varhese B. Banerjee A. Baker DP. Constantinescu SN. Pellegrini S. Fuchs SY. Basal ubiquitin-independent internalization of interferon alpha receptor is prevented by Tyk2-mediated masking of a linear endocytic motif. J Biol Chem. 2008;283:18566–18572. doi: 10.1074/jbc.M800991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar K. Barriere H. Carbone CJ, et al. Site-specific ubiquitination exposes a linear motif to promote interferon-alpha receptor endocytosis. J Cell Biol. 2007;179:935–950. doi: 10.1083/jcb.200706034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar K. Krolewski JJ. Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination of the IFNAR1 subunit of type I interferon receptor. J Biol Chem. 2004;279:46614–46620. doi: 10.1074/jbc.M407082200. [DOI] [PubMed] [Google Scholar]
- 28.Kumar K. Tang W. Ravindranath AK. Clark WA. Croze E. Fuchs SY. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J. 2003;22:5480–5490. doi: 10.1093/emboj/cdg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurent-Rolle M. Boer EF. Lubick KJ, et al. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol. 2010;84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X. Leung S. Kerr IM. Stark GR. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol. 1997;17:2048–2056. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin RJ. Chang BL. Yu HP. Liao CL. Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin RJ. Liao CL. Lin E. Lin YL. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J Virol. 2004;78:9285–9294. doi: 10.1128/JVI.78.17.9285-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindenbach BD. Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach BD. Rice CM. Flaviviridae: the viruses, their replication. In: Knipe HP, editor. Fields Virology. 4th. Lippincott, Williams, & Wilkins; Philadelphia: 2001. pp. 991–1041. [Google Scholar]
- 35.Ling LE. Zafari M. Reardon D. Brickelmeier M. Goelz SE. Benjamin CD. Human type I interferon receptor, IFNAR, is a heavily glycosylated 120-130 kD membrane protein. J Interferon Cytokine Res. 1995;15:55–61. doi: 10.1089/jir.1995.15.55. [DOI] [PubMed] [Google Scholar]
- 36.Liu J. HuangFu WC. Kumar KG, et al. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu WJ. Chen HB. Wang XJ. Huang H. Khromykh AA. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J Virol. 2004;78:12225–12235. doi: 10.1128/JVI.78.22.12225-12235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu WJ. Wang XJ. Mokhonov VV. Shi PY. Randall R. Khromykh AA. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol. 2005;79:1934–1942. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marciniak SJ. Yun CY. Oyadomari S, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marijanovic Z. Ragimbeau J. Kumar KG. Fuchs SY. Pellegrini S. TYK2 activity promotes ligand-induced IFNAR1 proteolysis. Biochem J. 2006;397:31–38. doi: 10.1042/BJ20060272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazzon M. Jones M. Davidson A. Chain B. Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- 42.Medigeshi GR. Lancaster AM. Hirsch AJ, et al. West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J Virol. 2007;81:10849–10860. doi: 10.1128/JVI.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller DM. Rahill BM. Boss JM. Lairmore MD. Durbin JE. Waldman JW. Sedmak DD. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz-Jordan JL. Laurent-Rolle M. Ashour J. Martinez-Sobrido L. Ashok M. Lipkin WI. Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munoz-Jordan JL. Sanchez-Burgos GG. Laurent-Rolle M. Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci USA. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nisenbaum C. Wallis K. Meningo-encephalitis due to West Nile fever. Reports of 2 cases. Helv Paediatr Acta. 1965;20:392–402. [PubMed] [Google Scholar]
- 47.Oyadomari S. Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 48.Park GS. Morris KL. Hallett RG. Bloom ME. Best SM. Identification of residues critical for the interferon antagonist function of Langat virus NS5 reveals a role for the RNA-dependent RNA polymerase domain. J Virol. 2007;81:6936–6946. doi: 10.1128/JVI.02830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen LR. Marfin AA. West Nile virus: a primer for the clinician. Ann Intern Med. 2002;137:173–179. doi: 10.7326/0003-4819-137-3-200208060-00009. [DOI] [PubMed] [Google Scholar]
- 50.Petersen LR. Roehrig JT. Hughes JM. West Nile virus encephalitis. N Engl J Med. 2002;347:1225–1226. doi: 10.1056/NEJMo020128. [DOI] [PubMed] [Google Scholar]
- 51.Ragimbeau J. Alcover EDA. Eid P. Uze G. Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 2003;22:537–547. doi: 10.1093/emboj/cdg038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richter MF. Dumenil G. Uze G. Fellous M. Pellegrini S. Specific contribution of Tyk2 JH regions to the binding and the expression of the interferon alpha/beta receptor component IFNAR1. J Biol Chem. 1998;273:24723–24729. doi: 10.1074/jbc.273.38.24723. [DOI] [PubMed] [Google Scholar]
- 53.Sampson BA. Ambrosi C. Charlot A. Reiber K. Veress JF. Armbrustmacher V. The pathology of human West Nile Virus infection. Hum Pathol. 2000;31:527–531. doi: 10.1053/hp.2000.8047. [DOI] [PubMed] [Google Scholar]
- 54.Sejvar JJ. Haddad MB. Tierney BC, et al. Neurologic manifestations and outcome of West Nile virus infection. JAMA. 2003;290:511–515. doi: 10.1001/jama.290.4.511. [DOI] [PubMed] [Google Scholar]
- 55.Sejvar JJ. Leis AA. Stokic DS, et al. Acute flaccid paralysis and West Nile virus infection. Emerg Infect Dis. 2003;9:788–793. doi: 10.3201/eid0907.030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi PY. Tilgner M. Lo MK. Kent KA. Bernard KA. Infectious cDNA clone of the epidemic West Nile virus from New York City. J Virol. 2002;76:5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su HL. Liao CL. Lin YL. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol. 2002;76:4162–4171. doi: 10.1128/JVI.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takaoka A. Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 59.Waris G. Tardif KD. Siddiqui A. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER-nucleus signal transduction pathway and activates NF-kappaB and STAT-3. Biochem Pharmacol. 2002;64:1425–1430. doi: 10.1016/s0006-2952(02)01300-x. [DOI] [PubMed] [Google Scholar]
- 60.Yamshchikov VF. Wengler G. Perelygin AA. Brinton MA. Compans RW. An infectious clone of the West Nile flavivirus. Virology. 2001;281:294–304. doi: 10.1006/viro.2000.0795. [DOI] [PubMed] [Google Scholar]
- 61.Yokota S. Yokosawa N. Kubota T, et al. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phosphorylation of STATs and janus kinases during an early infection stage. Virology. 2001;286:119–124. doi: 10.1006/viro.2001.0941. [DOI] [PubMed] [Google Scholar]
- 62.Yu CY. Hsu YW. Liao CL. Lin YL. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou J. Presky DH. Wu CY. Gubler U. Differential associations between the cytoplasmic regions of the interleukin-12 receptor subunits beta1 and beta2 and JAK kinases. J Biol Chem. 1997;272:6073–6077. doi: 10.1074/jbc.272.9.6073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.