Abstract
Brain metastasis (BM) can affect ~25% of non-small cell lung cancer (NSCLC) patients during their lifetime. Efforts to characterize patients that will develop BM have been disappointing. microRNAs (miRNAs) regulate the expression of target mRNAs. miRNAs play a role in regulating a variety of targets and, consequently, multiple pathways, which make them a powerful tool for early detection of disease, risk assessment, and prognosis. We investigated miRNAs that may serve as biomarkers to differentiate between NSCLC patients with and without BM. miRNA microarray profiling was performed on samples from clinically matched NSCLC from seven patients with BM (BM+) and six without BM (BM−). Using t-test and further qRT-PCR validation, eight miRNAs were confirmed to be significantly differentially-expressed. Of these, expression of miR-328 and miR-330-3p were able to correctly classify BM+ vs. BM− patients. This classifier was used on a validation cohort (n=15) and it correctly classified 12/15 patients. Gene expression analysis comparing A549 parental and A549 cells stably transfected to over-express miR-328 (A549–328) identified several significantly differentially-expressed genes. PRKCA was one of the genes over-expressed in A549–328 cells. Additionally, A549–328 cells had significantly increased cell migration compared to A549 cells, which was significantly reduced upon PRKCA knockdown. In summary, miR-328 has a role in conferring migratory potential to NSCLC cells working in part through PRKCA and with further corroboration in additional independent cohorts, these miRNAs may be incorporated into clinical treatment decision making to stratify NSCLC patients at higher risk for developing BM.
Keywords: microRNA, non-small cell lung cancer, brain metastasis, biomarker, migration
Introduction
In 2010, an estimated 222,520 cases of lung cancer will be diagnosed in the United States1. It is the leading cause of cancer deaths and ~88% of patients with primary lung malignancy have non-small cell lung cancer (NSCLC). Brain metastasis (BM) can affect up to 25% of these patients during their lifetime2. BM causes significant neurologic, cognitive, and emotional difficulties3 and negatively impacts survival4. Previous efforts to characterize patients that will develop BM have been fairly disappointing. Currently, prophylactic cranial irradiation (PCI) is offered to all small cell lung cancer patients (but not to NSCLC patients) with early-stage disease that have responded to therapy or have stable disease after initial systemic treatment5. However, better selection of patients to offer PCI will spare those patients unlikely to develop BM from PCI-related side effects. Currently, there are no common practice measures to reduce the risk of BM in NSCLC. In locally-advanced stage III NSCLC, a clinical trial to determine the benefit of PCI accrued slowly, was terminated early, and was not statistically powered to meet the primary endpoint of improvement in survival6.
Thus, there is a need for improvement in patient stratification. Molecular biomarkers could be of benefit to stratify these patients, but the ability to obtain adequate, quality tumor tissue in a standardized fashion for genomic profiling can be challenging and hence limiting. Recently, microRNAs (miRNAs) have been studied to characterize tumors7. miRNAs are small non-coding RNAs of 18–25 nucleotides that might impact various stages of development and progression of cancer. In general, one miRNA appears to regulate several hundred genes, and as a result, miRNA profiling could serve as a better classifier than gene expression profiling8.
miRNA profiling for lung cancer has been previously conducted by various groups to predict patient survival9. These profiles have also been correlated with clinico-pathological parameters of lung cancer patients9–11. Recently, Bishop et al, used this approach for classification of NSCLC12. Several of the miRNAs identified from these studies have been associated with the key regulatory pathways such as EGFR13 and KRAS 14 in lung cancer.
In this study, we hypothesized that miRNAs could serve as biomarkers to differentiate NSCLC patients with BM (BM+) from those without BM (BM−). We identified a classifier based on the expression of miR-328 and miR-330-3p, which could successfully differentiate BM− vs. BM+ cases in a separate validation cohort. In addition, our results show that miR-328 plays a role in the increased migratory potential in NSCLC cells.
Materials and Methods
Patients and Tumor Samples
This study was conducted under an Institutional Review Board (IRB) approved protocol. BM+ and BM− NSCLC patients identified through the Scottsdale Healthcare (SHC) tumor registry diagnosed during the periods January 1, 2003 to December 30, 2006 with formalin-fixed, paraffin-embedded (FFPE) tumor tissue available (SHC Discovery) were included in a previously published study 15. The pre-requisites to be included in this miRNA discovery study were performance of a computed tomography (CT) of the chest, whole-body 2-[18F]-Fluoro-2-Deoxy-D-Glucose positron emission computed tomography-CT (FDG-PET CT), and brain magnetic resonance imaging (MRI) study within 30 days of one another for each patient. Where possible, BM− NSCLC patients were matched to BM+ NSCLC patients by age, gender, histology, and stage at diagnosis. For this discovery cohort, seven patients with BM+ and six patients with BM− NSCLC were included (Table 1; Supplemental Table 1).
Table 1.
Clinical Characteristics of Patients and NSCLC tumors
| Discovery SHC Cohort | IOWA NSCLC Brain | Validation SHC Cohort | ||
|---|---|---|---|---|
| BM+ | BM− | Metastasis Tumor TissueX | ||
| No. of cases | 7 | 6 | 24 | 15 |
| Median Age years (range) | 58 (53–74) | 66.5 (60–71) | 60.5 (38–77) | 65 (46–78) |
| Gender | ||||
| Male | 1/7 (14.3%) | 3/6 (50%) | 13/24 (54.2%) | 5/15 (33.3%) |
| Female | 6/7 (85.7%) | 3/6 (50%) | 11/24(45.8%) | 10/15 (66.7%) |
| Stage at Diagnosis | ||||
| 1A or 1B | - | 2/6 (33%) | - | 4/15 (27%) |
| 2A or 2B | - | - | - | 11/15 (73%) |
| 3A or 3B | 4/7 (57.1%) | 1/6 (17%) | - | - |
| 4 | 3/7 (42.9%) | 3/6 (50%) | - | - |
| Histology | ||||
| Adenocarcinoma | 4/7 (57.1%) | 4/6 (66.6%) | 14/24 (58.4%) | 9/15 (60%) |
| Squamous cell carcinoma | 3/7 (42.9%) | 2/6 (33.3%) | 5/24 (20.8%) | 3/15 (20%) |
| NSCLC NOS#/Other | - | - | 5/24 (20.8%) | 3/15 (20%) |
“Stage at Diagnosis” information not available
Not otherwise specified or other histology is defined as neither adenocarcinoma nor squamous carcinoma or poorly differentiated and additional classification by pathologist was not available.
For validation of our miRNA profiling results, an independent cohort of 15 NSCLC patients with FFPE lung tumor tissue who underwent surgical resection at SHC between March 2001 and November 2006 were included under IRB approval (SHC Validation). These patients underwent preoperative evaluation per common practice guidelines including CT chest, FDG-PET CT, and either CT or MRI of the brain (Table 1).
Twenty-four NSCLC cases from the University of Iowa Hospitals and Clinics (IOWA) collected under an IRB approved protocol were also included, as an independent dataset for testing. These samples were all collected from metastatic brain lesions and flash frozen and stored in liquid nitrogen until the time of RNA extraction.
RNA extraction and miRNA microarray profiling
RNA was extracted from all FFPE NSCLC tumor specimens by manually scraping (macrodissection) the tumor tissue from the slides followed by de-paraffinization with xylene at 50°C. Three to six sections per tumor sample were used for RNA extraction. The pellet obtained after centrifugation was washed in 100% ethanol and digested with digestion buffer (RecoverAll Total Nucleic Acid Isolation, Part # 8788G, Ambion) and proteinase K solution at 50°C for 3 hours. Total RNA was extracted following standard protocols using the phenol method using the TRI reagent (Molecular Research Center Inc, Cincinnati, OH). The concentration and purity of isolated RNA was estimated using the ND-1000 micro-spectrophotometer (NanoDrop Technologies, Wilmington, DE). A minimum of 1 μg of total RNA was added to GenoExplorer™ microRNA Expression System (GenoSensor Corporation, Tempe, AZ) containing probes in triplicate for 678 validated human mature miRNAs with an additional 473 validated human pre-miRNAs (Sanger miRNA Registry, version 12.0 September 2008 http://www.mirbase.org) along with positive and negative control probes16. One miRNA microarray chip hybridization was performed per patient sample.
Identification of candidate miRNA biomarkers
miRNA expression was estimated from microarray profiling and was normalized to the reference gene 5S-rRNA after a thorough evaluation of several normalization techniques such as using a simple global scaling factor as in MAS 5.0 approach17, scale and median-shift method as used in18, 19, normalization with non-variant miRNA from the dataset20, and normalization with house-keeping genes21.
Differential expression analysis aims at finding genes or miRNAs that are significantly expressed in one condition in contrast to the other. A two-sided t-test was used to identify differentially-expressed miRNAs between the BM+ and BM− samples. The p-values generated from the t-test were corrected for multiple hypotheses testing using Benjamini-Hochberg correction22.
Correlation coefficients between BM+ and the clinical parameters: age, gender, stage at diagnosis, and histology were computed using Spearman's rank correlation. Similarly, correlation coefficients were also computed for miRNA with age, gender, stage at diagnosis, and histology.
Quantitative real-time PCR (qRT-PCR) analysis of miRNAs
Confirmation of the top 19 differentially-expressed miRNAs in SHC Discovery BM+ vs. BM− was performed using qRT-PCR by using the total RNA extracted from these samples. The GenoExplorer™ miRNAFirst-strand cDNA Core Kit (#2002–50, GenoSensor Corporation, Tempe, AZ) was used to generate miRNA first-strand cDNA. miRNA expression levels were measured using miRNA specific forward primers and a universal reverse primer (GenoSensor Corporation, Tempe, AZ) in a SYBR green assay (#04887352001, Roche, Indianapolis, IN)16.
The PCR reactions were carried out in triplicate in a 384-well plate format with 15 μl reaction volume using Lightcycler 480 (Roche, Indianapolis, IN). The PCR reaction conditions were 15 minutes denaturation at 94°C followed by 45 cycles of 94°C for 30 seconds, 59°C for 15 seconds, and 72°C for 30 seconds. Melting curve analysis was used to assess the specificity of the amplified product. Quantification of these miRNAs was carried out using delta-delta CT normalizing the result to the reference gene 5S-rRNA23.
Class prediction via strong-feature set algorithm
Strong-feature set algorithm
To identify the best and minimal features for predicting BM of NSCLC patients based on the expression of miRNAs, we used the “strong features method” developed previously, as a classifier model24. The model is a simple linear classifier with noise injection but modified to speed up the computation through analytical solution, instead of a Monte-Carlo simulation. The method has been developed to mitigate over-fitting problem in classifier design with small number of samples, by designing a classifier from a probability distribution resulting from spreading the mass of the sample points to make classification more difficult, while maintaining sample geometry.
Feature selection
As we focus on only eight miRNAs (hsa-miR-325, hsa-miR-326, hsamiR-328, hsa-miR-329-2-pre, hsa-miR-330-3p, hsa-miR-500*, hsa-miR-370, and hsa-miR-650-pre; henceforth hsa- prefix is removed when referring to these miRNAs), taken forward from the differential expression analysis to measure by qRT-PCR, we performed a full combinatorial search to identify best strong-features set. qRT-PCR measurements made from the same patient samples we used for miRNA microarray profiling (SHC Discovery cohort; BM data) were used for feature selection. For each combination of features (among 255 = 28−1), we used the leave-one-out method to estimate classification error. The feature set with the lowest leave-one-out error was deemed the best strong-feature set.
Classifier design for prediction
Once the best strong-feature set is identified as described above, all the samples in the SHC Discovery cohort were used to design a classifier to predict for BM in patients. In all classifier designs, the amount spread was set to 0.4, according to the suggestion made by Kim et al24.
Validation on an independent data set
The normalized PCR measurements were further processed by dividing the difference between its value and its mean across all the samples, i.e. z-score. Hence, each miRNA has zero mean and the standard deviation of one. This was done for both the SHC Discovery and SHC Validation data sets, separately.
Construction of stable lentiviral clones
A549 and H1703 NSCLC cell lines were obtained from ATCC (Manassas, VA) and maintained in RPMI-1640 medium supplemented with 10% FCS. Lentiviral constructs expressing GFP (Green Fluorescent protein)-empty vector (A549/H1703-empty) or GFP vector over-expressing miR-328 (A549/H1703-328) were obtained from Systems Biosciences Inc. (Mountain View, CA). Virus production and cell transduction in A549/H1703 cells was performed as described25 and for in vitro experiments cells were flow sorted to maintain >95% GFP positivity.
mRNA expression profiling
For each experimental sample, 3 × 105 cells were seeded in duplicate using standard growth conditions. After 24 hours, total RNA was isolated according to manufacturer's protocol (mirVana miRNA Isolation Kit, Ambion, Austin, TX). Total RNA yield was assessed using a NanoDrop 2000c (Thermo Scientific, Wilmington, DE), and quality was assessed on a BioAnalyzer 2400 using a BioAnalyzer RNA 6000 Nano LabChip Kit (Agilent Technologies, Palo Alto, CA).
RNA from A549-empty cells and A549-328 cells was analyzed for mRNA expression profiling. A quick-amplification kit (Agilent, Santa Clara, CA) was used to amplify and label 500 ng target mRNA species to complementary RNA (cRNA) for oligo microarrays according to the manufacturer's protocol. For each array, experimental samples were run in duplicate along with a commercial universal reference RNA (Stratagene, La Jolla, CA) that was labeled with cyanine 5-CTP and cyanine-3-CTP (Perkin Elmer, Boston, MA), respectively. cRNA concentration and labeling efficiency was measured spectrophotometrically. Approximately 800 ng of both Cy5-labeled experimental cRNA and Cy3-labeled universal reference RNA were hybridized to each microarray (adjusting for labeling efficiency).
Whole human genome 4 × 44K microarrays were hybridized and washed following Agilent's protocol. Images were captured using an Agilent DNA microarray scanner set at default settings for gene expression. Scanned images were processed using Feature Extractor v. 10.5.1.1. By applying a LOWESS (locally weighted linear regression) correction for dye-bias, background noise was subtracted from spot intensities. To filter the preprocessed data, genes with a background signal higher than feature signal were removed.
Differentially-expressed Genes and Pathway Analysis
To identify genes regulated by miR-328, we performed differential gene expression analysis on duplicate A549-empty and A549-328 cells. Differential expression analysis aimed at finding genes that were up-regulated or down-regulated for miR-328 over-expression and was done using two sided t-test. p-values generated from t-test were corrected for multiple hypotheses testing using Benjamini-Hochberg correction. The differentially-expressed gene list was used to identify significantly altered pathways using the GeneGo MetaCore (St. Joseph, MI).
Western Blot Analysis
Cell lysates were prepared for A549-empty and A549-328 cells. Protein concentration was determined by Pierce BCA assay (Thermo Scientific, Rockford, IL) and lysates were resolved by SDS-P A G E on 4–12% resolving gel. Proteins were transferred onto nitrocellulose membrane (Invitrogen, Carlsbad, CA) and PRKCA protein was identified using a rabbit-anti-PRKCA polyclonal antibody (Cell Signaling Technology, Danvers, MA) and an HRP-conjugated anti-rabbit secondary antibody (Promega Inc, Madison, WI). Bound antibodies were detected using Pierce SuperSignal West Dura Substrate kit (Thermo Scientific, Rockford, IL) and imaged. GAPDH were used as a loading control.
siRNA knockdown
All siRNAs were obtained from Qiagen Inc. (Valencia, CA). A549-328 cells were plated at a density of 3 × 104 cells/well in a 6 well plate. After 8 hours, cells were transfected with 20 nM of validated siRNA specific to PRKCA (PRKCA_1 or PRKCA_2). For controls, we transfected the cells with all-star negative (non-silencing control) and all-star positive (cell death control) siRNAs26. Seventy-two hours later, cells were collected either for Western blotting or for migration assay.
Migration Assay
A modified microwell Boyden chamber assay was used to determine the effect of miR-328 over-expression and PRKCA knockdown in migration of A549 and H1703 cells as described previously27. Briefly, RPMI 1640 medium supplemented with 10% FCS was added to the lower wells of a 12-well modified Boyden chamber (Neuroprobe, Cabin John, MD). Wells were covered with an 8-μm pore size Nucleopore filter (Neuroprobe) that had previously been coated with 50 μg/mL bovine collagen (Purecol®, Advanced Biomatrix, San Diego, CA). A549/H1703-empty cells and A549/H1703-328 cells, as well as, PRKCA knockdown A549-328 cells were suspended at the concentration of 4.8 × 104 cells in 100 μl of assay medium (RPMI 1640 medium supplemented with 10% FCS) and seeded into the upper wells. After incubation for five hours at 37°C, non-migrated cells were scraped off from the upper side of the filter, and filters were stained with 4', 6-diamidino-2-phenylindole (DAPI). Nuclei of migrated cells were counted in five high-power fields (HPF) with a 20× objective. These experiments were conducted in the absence of a chemoattractant and values were assessed in triplicate.
Results
Patient Samples
FFPE tumor specimens (n=13) were obtained for the SHC Discovery set and total RNA was extracted. The 13 FFPE tumor samples yielded approximately 0.5 to 5 μg of total RNA. Twelve of them (seven BM+ and five BM−) had sufficient total RNA yield to perform miRNA microarray profiling. All six BM− samples (including BM−2 which was not miRNA profiled) were evaluated by qRT-PCR. One BM+ sample that was miRNA microarray profiled had insufficient additional RNA for qRT-PCR analysis (sample BM+7). Fifteen samples were included in the SHC Validation set (Supplemental Table 1). All these samples had RNA a yield varying between 1.4 μg to 10 μg. RNA was also extracted from the 24 metastatic brain lesion samples (IOWA) and the samples yielded approximately 0.05 to 1 μg of total RNA. The clinical characteristics of the SHC Discovery patients (n=13), SHC Validation (n=15), and IOWA cases (n=24) are listed in Table 1, with details in Supplementary Table 1. To evaluate the expression of miR-328 and miR-330-3p in NSCLC brain metastasis samples, we extracted RNA from 24 IOWA cases. These results were compared with miR-328 and miR-330-3p expression in the SHC Validation cohort.
Expression profiling identifies differentially-expressed miRNAs in primary specimens of NSCLC patients with or without the development of BM
The raw and normalized data for all the tumor samples has been submitted to GEO (Accession # GSE23019). The data obtained by miRNA profiling of RNA isolated from the SHC Discovery samples was first analyzed using t-test. Table 2 shows the top 19 significantly differentially-expressed miRNAs in BM+ vs. BM− NSCLC samples. The fold change for the top 19 miRNA array expression for BM+ compared with BM− ranged from down-regulation of miR-92a in BM+ by 3.41 fold to 133 fold up-regulation of miR-329-1-pre, respectively.
Table 2.
Differentially Expressed miRNAs in BM+ vs. BM− NSCLC (adapted from (15)) This table shows the top best differentially-expressed miRNA identified by microarray analysis using t-test. The p-values are corrected using Benjamini-Hochberg method (22). Fold change here represents the fold change in BM+ vs. BM−.
| miRNA | Fold Change | p-value (Corrected) |
|---|---|---|
| hsa-miR-329-1-pre | 133.02 | 0.02174 |
| hsa-miR-326-pre | 94.24 | 0.02174 |
| hsa-miR-495-pre | 123.45 | 0.02174 |
| hsa-miR-500* | 106.19 | 0.02574 |
| hsa-miR-326 | 84.30 | 0.03198 |
| hsa-miR-370 | 57.52 | 0.03198 |
| hsa-miR-218 | 51.50 | 0.03198 |
| hsa-miR-330-3p | 53.02 | 0.03198 |
| hsa-miR-122a | 30.13 | 0.03198 |
| hsa-miR-325 | 61.25 | 0.03198 |
| hsa-miR-489-pre | 34.39 | 0.03658 |
| hsa-miR-599 | 40.42 | 0. 03658 |
| hsa-miR-328 | 64.19 | 0.03669 |
| hsa-miR-329-2-pre | 39.19 | 0.03669 |
| hsa-miR-346 | 25.31 | 0.03673 |
| hsa-miR-650-Pre | 13.58 | 0.03874 |
| hsa-miR-92a | −3.41 | 0.04126 |
| hsa-miR-193-b-pre | 16.12 | 0.04126 |
| hsa-miR-103 | 10.21 | 0.04901 |
To confirm the results of miRNA profiling, qRT-PCR was performed using specific miRNA primers on the cDNA prepared from RNA isolated from FFPE samples. Of the top 19 miRNA candidates identified by t-test analysis on miRNA profiling results, eight miRNAs were confirmed by qRT-PCR to be significantly differentially-expressed. The expression of miR-326, -370, -330-3p, -500*, -328, -325, -329-2-pre, and -650-pre was high in BM+ samples compared to BM− samples when measured by both miRNA profiling and qRT-PCR analysis (Supplementary Figure 1):
There was no significant correlation between BM+ and the clinical parameters: age, gender, stage at diagnosis, and histology. Similarly, correlation coefficients were also computed for the most discerning miRNAs. There was no significant correlation between the expression of these eight miRNAs and age, gender, stage at diagnosis, or histology (data not shown).
Design and validation of miRNA classifier for BM (Strong Feature Results)
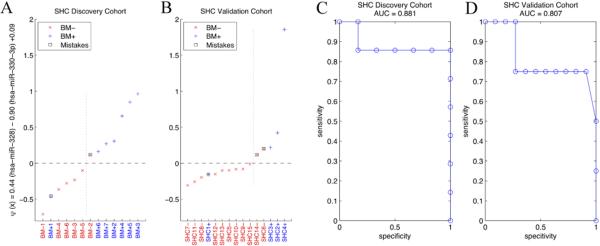
Upon testing all possible combinations for the feature set, the combination of miR-328 and miR-330-3p were deemed the best markers with the leave-one-out error of 3 out of 13 (0.2308), using the SHC Discovery data. Next, we designed a classifier with these two miRNAs from the same data set. Figure 1A shows the prediction of the classifier on the same data set, misclassifying BM+1 and BM−2, resulting in 84.6% accuracy (re-substitution error) which corresponds to sensitivity = 0.8571 and specificity = 0.8333. We then applied the same classifier to the SHC Validation data and the result is shown in Figure 1B. Three samples were misclassified, SHC1+, SHC6− and SHC14−, with 80% accuracy (sensitivity = 0.7500 and specificity = 0.8182). To evaluate the robustness of the classifier, we performed ROC analysis and the results are shown in Figure 1C and D. AUC were 0.881 and 0.807 for SHC Discovery and SHC Validation data sets, respectively.
Figure 1. Data analysis for SHC Discovery and SHC Validation cohorts.
(A and B) Discovery and validation for BM− and BM+ using miR-328 and miR-330-3p in the SHC Discovery and SHC Validation cohorts, respectively. Sensitivity and specificity are 0.8571 and 0.8333 for the SHC Discovery cohort and 0.7500 and 0.8182 for the SHC Validation cohort. (C and D) ROC curve for SHC Discovery and SHC Validation cohorts, respectively.
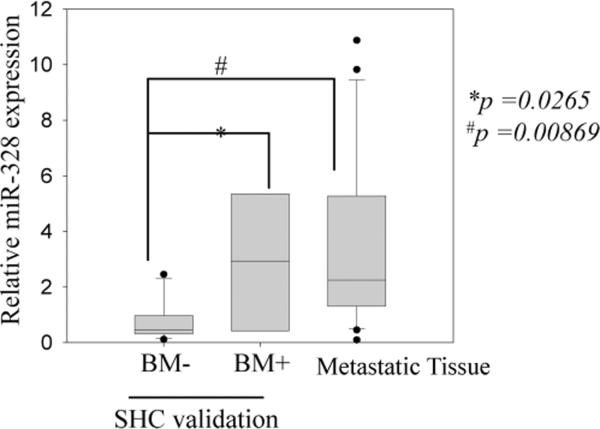
The expression of miR-328 in BM+ vs. BM− in the SHC Validation cohort is shown in Figure 2; it clearly shows that miR-328 is over-expressed in BM+ cases as compared to BM− cases (p=0.0265). In addition, metastatic NSCLC tissue extracted from the brain (IOWA samples) also show miR-328 over-expression as compared to BM− cases from SHC Validation samples (Figure 2; p = 0.00869). The expression of miR-330-3p was close to statistical significance in BM+ SHC Validation and IOWA metastatic samples as compared to BM− samples (p =0.07 and p =0.0537 respectively) but was not sufficient to distinguish BM+ vs. BM− cases (Supplemental Figure 2)
Figure 2. Relative expression of miR-328 in the validation cohort.
Box plot showing the relative expression of miR-328 in the validation set represents the interquartile range (25–75th percentile) and the line within this box is the median value. Bottom and top bars of the whisker indicate the 10th and 90th percentiles, respectively. Outlier values are indicated (closed circles). Significance between the indicated classes of specimens was tested using a two-sample t-test assuming unequal variances. The expression was normalized to the expression of 5S-rRNA in all samples plotted as fold change to A549 miR-328 expression (used as a reference).
Identification of miR-328 targets
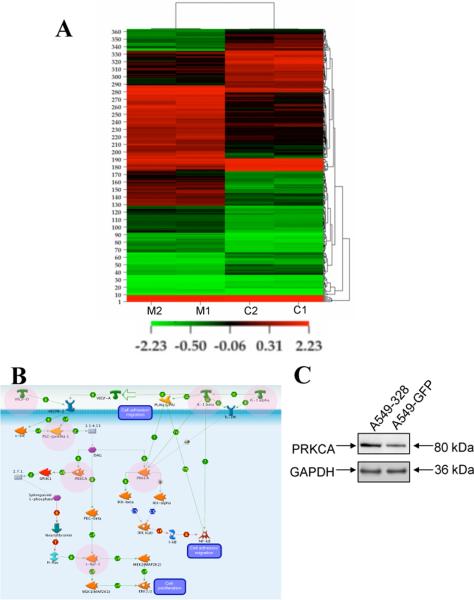
In order to identify the direct and indirect targets of miR-328 in NSCLC cells, we chose to work with two NSCLC cell lines, A549 and H1703. We checked the relative level of expression of miR-328 and found that H1703 expresses miR-328 at a slightly higher level as compared to A549 cells (Supplemental Figure 3). We stably transfected A549 and H1703 cells with either the GFP-empty vector or GFP-miR-328 over-expressing lentiviral construct (Supplemental Figure 4). RNA extracted from A549-328 and A549-empty cells was used for gene expression analysis using the Agilent platform. We generated a list of 363 genes with expression that was up- or down- regulated at least two-fold when the two conditions were compared. This list of 363 genes was used to perform Hierarchical Clustering of the datasets and the replicate samples were clustered very tightly together (Figure 3A; Supplemental Table 2). These differentially-expressed genes were then plugged into a pathway analysis tool to identify pathways that might be affected by these genes.
Figure 3. Gene and protein expression analysis in A549-empty and A549-328 cells.
(A) Hierarchical Clustering of 363 Genes Filtered at p value < 0.05 and fold change ≥ 2; 1 and 2 represent the duplicate runs. (B) A representative VEGF/IL1 signaling pathway leading to cellular migration/adhesion/proliferation that might be affected as a result of miR-328 over-expression. Protein products of several of the differentially-expressed genes are involved in this signaling cascade (these proteins are marked in pink circles). (C) Western blot showing over-expression of PRKCA in A549-328 cells as compared to A549-empty cells. GAPDH was used as a loading control.
We also performed the analysis for identifying the putative direct targets of miR-328 using microcosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/). We performed this analysis for all the downregulated genes from the 363 differentially-expressed genes based on the assumption that if a gene is a direct target of miR-328 then in miR-328 over-expressing cells, its expression will be downregulated. There were only three targets namely, PLCB3, PTGES2 and TLX3, which had miR-328 binding sites in their 3'UTR and were also downregulated in miR-328 over-expressing cells. But, as our clinical data showed that miR-328 might be involved in metastasis (as described below), we were interested in the differentially-expressed genes that play a role in cell migration and none of these three targets have a significant role in metastasis (from literature review) so we did not further study these targets.
Several signaling pathways such as interleukin-1 (IL-1) signaling, VEGF signaling, and PDGF signaling were shown to be affected by genes deregulated in miR-328 over-expressing cells (Supplemental Table 2). An example of a pathway that might be affected by miR-328 is shown in Figure 3B. This is the VEGF/IL1 signaling pathway, and several of the miR-328 target genes play a key role in controlling this signaling pathway that can lead to loss of cell adhesion and increased migration. These target genes included PRKCA, VEGF-D, NOTCH1, IL1-alpha, IL1-beta, and PLC-gamma amongst others. PRKCA (protein kinase C, alpha), which was one of the genes up-regulated in A549-328 cells, was also over-expressed at the protein level in A549-328 cells as compared to A549-empty cells (Figure 3C).
Role of miR-328 and PRKCA in NSCLC cell migration
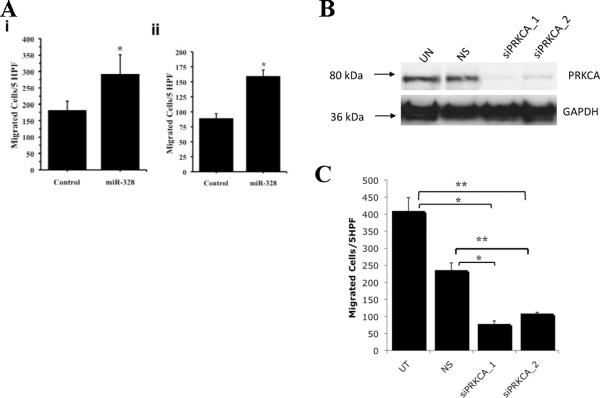
As miR-328 over-expression was seen in BM+ cases, we examined whether miR-328 had an effect on NSCLC cell migration using a modified Boyden chamber assay. Over-expression of miR-328 resulted in increased A549 cell migration (~two-fold) compared to A549-empty cells (p = 0.016) (Figure 4A). qRT-PCR results showed a 40-fold increase in miR-328 expression in A549-328 cells compared to A549-empty cells. Similarly, H1703-328 over-expressing cells also showed ~two-fold increase in migratory potential compared to H1703-empty cells (p = 0.0016; Figure 4A). These results suggest that miR-328 confers migratory potential to cancer cells and might be responsible for BM in NSCLC patients.
Figure 4. Role of miR-328 in NSCLC.
(A) Migration of NSCLC cells. Graph showing migration of GFP-empty vector (Control) and miR-328 over-expressing cells for cell line (i) A549 and (ii) H1703 in modified microwell Boyden chamber assay. Both cell lines demonstrated significant increase in cell migration in miR-328 over-expression as compared to Control [p = 0.014 (A549) and 0.0006 (H1703)]. Plotted values are mean ± standard deviation of triplicate determinations. HPF: High Power Fields. (B) siRNA mediated knockdown of PRKCA. A549-328 cells were treated with specific PRKCA siRNAs (siPRKCA_1 or siPRKCA_2) or non-silencing control (NS) along with an Untreated (UT) control. Seventy-two hours later protein lysates were prepared and Western blot was performed. The results show successful knockdown of PRKCA protein expression. GAPDH was used as a loading control. (C) Effect of PRKCA knockdown on the migration of NSCLC cells. Graph showing migration of A549-328 cells vs. PRKCA silenced A549-328 cells in modified microwell Boyden chamber assay. PRKCA silencing in A549-328 cells demonstrated significant reduction in cell migration as compared to Untreated as well as Non-silencing control [*p < 0.005; #p<0.0001]. Plotted values are mean ± standard deviation of triplicate determinations. HPF: High Power Fields
We next looked at the differentially-expressed genes (in miR-328 over-expressing cells) involved in metastasis of cancer and/or migration of cancer cells and found that PRKCA was upregulated in the miR-328 over-expressing cells. PRKCA (also known as PKC-alpha) has been shown to play a significant role in invasion and migration of cancer cells and hence it was chosen for further analysis28, 29.
Following this, we conducted siRNA knockdown experiments using two validated siRNA sequences against PRKCA (Figure 4B confirms PRKCA knockdown using these sequences) and tested their migration potential using the modified Boyden chamber assay (Figure 4C). The results showed that PRKCA knockdown leads to reduced migration in A549-328 cells compared to control cells thus confirming that the increased migratory potential of NSCLC cells might in part be due to its downstream indirect target, PRKCA.
Discussion
BM has a lifetime prevalence approaching 25% in NSCLC. In these patients, BM can cause neurologic, cognitive, and emotional difficulties. The ability to identify patients at risk for developing BM may lead to new prophylactic intervention that may mitigate morbidity and mortality; however, previous efforts to characterize NSCLC patients that will develop BM have been fairly disappointing.
There have been several previous studies on miRNAs that could have predictive and prognostic relevance in lung cancer. One of the initial studies showed suppression of NSCLC development by the let-7 miRNA family30. Expression of several different miRNAs, namely, miR-221, let-7a, miR-137, -182*, -372a, -1, -30d, -486 and -499 have been associated with survival of lung cancer patients previously31, 32. Despite these reports, there have been no studies looking for miRNA biomarkers to stratify NSCLC patients for BM prediction. Thus, we sought to identify miRNA candidate biomarkers that could successfully differentiate between BM+ and BM− patients. For this, we used miRNA microarray profiling on a discovery cohort of 12 NSCLC patients (seven BM+ and five BM−). Our microarray platform consisted of both pre- and mature- miRNA probes. Pre-miRNA probes were included as it has been previously shown that the expression of miRNAs can be regulated at the post-transcriptional level33. Such differential processing of precursor miRNAs into mature miRNAs leads to tissue- and developmental-specific miRNA expression. Common genetic polymorphisms also play a role in the tumorigenesis process through the somatic mutations as illustrated in a study by Jazdzewski et al. These authors demonstrated that a common polymorphism in pre-miR-146a affects the amount of mature miRNA and contributes to the genetic predisposition to papillary thyroid carcinoma34. In these contexts, looking exclusively at the level of mature miRNAs might not be sufficient to correlate the phenotype of a cell with its miRNA signature and it is useful to include studying the level of pre-miRNA. Another reason to profile pre-miRNAs is that mature miRNAs of identical sequence can be derived from different pre-miRNAs, each located at a different genomic location and each under control of a different promoter. Thus, profiling pre-miRNA levels provides a stepping-stone towards the identification of miRNA regulatory elements.
In our earlier publication on the SHC Discovery set15, we used two well-established methods: in-silico conditioning35 and SAM to find discriminating miRNAs between BM+ and BM− tumor samples. In this paper, we were able to find similar results using simple t-tests. All the three methods have shown consensus in the discerning miRNA between BM+/−(one case was different from in-silico conditioning results). Even though, clustering methods like PAM could be a promising way to find discerning miRNAs, the number of samples (n=12) in our study are extremely small for a supervised classification approach.
We confirmed the expression of eight miRNAs that were significantly differentially-expressed between BM+ and BM− by qRT-PCR. Some of these differential miRNAs were the pre-miRNAs. But, on comparing the expression levels of pre-miRNA vs. mature miRNAs on the microarray experiment, we observed that even though the mature miRNAs were not significantly differentially-expressed as compared to respective pre-miRNAs, the trend of their expression was similar across patient samples.
Upon statistical analysis, we identified a classifier based on miR-328 and miR-330-3p expression, which could successfully distinguish BM+ vs. BM− cases in a validation cohort of 15 patients. This classifier performed well (80% accuracy) on the SHC Validation cohort and successfully classified 12 of 15 NSCLC patients. The clinical parameters namely, age, gender, stage at diagnosis, or histology did not show significant correlation with BM+. Interestingly, these eight miRNAs also did not significantly correlate with age, gender, stage at diagnosis, or histology, suggesting, that these miRNAs may be independent risk factors for BM development.
In the BM positive condition, the elevated expression of miR-328 in both thoracic and brain NSCLC samples, suggests this miRNA may be involved in “brain-seeking” metastatic potential. This hypothesis is supported by our observation that miR-328 is over-expressed in both primary lung cancer specimens (SHC Discovery and Validation cohorts), as well as, brain metastasis samples (IOWA cohort). We are aware that the small number of patients included in the study is a significant limitation but since additional well-annotated samples were not readily available to us (we made several inquiries to thoracic oncology and pathology investigators), we hope that publishing our findings will lead other investigators to independently validate our findings in their specimen collections. Additionally, we are embarking on a prospective collection of tissue specimens that will have adequate follow up so that in the future we will have larger number of tumor samples to validate our findings.
As the implication of miRNAs in carcinogenesis is relatively new, the biological importance of miR-328 has not been fully elucidated. There are no previous reports on the association of lung cancer and miR-328. In a recent report, Eiring et al showed that loss of miR-328 occurs in blast crisis chronic myelogenous leukemia in a BCR/ABL dose- and kinase-dependent manner through the MAPK-hnRNP E2 pathway. Their data revealed the dual ability of miR-328 to control cell fate through base pairing with mRNA targets and through decoy activity that interferes with the function of regulatory proteins36. ABCG2, CD44 and β-amyloid precursor protein-converting enzyme 1 are the other genes that have been shown to be miR-328 targets37–39. In order to identify novel targets of miR-328 in NSCLC cells, we conducted a gene expression analysis comparing miR-328 over-expressing cells with parental NSCLC cells. Our results identified 363 differentially-expressed genes in A549-328 cells. Upon pathway analysis, we observed that several of these target genes are implicated in migration of cancer cells including PRKCA, IL-1beta, and c-Raf1.
Recently, investigators have examined clinical and histological characteristics that could stratify breast cancer patients to predict development of BM. In a study by Graesslin et al, the authors show the development of a robust nomogram that is able to predict the development of subsequent BM in patients with proven metastatic breast cancer40. In addition, there have been at least two different studies looking for biomarkers that predict BM in breast cancer patients using gene expression analysis. Examining the differentially-expressed genes associated with miR-328 over-expression in vitro from our work, and comparing them to the breast cancer gene candidates associated with BM41, 42, we observed that IL1-beta and KCNMA1 (potassium large conductance calcium-activated channel, subfamily M, alpha member 1) are common to both lists. In addition, several genes of the SERPIN and ZNF families are also present on both the lists. Although we have not tested this hypothesis, it is possible that these genes are partly responsible for predisposition to BM in either breast and/or NSCLC and the expression of these genes may be regulated directly or indirectly by miR-328.
To determine the functional significance of miR-328 over-expression in NSCLC, we performed a migration assay and showed that miR-328 over-expression leads to increased migration potential of A549 and H1703 cells thus showing that miR-328 over-expression offers a migratory advantage to NSCLC. Next, we wanted to test whether the miR-328 target genes identified in our study play a role in this migration potential. For this, we knocked down the expression of PRKCA in A549-328 cells and demonstrated that the increased migration of A549-miR328 cells was in part to due increased PRKCA expression. Interestingly there have been several reports on the role of PRKCA in migration of lung cancer cells and the report by Cheng et al has shown that IL1-beta via PRKCA induces the expression of uPA (urokinase plasminogen activator), which leads to migration of A549 NSCLC cells28, 43. PRKCA and IL1-beta were both identified as targets of miR-328 in our gene expression studies supporting our result that miR-328 leads to increased migration of NSCLC cells in part through this pathway.
With a clearer understanding of how these miRNAs influence BM development and independent validation by other investigators in other clinically well-characterized patient samples sets, assessment of these miRNAs may potentially be incorporated into clinical treatment decision-making. We believe that such a trial is feasible with the backing of like-minded investigators and industry partners. We acknowledge the limited sample size reported here, but hope that these results can be corroborated in independent sample sets of matched thoracic and brain NSCLC. Ideally, these miRNA biomarkers may assist clinicians in stratifying the high-risk patients on a clinical trial for either PCI or a new intervention that may mitigate BM development, ultimately leading to a new standard of care for NSCLC patients.
Supplementary Material
Acknowledgments
We thank the patients and treating physicians and staff at Scottsdale Healthcare and University of Iowa Hospitals and Clinics (IOWA). We thank the generous support from the IBIS Foundation of Arizona (ARR, SN, GJW), TGen Foundation (ARR, GJW), and the Scottsdale Healthcare Foundation (GJW). This research was supported in part by funds from the Howard Hughes Medical Institute through the Undergraduate Science Education Program and from the ASU School of Life Sciences to SS.
We thank David K. Edwards V and Carlos D. Lorenzo for assistance with sample preparation. NLT was partially supported by NIH R01CA130940 and SFAZ CAA 0244-08. SK was partially supported by NIH 1R21LM009706-01 and SFAZ CAA 0243-08, SN was partially supported by SFAZ CAA 0243-08.
Footnotes
Potential conflict of interest: Dr. Glen J. Weiss, Patent filed for use of microRNAs as theranostic tools. Research funded by IBIS Foundation of Arizona, TGen Foundation and SHC Foundation
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajduch M, Jacob-Hirsch J, Amariglio N, Krupsky M, Simansky DA, Ram Z, Pfeffer R, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 2009;15:1755–61. doi: 10.1158/1078-0432.CCR-08-2124. [DOI] [PubMed] [Google Scholar]
- 3.Laack NN, Brown PD. Cognitive sequelae of brain radiation in adults. Semin Oncol. 2004;31:702–13. doi: 10.1053/j.seminoncol.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Oh Y, Taylor S, Bekele BN, Debnam JM, Allen PK, Suki D, Sawaya R, Komaki R, Stewart DJ, Karp DD. Number of metastatic sites is a strong predictor of survival in patients with nonsmall cell lung cancer with or without brain metastases. Cancer. 2009;115:2930–8. doi: 10.1002/cncr.24333. [DOI] [PubMed] [Google Scholar]
- 5.Pugh TJ, Gaspar LE. Prophylactic cranial irradiation for patients with lung cancer. Clin Lung Cancer. 2007;8:365–8. doi: 10.3816/CLC.2007.n.016. [DOI] [PubMed] [Google Scholar]
- 6.Gore EMBK, Wong S. A phase III comparison of prophylactic cranial irradiation versus observation in patients with locally advanced non-small cell lung cancer: Initial analysis of Radiation Therapy Oncology Group 0214. J Clin Oncol. 2009;27(15S):7506. doi: 10.1200/JCO.2010.29.1609. Meeting Abstracts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3:563–78. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 9.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola FM, Tucker MA, Bertazzi PA, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao W, Yu Y, Cao H, Shen H, Li X, Pan S, Shu Y. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64:399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–9. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 12.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–9. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 13.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, Gemma A, Kudoh S, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasser S, Ranade A, Sridhar S, Haney L, Korn RL, Gotway MB, Weiss GJ, Kim S. Identifying MiRNA and Imaging Features Associated with Metastasis of Lung Cancer to the Brain. IEEE International Conference on Bioinformatics & Biomedicine; 2009. pp. 246–51. [DOI] [PubMed] [Google Scholar]
- 16.Ranade AR, Cherba D, Sridhar S, Richardson P, Webb C, Paripati A, Bowles B, Weiss GJ. MicroRNA 92a-2*: A Biomarker Predictive for Chemoresistance and Prognostic for Survival in Patients with Small Cell Lung Cancer. J Thorac Oncol. 2010;5:1273–8. doi: 10.1097/JTO.0b013e3181dea6be. [DOI] [PubMed] [Google Scholar]
- 17.Microarray Suite 5.0 User's Guide. Affymetrix edition 2002. Affymetrix. [Google Scholar]
- 18.Gene Spring. http://www.chem.agilent.com/Library/usermanuals/Public/GeneSpringGX10_QuickStartG uide.pdf.; [Last accessed July 28, 2010]
- 19.Sanzari JK, Nuth M, Kennedy AR. Induction of cytokine gene expression in human thyroid epithelial cells irradiated with HZE particles (iron ions) Radiat Res. 2009;172:437–43. doi: 10.1667/RR1363.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 21.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Dougherty ER, Barrera J, Chen Y, Bittner ML, Trent JM. Strong feature sets from small samples. J Comput Biol. 2002;9:127–46. doi: 10.1089/10665270252833226. [DOI] [PubMed] [Google Scholar]
- 25.Coleman JE, Huentelman MJ, Kasparov S, Metcalfe BL, Paton JF, Katovich MJ, Semple-Rowland SL, Raizada MK. Efficient large-scale production and concentration of HIV-1-based lentiviral vectors for use in vivo. Physiol Genomics. 2003;12:221–8. doi: 10.1152/physiolgenomics.00135.2002. [DOI] [PubMed] [Google Scholar]
- 26.Azorsa DO, Gonzales IM, Basu GD, Choudhary A, Arora S, Bisanz KM, Kiefer JA, Henderson MC, Trent JM, Von Hoff DD, Mousses S. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J Transl Med. 2009;7:43. doi: 10.1186/1479-5876-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamszus K, Schmidt NO, Jin L, Laterra J, Zagzag D, Way D, Witte M, Weinand M, Goldberg ID, Westphal M, Rosen EM. Scatter factor promotes motility of human glioma and neuromicrovascular endothelial cells. Int J Cancer. 1998;75:19–28. doi: 10.1002/(sici)1097-0215(19980105)75:1<19::aid-ijc4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Cheng CY, Hsieh HL, Sun CC, Lin CC, Luo SF, Yang CM. IL-1 beta induces urokinase-plasminogen activator expression and cell migration through PKC alpha, JNK1/2, and NF-kappaB in A549 cells. J Cell Physiol. 2009;219:183–93. doi: 10.1002/jcp.21669. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh YH, Wu TT, Huang CY, Hsieh YS, Hwang JM, Liu JY. p38 mitogen-activated protein kinase pathway is involved in protein kinase Calpha-regulated invasion in human hepatocellular carcinoma cells. Cancer Res. 2007;67:4320–7. doi: 10.1158/0008-5472.CAN-06-2486. [DOI] [PubMed] [Google Scholar]
- 30.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–6. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 33.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–7. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasser S, Weiss GJ, Ranade AR, Kim S. In-Silico Conditioning Of Microrna To Identify Potential Biomarkers From Serum For Pancreatic Cancer. IEEE International Workshop on Genomic Signal Processing and Statistics, GENSIPS.2009. pp. 1–4. [Google Scholar]
- 36.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–65. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75:1374–9. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, Lu WY, Zhang Y, Yang BB. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS One. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–81. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graesslin O, Abdulkarim BS, Coutant C, Huguet F, Gabos Z, Hsu L, Marpeau O, Uzan S, Pusztai L, Strom EA, Hortobagyi GN, Rouzier R, et al. Nomogram to predict subsequent brain metastasis in patients with metastatic breast cancer. J Clin Oncol. 2010;28:2032–7. doi: 10.1200/JCO.2009.24.6314. [DOI] [PubMed] [Google Scholar]
- 41.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaitan D, Sankpal UT, Weksler B, Meister EA, Romero IA, Couraud PO, Ningaraj NS. Role of KCNMA1 gene in breast cancer invasion and metastasis to brain. BMC Cancer. 2009;9:258. doi: 10.1186/1471-2407-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Yang H, Liu H, Huang J, Song X. Effect of staurosporine on the mobility and invasiveness of lung adenocarcinoma A549 cells: an in vitro study. BMC Cancer. 2009;9:174. doi: 10.1186/1471-2407-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.