Abstract
The ETS family transcription factor PU.1 is a key regulator of haematopoietic differentiation. Its expression is dynamically controlled throughout haematopoiesis in order to direct appropriate lineage specification. Elucidating the biological role of PU.1 has proved challenging. This paper will discuss how a range of experiments in cell lines and mutant and transgenic mouse models have enhanced our knowledge of the mechanisms by which PU.1 drives lineage-specific differentiation during haematopoiesis.
1. Introduction
Haematopoiesis is a lifelong process that generates the range of blood cell types that exhibit distinct and specialised functions. Transcription factors play a critical role in this complex and highly orchestrated process, directing multipotent haematopoietic stem cells (HSCs) towards lineage commitment by regulating lineage-specific gene expression, proliferation, and differentiation. The ETS family member PU.1 is one such transcription factor
The PU.1 gene was first identified as a proviral integration site for the spleen focus forming virus (SFFV) in erythroleukaemias [1]. SFFV integration in the PU.1 locus leads to increased PU.1 transcription and subsequent erythroleukaemic transformation. It has since emerged that PU.1 is one of the major haematopoietic regulators, with a particular role in directing differentiation within the myeloid and lymphoid pathways [2]. Several PU.1 null and mutant mouse lines have been generated and exhibit varied phenotypes depending on the nature of PU.1 defect [3]. PU.1 knockout mice succumb to neonatal death and show a marked lack of myeloid cells, T and B cells [4, 5]. Erythropoiesis is also altered in the foetal livers of PU.1 null mice with erythroid progenitors displaying reduced self-renewal capacity and a propensity to differentiate prematurely [6].
PU.1 is thus crucial in directing many facets of haematopoiesis and concordant with this, its expression fluctuates dynamically in the various haematopoietic differentiation pathways (Figure 1). Importantly, the regulation of differentiation by PU.1 is not merely via a “presence or absence of expression” mechanism but by a dose-dependent effect. For instance, the expression of PU.1 is low in long-term reconstituting (LT)-HSCs but rises as these progenitors become more lineage restricted and form precursor cells known as common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs). Upon further lineage differentiation and maturation, PU.1 is expressed at varied levels in mature blood cells, with higher levels found in macrophages than B cells and low levels in mature erythroid cells, megakaryocytes, and T cells [7–9]. Moreover, not only is PU.1 differentially expressed in the various haematopoietic cells, but also lineage specification is sensitive to, and directed by, the varied dosage of PU.1 in differentiating progenitor cells. In addition, inappropriate expression of PU.1 in specific haematopoietic cells can result in leukaemic transformation, as in the case of T-cell lymphomas and, as mentioned previously, erythroleukaemias [10, 11].
Figure 1.
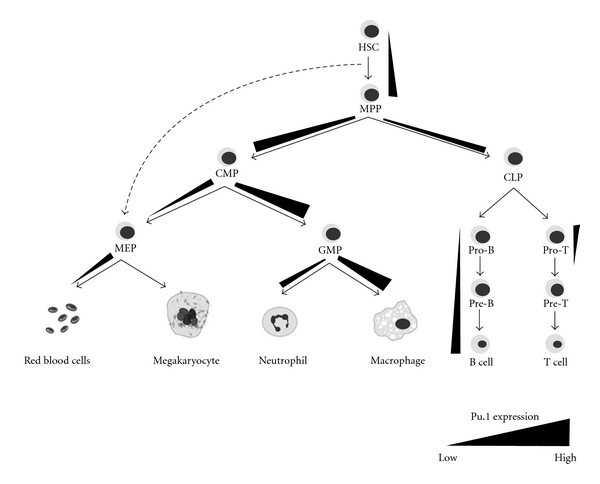
Schematic showing the changing expression of PU.1 during haematopoiesis. PU.1 levels, where known, are represented by gradient bars. Gradient bars are not drawn to scale. Differentiation pathways are denoted by arrows. Abbreviations: HSC, haematopoietic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MEP, megakaryocyte-erythroid progenitor; GMP, granulocyte-macrophage progenitor.
This paper focuses on how the expression pattern of PU.1 differs between different committed precursors and how this serves to determine cell fate. The interplay between PU.1 and other antagonistic haematopoietic regulators will also be discussed.
2. PU.1 Levels Are Important in Directing Haematopoietic Progenitor Cell Fate
PU.1 expression in HSCs is important for their self-renewal and for their development into CMPs and CLPs [12]. These two progenitor pools then further differentiate to form mature blood cells including megakaryocytes, red blood cells (RBCs), neutrophils and macrophages (all CMP derived), and B and T cells (CLP derived) (Figure 1). Iwasaki et al. showed that ablating PU.1 expression results in a decrease in HSC numbers by an order of magnitude and that CMPs, CLPs, and CMP progeny GMPs (granulocyte-monocyte progenitors; Figure 1) are all undetectable in PU.1 knockout foetal livers [12]. These haematopoietic defects are cell autonomous as PU.1 null HSCs fail to generate granular or myeloid colonies when cultured in vitro [12, 13]. Similarly, in competitive repopulation assays, PU.1 knockout foetal liver HSCs that were injected into lethally irradiated mice were barely detectable in peripheral blood, and the low numbers that were found within the bone marrow (BM) failed to generate monocytes and B and T lymphocytes [12, 14, 15]. Interestingly, the formation of megakaryocyte-erythroid progenitors (MEPs), however, remains intact in PU.1 knockout mice [12]. MEPs were conventionally thought to arise only from CMPs (Figure 1). However, the lack of CMPs and abundance of MEPs in PU.1 null mice suggest that MEPs may bypass the CMP stage and develop directly from HSCs, a hypothesis that is supported by several other studies [16–19]. Taken together, these data suggest that PU.1 is crucially important for normal development of HSCs into CMPs and CLPs but is dispensable for production of MEPs.
In order to explore the role of PU.1 in haematopoietic lineages further, studies have been performed using reporter systems in which green fluorescent protein (GFP) has been knocked into the PU.1 locus to allow its expression pattern to be tracked throughout haematopoietic differentiation [6–8, 12]. While a low level of PU.1 is detected in LT-HSCs (Lin− Sca-1+ c-kit+ CD34−), its expression increases as these cells become multipotent progenitors (MPPs; Lin− Sca-1+ c-kit+ CD34+) and subsequently develop into both CMPs and CLPs [7, 12, 20]. At this stage, the level of PU.1 expression is decisive in permitting progression towards either the myeloid or lymphoid lineage [21, 22]. Culturing PU.1+/− foetal liver progenitor cells (Lin−) in cytokines which favour lymphopoiesis predominantly generates pro-B-cells, while overexpression of PU.1 in these cells dramatically skews commitment towards the myeloid lineage and results in a marked increase in macrophage production [2]. Therefore, high PU.1 expression promotes macrophage generation, while a lower level is important for B-cell development. Consistent with this, PU.1 levels have been estimated to be approximately eightfold higher in macrophages compared to B cells [2, 8].
PU.1 not only influences the myeloid-lymphoid decision of MPPs but also regulates the potential differentiation pathways to which CMPs and CLPs can commit. For instance, fine tuning of PU.1 levels in CMPs is important in directing differentiation towards MEPs or GMPs. PU.1 is expressed at high levels in GMPs and directs commitment towards the neutrophil and monocyte lineages. In contrast, PU.1 expression is downregulated in MEPs, and this is essential for the development of megakaryocytes and RBCs [7, 8, 20]. Within the lymphoid pathway, differentiation of CLPs to form B cells or T cells is accompanied by a decrease in PU.1 levels, and PU.1 is further silenced as T cells develop. Conversely, PU.1 expression in B cells gradually increases as they mature, although not to the extent observed in macrophages [2, 7, 8]. Together, these data collectively illustrate the importance of PU.1 in maintaining the HSC pool and in directing differentiation towards the myeloid and lymphoid pathways. The role of PU.1 in specific lineage differentiation programs will now be discussed in more detail.
3. PU.1 Is a Negative Regulator of Erythropoiesis
Downregulation of PU.1 expression in committed MEPs is important for erythroid differentiation. Low levels of PU.1 in early erythroid precursors are essential for them to undergo proliferation before terminally differentiating into RBCs. Using a PU.1 null mouse model, Back et al. showed that erythroid cells from these mice differentiate prematurely and are susceptible to apoptosis [6]. Another interesting transgenic model generated by Tavitian's group overexpresses PU.1 in all haematopoietic lineages except T cells [11]. This results in splenic hyperplasia and anaemia in approximately half of the transgenic mice, characterised by high numbers of immature blast cells and poorly haemoglobinised erythroblasts. The respective phenotypes from these two transgenic lines complement each other well. Whereas under normal physiological conditions PU.1 expression is diminished to allow terminal differentiation of erythroblasts, the persistent expression of PU.1 in Tavitian's mouse line results in a maturation block with an overrepresentation of immature erythroblasts [11, 20, 23]. Conversely, elimination of PU.1 results in premature differentiation of these cells.
In fact, several groups have clearly demonstrated this point utilising the widely employed MEL (murine erythroleukemia) cell line. This cell line is derived from mice infected with Friend virus where the SFFV component has integrated into the Spi-1/PU.1 locus and transcriptionally activates the gene, resulting in erythroblast transformation [1, 24]. MEL cells, which exhibit ectopic PU.1 expression, are arrested at the erythroblast stage and undergo continual proliferation without differentiation [25, 26]. Treatment with chemical agents such as DMSO (dimethyl sulfoxide) or HMBA (hexamethylene bisacetamide) results in growth arrest, erythroid differentiation, and haemoglobin production, along with a concomitant decrease in PU.1 expression [27, 28]. The reduction in PU.1 expression is crucial for the differentiation of these cells. Forced expression of PU.1 in MELs inhibits DMSO- and HMBA-induced differentiation, while silencing PU.1 in these cells has been shown to be sufficient to drive terminal differentiation in the absence of any chemical inducers [25, 27–29].
PU.1 thus promotes erythroid cell proliferation and prevents differentiation. It has been proposed that one mechanism by which PU.1 does this is by regulating controllers of the cell cycle [25]. PU.1 directly activates expression of Cdk6 (cyclin-dependent kinase 6), a G1 phase-specific cell cycle kinase known to inhibit MEL cell differentiation [25, 30]. Cdk6 associates with D-cyclins to regulate cell cycle progression through the G1 phase, and like PU.1, Cdk6 is constitutively expressed in proliferating MELs [30]. After chemical induction, downregulation of PU.1 results in decreased Cdk6 levels, thus arresting proliferation and permitting differentiation.
Another established mechanism by which PU.1 inhibits erythroid differentiation is by antagonising the erythroid regulator Gata-1. These two transcription factors are known to exert opposing effects on each other, and the interplay between these two master regulators is instrumental in the decision for CMPs to commit towards either the myeloid or erythroid lineage [31, 32]. PU.1 interacts with Gata-1 and prevents its transcriptional activity while conversely, Gata-1 inhibits PU.1 function by disrupting its interaction with the coactivator c-Jun [32–34]. Hence, these data collectively suggest that PU.1 inhibits erythroid differentiation by upregulating Cdk6 to promote proliferation and by antagonising the master erythroid regulator Gata-1. This may also explain why PU.1 expression must be downregulated during erythropoiesis to allow normal production of RBCs.
4. PU.1 Is a Master Myeloid Regulator
Both neutrophils and monocytes are generated from GMPs. Differentiation towards either of these cell fates is highly dependent on PU.1 [7, 8]. Multiple knockout models have demonstrated the importance of PU.1 in myelopoiesis; abrogation of PU.1 results in a marked lack of CMPs, defective granulocytic neutrophil production, and an absence of mature macrophages [4, 5, 12, 35]. Because PU.1 is absolutely required early during myelopoiesis at the CMP stage, questions have been asked as to whether it is dispensable once cells have committed to the CMP pathway. In answering this, Iwasaki et al. purified PU.1 null CMPs and GMPs from a conditional knockout mouse and assessed their capacity to form myeloid colonies in culture [12]. Both CMPs and GMPs were found to be unable to contribute to the mature myeloid fraction, indicating that PU.1 is further required following commitment to the myeloid lineage to promote differentiation to generate granulocytes and macrophages. PU.1 has since been shown to enable committed cells to respond to a variety of myeloid growth factors by regulating the expression of a number of myeloid-specific genes, including the cytokine receptors granulocyte/macrophage colony-stimulating factor receptor α (GM-CSFRα), granulocyte-CSFR (G-CSFR), macrophage-CSFR (M-CSFR), and interleukin-7 receptor α (IL-7Rα) [36, 37].
4.1. Regulation of Macrophage versus Neutrophil Production
As a master myeloid regulator, PU.1 does not only regulate GMP development but also directs the differentiation pathways that give rise to both neutrophils and macrophages. Again, precise control of PU.1 levels in GMPs is required to direct these distinct differentiation programs. Different dosage of PU.1 expression in GMPs modulates distinct regulatory networks which involve a number of lineage-specific transcription factors such as Egr-2 (early growth response-2), Gfi-1 (growth factor independent-1), and C/ebpα (CCAAT enhancer-binding protein α) [21, 38, 39].
Using myeloid cell lines with an inducible transgene allowing high or low levels of PU.1, Laslo et al. revealed that low expression of PU.1 activates a mixed lineage of macrophage and neutrophil genes [39]. When PU.1 levels exceed a certain threshold, this induces the expression of Egr-2 and the transcriptional repressor Nab-2. Together, Egr-2 and Nab-2 repress the expression of Gfi-1, a transcription factor that promotes neutrophil differentiation. This results in the silencing of neutrophil genes and promotes macrophage differentiation [21, 39]. PU.1 thus indirectly represses Gfi-1 expression and conversely, PU.1 levels are elevated in Gfi-1 knockout mice [21]. Similar to Gata-1 and PU.1 antagonising each other's function in the myeloid-erythroid decision of CMPs, it appears that Gfi-1 and PU.1 negatively regulate each other's expression to determine the macrophage-neutrophil decision of GMPs [21].
Like Gfi-1, the neutrophil transcription factor C/ebpα is also expressed in myeloid cells and antagonises PU.1 to direct neutrophil differentiation. Dahl et al. showed that haematopoietic progenitors expressing high levels of inducible PU.1 predominantly develop into macrophages when cultured in IL-3 (interleukin-3) [38]. However, when these cells are pretreated with G-CSF (which promotes granulocyte development) prior to PU.1 induction, they display an upregulation of C/ebpα and instead form neutrophils. Thus, it can be deduced that with low levels of PU.1 in GMPs, the expression of Gfi-1 and C/ebpα are sufficient to antagonise PU.1 function by repressing macrophage genes and promoting neutrophil development. In contrast, when lineage commitment is directed towards macrophage development, PU.1 expression is upregulated to overcome the antagonism imposed by the two neutrophil regulators.
5. Regulation of Lymphopoiesis: PU.1 in B-Cell and T-Cell Maturation
In addition to being a major myeloid regulator, PU.1 has also been shown to modulate lymphopoiesis, the process that gives rise to B cells and T cells. PU.1 mutant mice exhibit a loss of the B and T cell compartments and develop fatal septicaemia within 2 days of birth owing to a lack of mature and functional immune cells [5, 35]. Studies investigating the pattern of PU.1 expression using GFP-reporter mice have revealed that PU.1 levels increase as B-cells mature, while PU.1 is completely silenced in mature T cells [7, 8]. Despite the requirement for PU.1 for B-cell production, its expression is dispensable once progenitors are committed to the lymphoid lineage as revealed by Iwasaki et al. [12]. PU.1 null CLPs generate B cells and express B-cell genes at comparable levels to wildtype CLPs in culture. In vivo, targeted deletion of PU.1 after the pre-B-cell stage does not disrupt immunoglobulin expression or the cells' response to a variety of mitogenic agents. This suggests that PU.1 is not essential for B-cell maturation once it has directed progenitors to the CLP stage. One possible explanation for this is that other members of the ETS family of transcription factors, such as Spi-B, may have functional redundancy with PU.1 in B cells [40, 41]. Similar to in B cells, PU.1 is only required at the early CLP stage for T-cell generation. Once cells develop to pro-T-cells, PU.1 expression is dramatically silenced. Silencing of PU.1 is required for T cell maturation and in fact, forced overexpression of PU.1 results in growth arrest and a T-cell maturation block [42].
6. Dosage-Dependent Regulation and Pathogenesis
It is clear that precise levels of PU.1 expressed at different stages within the haematopoietic lineage are crucial in directing proper differentiation and cell fate commitment (Figure 1). Hypomorphic mouse lines expressing varied doses of PU.1 have been particularly helpful in providing a better picture on how PU.1 activates and represses specific sets of genes. The PU.1BN and PU.1Blac mice have been estimated to express approximately 20% (“high” concentration) and 2% (“low” concentration) of PU.1 compared to wildtype mice, respectively, [22, 35]. By performing microarrays on PU.1BN, PU.1Blac, and PU.1−/−myeloid cell lines, it was revealed that PU.1 can regulate its targets in four distinct modes [22]. Some target genes are activated or repressed equally at both “high” and “low” concentrations of PU.1; others are exclusively activated or repressed at either “high” or “low” levels of PU.1 but not both, and the final group of targets can be activated or repressed in a dose-dependent manner by PU.1, that is, the degree of their transcriptional regulation is dependent on the level of PU.1. Targets which are repressed in such a “gradient” manner include erythroid and T-cell-specific genes, while genes which are activated include the myeloid genes. This illustrates the dependence on PU.1 levels in the regulation of these lineage-specific genes and may in part explain why PU.1 expression varies in certain cell types during specific stages of haematopoiesis.
In fact, when this delicate expression control is disrupted, the balance between different haematopoietic regulators is upset, and in many occasions, this results in oncogenic transformation [11, 33, 43]. A notable example is the SFFV-induced transcriptional activation of PU.1 in Friend virus infected mice which develop acute erythroleukaemia [1]. In this instance, the high level of PU.1 expression disrupts the stoichiometry between PU.1 and Gata-1. Zhang et al. showed by EMSA (electrophoretic mobility shift assays) that PU.1 directly interacts with the Gata-1 DNA recognition motif thereby blocking its DNA-binding activity [33, 34]. Gata-1 transactivation which ordinarily promotes normal erythroid differentiation is consequently inhibited and allows erythroleukaemic transformation [33, 44]. Studies have also revealed that deletion of a −14 kb upstream regulatory region (URE) reduces PU.1 expression by 80% in mice and results in acute myeloid leukaemia (AML) and T-cell lymphoma [10, 13]. These mice have an accumulation of immature myeloblasts and neutrophils in both the bone marrow and spleen accompanied by an increased number of immature white blood cells in peripheral blood. Persistent PU.1 expression also induces transformation of early T-cell progenitors, which subsequently develop into aggressive lymphomas [10]. Lastly, a series of PU.1 mutations have been shown to associate with AML in humans [43]. These mutations are thought to abrogate PU.1 function by a number of mechanisms, for example, by reducing its DNA-binding capacity or by disrupting its interaction with coregulators or other transcription factors. However, attempts by a number of other groups have failed to demonstrate an association between PU.1 mutations and AML [45–47], suggesting that the mutations identified in the Mueller study may be linked to rare subsets of the disease. Nonetheless, this does not exclude the possibility that PU.1 haploinsufficiency can promote leukaemogenesis in humans, further emphasising the importance of maintaining precise PU.1 expression levels in haematopoietic cells.
7. Conclusion
Through the study of a variety of transgenic mouse models and cell lines, PU.1 has emerged as a key regulator of haematopoiesis. The expression of PU.1 is exquisitely and dynamically controlled throughout the various haematopoietic differentiation pathways. Precise PU.1 levels direct cell fate by regulating gene expression in a graded manner and by antagonising the function of other haematopoietic regulators, such as Gata-1. Disturbing the normal expression pattern of PU.1 in haematopoietic cells can lead to skewed lineage commitment and in some instances, oncogenesis. Indeed, PU.1 haploinsufficiency has been linked with AML in humans. Understanding the mechanisms by which PU.1 and other factors regulate haematopoietic differentiation is of significant importance, as manipulation of these factors may offer a therapeutic option for the treatment of leukaemias and other haematopoietic disorders.
References
- 1.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331(6153):277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 2.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288(5470):1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 3.Kastner P, Chan S. PU.1: a crucial and versatile player in hematopoiesis and leukemia. International Journal of Biochemistry and Cell Biology. 2008;40(1):22–27. doi: 10.1016/j.biocel.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 4.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 5.McKercher SR, Torbett BE, Anderson KL, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO Journal. 1996;15(20):5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 6.Back J, Dierich A, Bronn C, Kastner P, Chan S. PU.1 determines the self-renewal capacity of erythroid progenitor cells. Blood. 2004;103(10):3615–3623. doi: 10.1182/blood-2003-11-4089. [DOI] [PubMed] [Google Scholar]
- 7.Back J, Allman D, Chan S, Kastner P. Visualizing PU.1 activity during hematopoiesis. Experimental Hematology. 2005;33(4):395–402. doi: 10.1016/j.exphem.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. Journal of Experimental Medicine. 2005;201(2):221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klemsz MJ, McKercher SR, Celada A, van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61(1):113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 10.Rosenbauer F, Owens BM, Yu L, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nature Genetics. 2006;38(1):27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 11.Moreau-Gachelin F, Wendling F, Molina T, et al. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Molecular and Cellular Biology. 1996;16(5):2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki H, Somoza C, Shigematsu H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106(5):1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbauer F, Wagner K, Kutok JL, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nature Genetics. 2004;36(6):624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 14.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. Journal of Experimental Medicine. 2005;201(9):1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher RC, Lovelock JD, Scott EW. A critical role for PU.1 in homing and long-term engraftment by hematopoietic stem cells in the bone marrow. Blood. 1999;94(4):1283–1290. [PubMed] [Google Scholar]
- 16.Akashi K, Traver D, Zon LI. The complex cartography of stem cell commitment. Cell. 2005;121(2):160–162. doi: 10.1016/j.cell.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 19.Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2010;2(6):640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arinobu Y, Mizuno S, Chong Y, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1(4):416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31(4):576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamath MB, Houston IB, Janovski AJ, et al. Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia. 2008;22(6):1214–1225. doi: 10.1038/leu.2008.67. [DOI] [PubMed] [Google Scholar]
- 23.Pop R, Shearstone JR, Shen Q, et al. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biology. 2010;8(9) doi: 10.1371/journal.pbio.1000484. Article ID e1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starck J, Doubeikovski A, Sarrazin S, et al. Spi-1/PU.1 is a positive regulator of the Fli-1 gene involved in inhibition of erythroid differentiation in friend erythroleukemic cell lines. Molecular and Cellular Biology. 1999;19(1):121–135. doi: 10.1128/mcb.19.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choe KS, Ujhelly O, Wontakal SN, Skoultchi AI. PU.1 directly regulates cdk6 gene expression, linking the cell proliferation and differentiation programs in erythroid cells. Journal of Biological Chemistry. 2010;285(5):3044–3052. doi: 10.1074/jbc.M109.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997;14(1):123–131. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- 27.Papetti M, Skoultchi AI. Reprogramming leukemia cells to terminal differentiation and growth arrest by RNA interference of PU.1. Molecular Cancer Research. 2007;5(10):1053–1062. doi: 10.1158/1541-7786.MCR-07-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atar O, Levi BZ. PU.1 silencing leads to terminal differentiation of erythroleukemia cells. Biochemical and Biophysical Research Communications. 2005;329(4):1288–1292. doi: 10.1016/j.bbrc.2005.02.109. [DOI] [PubMed] [Google Scholar]
- 29.Yamada T, Kondoh N, Matsumoto M, Yoshida M, Maekawa A, Oikawa T. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood. 1997;89(4):1383–1393. [PubMed] [Google Scholar]
- 30.Matushansky I, Radparvar F, Skoultchi AI. CDK6 blocks differentiation: coupling cell proliferation to the block to differentiation in leukemic cells. Oncogene. 2003;22(27):4143–4149. doi: 10.1038/sj.onc.1206484. [DOI] [PubMed] [Google Scholar]
- 31.Burda P, Curik N, Kokavec J, et al. PU.1 activation relieves GATA-1-mediated repression of Cebpa and Cbfb during leukemia differentiation. Molecular Cancer Research. 2009;7(10):1693–1703. doi: 10.1158/1541-7786.MCR-09-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA- 1: functional antagonism in erythroid cells. Genes and Development. 1999;13(11):1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P, Zhang X, Iwama A, et al. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96(8):2641–2648. [PubMed] [Google Scholar]
- 34.Zhang P, Behre G, Pan J, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houston IB, Kamath MB, Schweitzer BL, Chlon TM, DeKoter RP. Reduction in PU.1 activity results in a block to B-cell development, abnormal myeloid proliferation, and neonatal lethality. Experimental Hematology. 2007;35(7):1056–1068. doi: 10.1016/j.exphem.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hohaus S, Petrovick MS, Voso MT, Sun Z, Zhang DE, Tenen DG. PU.1 (Spi-1) and C/EBPα regulate expression of the granulocyte-macrophage colony-stimulating factor receptor α gene. Molecular and Cellular Biology. 1995;15(10):5830–5845. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Molecular and Cellular Biology. 1994;14(1):373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahl R, Walsh JC, Lancki D, et al. Regulation of macrophage and neutrophil cell fates by the PU.1: C/EBPα ratio and granulocyte colony-stimulating factor. Nature Immunology. 2003;4(10):1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 39.Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 40.Hu CJ, Rao S, Ramirez-Bergeron DL, et al. PU.1/Spi-B regulation of c-rel is essential for mature B cell survival. Immunity. 2001;15(4):545–555. doi: 10.1016/s1074-7613(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 41.Su GH, Ip HS, Cobb BS, Lu MM, Chen HM, Simon MC. The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. Journal of Experimental Medicine. 1996;184(1):203–214. doi: 10.1084/jem.184.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16(2):285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 43.Mueller BU, Pabst T, Osato M, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100(3):998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]
- 44.Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24(7):1249–1257. doi: 10.1038/leu.2010.104. [DOI] [PubMed] [Google Scholar]
- 45.Suraweera N, Meijne E, Moody J, et al. Mutations of the PU.1 Ets domain are specifically associated with murine radiation-induced, but not human therapy-related, acute myeloid leukaemia. Oncogene. 2005;24(22):3678–3683. doi: 10.1038/sj.onc.1208422. [DOI] [PubMed] [Google Scholar]
- 46.Vegesna V, Takeuchi S, Hofmann WK, et al. C/EBP-β, C/EBP-δ, PU.1, AML1 genes: mutational analysis in 381 samples of hematopoietic and solid malignancies. Leukemia Research. 2002;26(5):451–457. doi: 10.1016/s0145-2126(01)00150-3. [DOI] [PubMed] [Google Scholar]
- 47.Ley TJ, Minx PJ, Walter MJ, et al. A pilot study of high-throughput, sequence-based mutational profiling of primary human acute myeloid leukemia cell genomes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):14275–14280. doi: 10.1073/pnas.2335924100. [DOI] [PMC free article] [PubMed] [Google Scholar]


