Abstract
Composting is a way of transforming the organic waste into fertilizer, minimizing the use of inorganic compounds that may contaminate the environment. This transformation is the result of the microorganism action, converting complex carbon sources into energy. Enzymes that are exported by the microorganisms to the surrounding environment mediate this process. The aiming of the present work is to prospect the compost produced by the organic composting unit (OCU) of the Fundação Parque Zoológico de São Paulo (FPZSP) to find novel starch hydrolyzing organisms (SHO) that secrete large amounts of amylases under harsh conditions, such as high temperature. We found five bacterial isolates that have amylolytic activity induced by soluble starch and 39°C temperature of growth. These bacterial strains were identified by MALDI-TOF (Matrix-assisted laser desorption/ionization-Time of Flight) analysis, a rapid and efficient methodology for microbe identification in large scale. Our results present amylolytic strains that belong to diverse taxonomic groups (Solibacillus silvestris, Arthrobacter arilaitensis, Isoptericola variabilis, and Acinetobacter calcoaceticus); some of them have never been associated with this kind of hydrolytic activity before. The information regarding enzyme induction will be important to optimize the production by the bacterial isolates, which may be a great value for biotechnological applications.
1. Introduction
Amylases produced by plants, animals, and microorganisms have a wide range of industrial applications, such as pharmaceutical, animal feed, paper, textile, food industry, and renewable energy [1–3]. Most amylases used in industry are from microbial source due to several factors, for example, the great microbial genetic diversity present in the environment, high enzymatic activity in a wide range of conditions (extreme pH, temperature, osmolarity, pressure, etc.), and simple and cost effective production [4]. Industrial processes require robust enzymes, especially because large amounts of heat are generated during the process, accumulation of inhibitors, high salt concentrations, and so forth; therefore, hydrolytic microbes, which have their hydrolases induced by temperature, may produce thermotolerant enzymes and, consequently, are highly desirable [5, 6].
Composting is the transformation of residual organic matter into fertilizer accomplished by microorganism, which secrete, among other things, hydrolytic enzymes to assimilate diverse energy sources available in the substrate in order to survive. Compost piles can reach up to 80°C due to microbial metabolism, favoring the development of thermophylic organisms. In this process, the interaction between biotic and nonbiotic factors leads to constant transformation of the complex microbial community over time, which is the mechanism underlining organic matter transformation [7–10].
In Brazil, the São Paulo zoo has an organic compost production unit (OCU), where all the waste from about 3000 native and exotic animals are mixed with plant debris from the Atlantic Forest in a ratio of 30 parts of carbon to 1 part of nitrogen in aerobic condition to produce 600 tons compost per year. This is a self-sustainable process since the final compost generated is used in the zoo farm to fertilize the land, which produces most of the food consumed by the animals in the park. In this work, we used this compost as the starting material to prospect amylolytic enzymes produced by novel thermophylic starch-hydrolyzing organisms (SHOs). To our knowledge, compost comprising these properties has never been screened for amylolytic microorganisms. Here, we describe the isolation and screening of a compost microbe collection (387 entries) for amylolytic activity. Five bacterial isolates from diverse taxonomic groups, identified by MALDI-TOF spectrometry and 16S gene sequencing, were capable of using starch as the major carbon source and secrete amylolytic enzymes to the extracellular environment in response to a combination of soluble starch and 39°C growth temperature. Since many hydrolyzing enzymes are subjected to catabolism repression [11], characterization of induction profile is important for amylase production, fermentation strategies, enzyme purification, and SHO utilization in biotechnology.
2. Material and Methods
2.1. Compost Sampling
Samples were collected according to the method described by Bitencourt et al. [12]. In brief, the temperature of several piles (8 m3 each) was measured in five different points. Those that displayed temperatures higher than 45°C were probed with a soil auger at 1 meter depth. Organic wastes present in sampled compost piles were food waste, droppings and excreta, beds of native and exotic wild animals, animals carcasses, wood chips from gardening and Atlantic Forest plant waste. Phases of decomposition were considered active degradation (before aeration) and mature compost (after aeration). Pile aeration was achieved by mechanical revolving of the material after 50 to 60 days after composting started.
2.2. Microbial Isolation and Morphological Characterization
All methods of isolation used in this work are suitable to aerobic organisms. Once the samples were removed from the piles at the desired temperature, one gram of compost was weighted and diluted in 5 mL of 0.9% sterile saline, vortex, and allowed to decant for 2 hours. Serial dilutions were prepared and spread on nutrient agar plates (Himedia, India) and incubated at 30°C. After 24 and 48 hours, plates containing isolated colonies were selected, and the isolated colonies were transferred individually to obtain single purified colonies. In order to classify the isolated microorganisms in the main groups of bacteria, yeast and fungi, microscopic analysis of purified colonies was carried out in fresh preparations analyzed in optic microscope (Axiscope, Zeiss). Gram staining was conducted with commercial kit (LaborClin, Brazil) and confirmed by growth on MacConkey agar (Himedia, India) plates. Bacteria capable of growth in this selective medium were considered Gram negative.
2.3. Enzymatic Activity on Agar Plates
The purified colonies were point inoculated on starch induction medium containing 0.6% peptone, 0.5 g/L MgSO4, 0.5 g/L KCl, 1% soluble starch, 2% agar, pH 7.0 for bacteria and 1.4 g/L de KH2PO4, 10 g/L de NH4NO3, 0.5 g/L de KCl, 0.1 g/L de MgSO4.7H2O, 0.01 g/L de FeSO4.7H2O, 2% soluble starch, and 2% agar, pH 5.5 for yeast and filamentous fungi [13], incubated at 30°C, 39°C, and 45°C for 48 to 72 hours. The starch present in the bacteria induction medium was considered the major carbon source. However, it was not the sole carbon source, since peptone, which is considered a complex additive, was added to the medium. After the incubation period, the plates were exposed to iodine crystals during 5 minutes to reveal the starch degradation zone [14]. Microorganisms were considered amylolytic when they were able to grow in the presence of starch as the major carbon source and formed a degradation zone around the colony. All experiments were done in triplicate at 3 temperatures (30, 39, and 45°C), and a starch hydrolytic index (SHI) was assigned to each isolate. SHI was calculated as the sum of colony and degradation zone diameter (cm) divided by colony diameter.
2.4. Growth Evaluation and Amylase Production
each isolate under investigation was cultured for 16 hours in 50 mL of LB (Luria Bertani) complex medium (10 g/L NaCl, 10 g/L Tryptone and 5 g/L Yeast Extract, pH 7.0) at 150 rpm at 30°C. Cells were harvested by centrifugation, washed twice in sterile distilled water, and inoculated (OD600 0,3) in 50 mL of starch medium (0.6% peptone, 0.5 g/L MgSO4, 0.5 g/L KCl, 1% soluble starch) or LB medium. The cultures were incubated on a rotary shaker (150 rpm) at 30, 39 and 45°C. Samples of the supernatant were collected at specific time points (0, 2, 4, 6, 16, 18, 24, and 26 hours) to evaluate the growth pattern at OD600. The aliquots drawn for the enzymatic assays were centrifuged to remove the cells followed by filter sterilization (0,22 μm).
2.5. Determination of Enzymatic Activity
The enzymatic activity was determined according to Smith & Roe [15] adapted by Medda & Chandra [16]. Briefly, a total of 0.5 mL of cell-free supernatant of the cultures were assayed for amylolytic activity in 1 mL of 1% soluble starch diluted in 50 mM acetate buffer pH 5.6 during 15 minutes at 96°C for all samples taken from all time points. The cell pellet extract (0.5 mL) was also assayed for the presence of intracellular amylolytic enzymes. In brief, pellet was washed two times in water, resuspended in the reaction buffer, and submitted to 3 cycles of 1 minute ultrasonic liquid processing (LKB Instruments, Australia) under ice bath during the entire procedure. A sample of the cell extract was observed under the microscope to ensure cell lyses. Reducing sugar for supernatant and cellular extract was assayed by dinitrosalicylic acid (Sigma, USA) method according to standard protocol by Miller [17]. Protein concentration of each time point was determined by Bradford method [18], using the Bio-Rad protein assay kit (BioRad, USA) according to the instructions provided by the manufacturer. All growth curves and reducing sugar assays were performed in triplicates (n = 9). One unit was considered the amount of enzyme required to generate 1 μmol of glucose per minute at 96°C. Specific activity was defined as U/mg of protein. Maximum total units refer to all units produced in 50 mL culture at the time point in which the highest specific activity was achieved.
2.6. Bacterial Identification by Mass Spectrometry
Taxonomic identification was done by MALDI-TOF analysis [19], which provides protein profiles from each isolate. The methodology employed was the ethanol/formic acid extraction, which is suggested by Bruker Daltonics. In brief, cells from single colonies were recovered by scraping the plate and transferring the material into a tube with 300 μL of sterile water and mixing carefully. Absolute ethanol (900 μL) was added, mixed, and centrifuged at 10.000 g for 2 minute at room temperature (25°C). The supernatant was discarded and the pellet air dried. The dry pellets were mixed thoroughly with 50 μL 70% formic acid (Merck, USA) and 50 μL acetonitrile (Merck, USA). The suspension was centrifuged at 10.000 g for 2 minutes, and 1 μL of the supernatant was place onto an Micro Scout Plate (MSP) 96 polished steel target (Bruker Daltonics GmbH, Germany) and dried at room temperature (25°C). Each sample was overlaid with 1 μL of the matrix solution which consisted of a saturated solution of a-cyano-4-hydroxy-cinnamic acid (Sigma, USA) in 50% acetonitrile—2.5% trifluoroacetic (Sigma, USA), and the matrix/sample was cocrystallized by air drying at room temperature (25°C). Measurements were performed with a Microflex LT mass spectrometer (Bruker Daltonics) using FlexControl software (version 3.0, Bruker Daltonics). Spectra were recorded in the positive linear mode (laser frequency, 20 Hz; ion source 1 voltage, 20 kV; ion source 2 voltage, 18.6 kV; lens voltage, 7.5 kV; mass range, 2000 to 20 000 Da). For each spectrum, 240 shots in 50-shot steps from different positions of the target spot (automatic mode) were collected and analyzed. Spectra were internally calibrated by using Escherichia coli ribosomal proteins. The raw spectra were imported into the BioTyper software (version 2.0, Bruker Daltonics), processed by standard pattern matching with default settings, and the results reported in a ranking table. Outcomes of the pattern-matching process were expressed as proposed by MALDI-TOF biotyper (MT) manufacturer with ID scores ranging from 0 to 3. Scores < 1.70 were considered not to have a generated reliable ID; a score of 1.7 < ID < 1.9 was considered ID to genus, and a score > 1.9 was used for reliable species ID.
2.7. Bacterial Identification by 16S DNA Sequencing
gDNA was extracted from 10 mL overnight cultures with Axygen bacterial genomic DNA extraction kit (Axygen Scientific, USA). Small Subunit RNA (16S) was amplified by PCR (polymerase chain reaction), using 1 U Taq Polymerase (New England Biolabs, USA), 1x Buffer (10 mM Tris-HCl, 50 mM KCl, pH 8.3), 2 mM MgCl2, 200 μM dNTP's (Fermentas, Finland), 0.2 μM of each primer (PRCP005 5′ GTGCCAGCMGCCGCGG 3′ and PRCP006 5′ ACGGGCGGTGTGTRC 3′, IDT, USA). Amplification condition: 1 cycle 94°C for 5 minutes, 35 cycles: denaturing at 94°C for 30 seconds, primers annealing at 54°C for 30 seconds, and extension at 72°C for 1 minute, a final extension of 72°C for 7 minutes. Single-band amplicons were purified with PCR Clean up Kit (Axigen Scientific, USA) and double stranded sequenced with BigDye Terminator (Applied Biosystems, USA) according to the manufacturer's instructions. The sequences were aligned using SeqMan module of software Lasergene (DNAStar, USA) and blasted against Genebank and Ribosomal Database Project (RDP). Identification at species level was considered for sequence similarities above 97%.
3. Results and Discussion
3.1. Microorganism Isolation from São Paulo Zoo Park Organic Compost Unit
Three samples of different compost piles were removed from São Paulo zoo OCU over an approximately 5-month period. The properties (composition, temperature, pH and decomposition phase) of the compost pile are summarized in Table 1.
Table 1.
Compost pile properties.
| Properties | Pile 1 | Pile 2 | Pile 3 |
|---|---|---|---|
| Date | 19.02.2009 | 12.05.2009 | 18.07.2009 |
| Average temperature | 48.7 | 72.6 | 55.2 |
| pH | — | 6.1 | 6 |
| Decomposition phase | *Active degradation | Active degradation | **Mature |
| Number of isolates | 183 | 38 | 166 |
| SHO | 15% | 13.1% | 12.6% |
*Active degradation: before aeration.
**Mature: after aeration.
A total of 387 microorganisms were isolated at 30°C from these samples as described in materials and methods. Among these, under microscopic examination, 67% were classified as bacteria, 20% as filamentous fungi, 11% as yeast, and 2% remains undetermined, due to ambiguous morphology, which impaired positive classification. Table 1 shows that, as the temperature in the compost pile rises, there is a decrease in microbial population, which is reflected by the lower number of isolates in compost pile 2 (n = 38) compared to piles 1 and 3 (n = 183 and n = 166, resp.).
3.2. Amylolytic Microorganism Screening
Fifty-five microorganisms were classified as SHO (14%). Considering each compost pile individually, the SHO percentage is very similar (Table 1). Among these SHO isolates, 10 amylolytic bacteria were chosen for further studies. The selection criteria was (i) the ability to grow in 1% starch medium as the major carbon source at 30, 39, and 45°C and (ii) the production of a SHI of, at least, 1.5 in total average (Table 2). Also, gram staining and colony macromorphology were taken into account, in order to guarantee the analysis of a greater diversity of microorganisms (data not shown).
Table 2.
Starch hydrolytic index (SHI) on 1% starch solid medium at 3 different temperatures for 10 SHO chosen for further studies.
| Isolate | SHI 30°C | SHI 39°C | SHI 45°C | Average |
|---|---|---|---|---|
| URX291 | 1.6 ± 0.2 | 1.9 ± 0.5 | 2 ± 0.8 | 1.8 ± 0.2 |
| URX303 | 1.7 ± 0.3 | 1.8 ± 0.8 | 1.9 ± 1.0 | 1.8 ± 0.1 |
| URX336 | 1.5 ± 0.3 | 1.7 ± 0.1 | 1.5 ± 0.5 | 1.6 ± 0.1 |
| URX350 | 1.8 ± 0.4 | 1.9 ± 0.5 | 1.8 ± 0.5 | 1.8 ± 0.1 |
| URX356 | 1.6 ± 0.2 | 1.9 ± 0.3 | 2.0 ± 0.8 | 1.8 ± 0.2 |
| URX377 | 2.1 ± 0.1 | 2.0 ± 0.3 | 1.8 ± 0.2 | 2.0 ± 0.2 |
| URX379 | 1.9 ± 0.3 | 1.7 ± 0.2 | 2.0 ± 0.7 | 1.9 ± 0.2 |
| UED487 | 1.6 ± 0.3 | 1.9 ± 0.5 | 2.0 ± 0.6 | 1.8 ± 0.2 |
| UED 641 | 2.3 ± 1.1 | 2.0 ± 0.5 | 1.6 ± 0.2 | 2.0 ± 0.3 |
| UED 644 | 2.6 ± 1.2 | 1.7 ± 0.4 | 1.9 ± 0.5 | 2.1 ± 0.5 |
Each data point is the result of 3 replicates for each temperature.
None of the yeast and filamentous fungus isolated was classified as SHO. It is possible that the high temperature observed at the collection points taken for this work (Table 1) exterminated most of these organisms, or the isolation conditions did not favor them. Therefore, samples collected from piles with a lower temperature and a different isolation strategy may be necessary to augment the number of filamentous fungus and yeast capable of starch degradation. An enrichment liquid media containing soluble starch as the sole carbon source, low pH and the antibiotics addition to the media maybe a way to preferentially isolate amylolytic yeasts and fungi. The compost sample could be directly added to this media and cultured overtime. After several passages in liquid media, the enriched culture could be inoculated in solid starch media to obtain single colonies.
3.3. Quantification of the Amylolytic Activity in Liquid Culture
Growth curves in starch liquid medium were performed at 30, 39, and 45°C for ten selected isolates; however Figure 1 shows the growth pattern for five isolates chosen based on their enzymatic activity in response to temperature. Figures 1(a) and 1(b) show that the performance of each isolate follows a similar trend of growth under these conditions (30°C versus 39°C). At 45°C, all isolates presented a reduce growth and isolates URX303 and URX350 grew to a lower extent compared to other three isolates, especially at the end of the growth period (Figure 1(c)).
Figure 1.
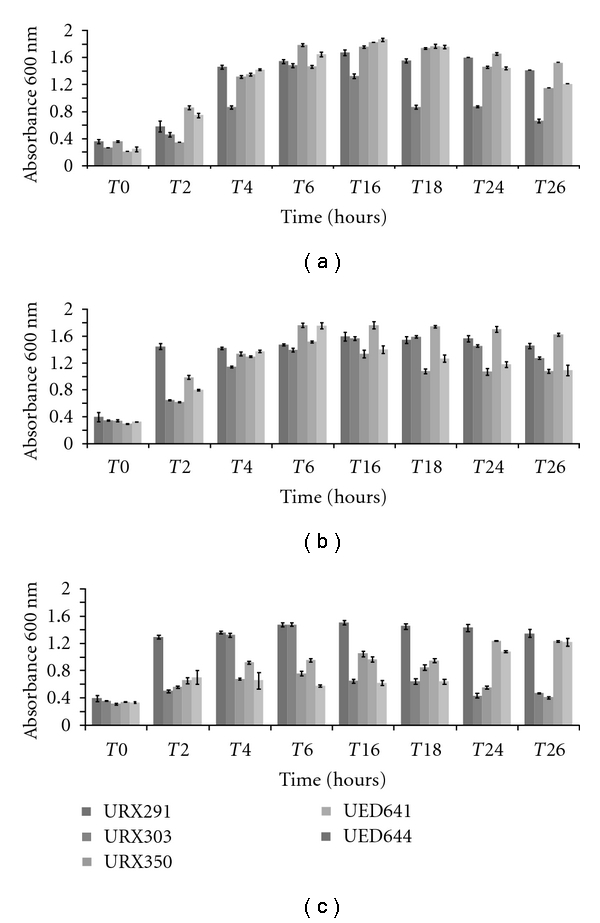
Growth pattern of five isolates over 26-hour period. (a) 30°C, (b) 39°C, and (c) 45°C. All growth curves were done in triplicates for all three temperatures, the bars represent standard deviation.
The amylolytic activity in the supernatant of the cultures grown at three temperatures (30, 39, and 45°C) was measured over a 26-hour period (8 time points) in order to establish the temperature induction pattern for the extracellular starch degrading enzymes. Activity was determined by measuring the reducing sugar released during the incubation of cell-free supernatant with soluble starch, according to the conditions described in Material and Methods. Also, the amount of reducing sugar released by enzymes present in the cell pellet extract was assayed, but no activity was detected (data not shown), suggesting that most of the amylases produced are exported to the extracellular fraction and any active enzyme that remained inside the cells was probably at concentrations bellow the sensitivity of this assay. In accordance to our results, Ryan et al. [20] reported the analysis of Bifidobacteria strains from the Japan Collection of Microorganisms (JCMs) that produces extracellular starch hydrolyzing enzymes. In this report, cell-free supernatants and cellular extracts were qualitative assayed for amylolytic activity on plates containing starch, amylopectin, or pullulan. They found that the amylolytic activity was present only in the extracellular fraction. In our assays cellular extracts showed no activity.
Out of the 10 isolates analyzed for extracellular amylolytic activity, 5 isolates (URX291, URX303, URX350, UED641, and UED644) clearly showed induction in the presence of starch as the major carbon source at 39°C growth temperature (Figures 2(a)to 2(e) and Table 3). Moreover, none of these 5 chosen isolates showed significant extracellular enzymatic induction at 30°C and 45°C, indicating that the optimum temperature for amylase secretion in this group of isolates is 39°C (Figures 2(a)–2(e)). Therefore, all subsequent enzymatic analysis was performed with supernatant harvested at 39°C and 30°C. The latter temperature of cultivation was employed as a control.
Figure 2.
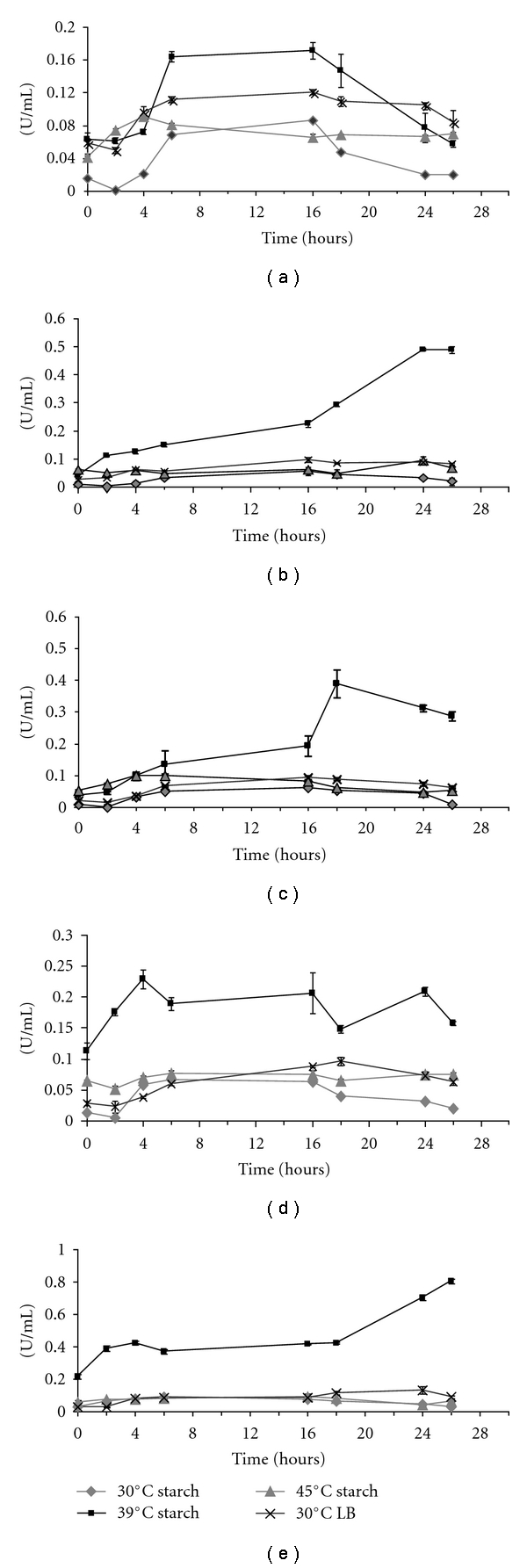
Kinetics of amylase activity in the culture supernatants of five isolates (a) URX291, (b) URX303, (c) URX350, (d) UED 641, and (e) UED644 in starch medium for 26 hours at 30 (diamonds), 39 (squares), 45°C (triangles), and in LB rich medium at 30°C (crosses). n = 9. Vertical lines represent error bars.
Table 3.
Maximum specific activity (U/mg of protein) and the total units produced at the time point (hours) where the highest specific activity was found for each of the 5 isolates that showed enzymatic induction. 30°C and 39°C are the temperatures of growth where supernatant samples were collected to conduct the enzymatic assays. LB indicates the complex medium used, and starch refers to the induction medium containing starch as the major carbon source, in addition to a trace of peptone. All data points are average of 3 replicates of growth curves and 3 replicates of enzyme assay per time point (equals to 9 replicates).
| Strain | 30°C | 39°C | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LB | Starch | LB | Starch | |||||||||
| U/mg | Total units | Time point | U/mg | Total units | Time point | U/mg | Total units | Time point | U/mg | Total units | Time point | |
| URX291 | 0.079 ± 0.007 | 4 | 6 | 0.022 ± 0.001 | 4.3 | 16 | 0.043 ± 0.002 | 5.9 | 18 | 0.114 ± 0.016 | 8.2 | 6 |
| URX303 | 0.098 ± 0.002 | 6.8 | 18 | 0.015 ± 0.002 | 2.7 | 16 | 0.036 ± 0.011 | 4.5 | 16 | 0.278 ± 0.024 | 25.2 | 26 |
| URX350 | 0.097 ± 0.007 | 6.5 | 16 | 0.022 ± 0.004 | 2.2 | 16 | 0.037 ± 0.001 | 4.7 | 16 | 0.134 ± 0.014 | 19.4 | 18 |
| UED641 | 0.094 ± 0.005 | 6.1 | 16 | 0.029 ± 0.002 | 2.9 | 4 | 0.035 ± 0.001 | 4.4 | 16 | 0.120 ± 0.006 | 8.7 | 2 |
| UED644 | 0.160 ± 0.027 | 9.2 | 24 | 0.037 ± 0.003 | 3.2 | 2 | 0.035 ± 0.001 | 4.3 | 16 | 0.230 ± 0.024 | 19.4 | 2 |
In order to verify if expression of the amylolytic enzymes is dependent upon substrate induction, besides temperature, the specific activity (U/mg of protein) was calculated for all 5 isolates in starch induction medium and LB at all 8 time points collected during growth at 30 and 39°C. Table 3 shows the time points in which the highest specific activity can be found for the 5 isolates in each cultivation condition (maximum specific activity). All isolates had higher specific activity in starch at 39°C. Isolates URX303 and UED644 had the highest specific activity (0.278 at 26 hours and 0.230 at 2 hours, resp.), showing that URX303 has the longest time of induction response whereas UED644 had the fastest among these isolates.
Orlando et al. [21] reported a Bacillus subtilis strain that presented a specific activity of 0.83 U/mg of protein in crude extract. However, the authors used much larger culture volume (50 liter tank) and longer fermentation (42 hours). Mabrouk et al. [22] also reported the cloning and expression of a Bacillus sp. amylase in heterologous system and was able to achieve 0,93 U/mg of protein in crude extract. Carvalho et al. [23] also studied an unidentified bacteria isolated from petroleum contaminated soil that presented the maximum amyloglucosidase activity at 0.18 U/mL, using starch as the sole carbon source. A screen for starch degrading Bifidobacterial strains by Ryan et al. [20] reported that 11 of these microorganisms belonging to various species are able to produce a range of specific activities (from 0.84 to 1.35 U/mg of protein).
Kubrak et al. [24] reported a Bacillus sp. BKL20 strain isolated from natural can reach up to 7 U/mg of enzyme specific activity. However, the cultivation parameters (starch, peptone and yeast extract concentration and temperature) have been optimized for this strain reflection an increment of activity.
Table 3 shows that the amylase induction is dependent on the combination of starch-containing media and 39°C temperature of culture for isolates URX303, URX350 and UED644 (9.3, 8.8, and 6.0 fold induction over 30°C, resp.). On the other hand, the induction for these isolates is less pronounced when they are switched from LB to starch at 39°C (5.6, 4.1, and 4.5 fold induction, resp.). For isolates URX291 and UED641, the fold-induction for amylase production on starch-containing medium at 30°C versus 39°C was 1.9 and 3, respectively.
The enzyme activities reported in this work are relevant once compared to the activities available in the literature and cited above, especially considering that no optimization of the growth conditions has been performed for each isolate. Furthermore, these results are of great interest in other aspects, such as for the optimization of fermentation procedure, protein purification, and microbial characterization. The knowledge of the induction kinetics regarding substrate and temperature will allow the harvesting of the largest amounts of protein, which is an important factor regarding production and often a limiting step in protein chemistry. Moreover, this information will be valuable for enzyme production by SHO, considering that they can be used in biotechnological processes.
3.4. SHO Identification
We believe that the São Paulo Zoo OCU has the potential to host a large number of novels, not yet described microorganisms, which could be useful for hydrolytic enzyme search. In order to evaluate this possibility, we used two different tools to identify the selected amylolytic microbes and attribute at least genus level identification to each one of them. As a first screen, we generated mass spectrometry profiles and compared them to the Bruker Daltonics database using Biotyper software. Table 4 shows the results found by this technology. Five out of ten microorganisms could not be identified because no similar profiles were found in the database. Since the database is composed roughly of 4000 entries from clinical origin, and we are prospecting environmental microbes, this outcome is not surprising. In order to validate MALDI-TOF results and attempt positive identification of the unidentified isolates, we conducted molecular identification by 16S gene sequencing (Table 4).
Table 4.
Microorganism identification based on 16S gene sequence and protein profile by MALDI-TOF.
| Isolate | Biotyper | 16S (SSU RNA) | Accession # |
|---|---|---|---|
| URX291 | Bacillus sp. | Bacillus megaterium DSM319 (99%) | CP001982.1 |
| URX303 | (−) | Bacillus safensis DSM 19292 (99%) | JF798363.1 |
| URX336 | (−) | Bacillus subtilis CCGB:1266 (99%) | GU434357.1 |
| URX350 | (−) | Enterobacter asburiae DSM 17506 (100%) | HQ242717.1 |
| URX356 | (−) | Solibacillus silvestris BCHCNZ317 (98%) | GU188937.1 |
| URX377 | Arthrobacter sp. | Arthrobacter arilaitensis RE117 (97%) | FQ311875.1 |
| URX379 | Arthrobacter sp. | Arthrobacter arilaitensis RE117 (96%) | FQ311875.2 |
| UED487 | (−) | Isoptericola variabilis NBRC 104115 (99%) | AB489221.1 |
| UED641 | Acinetobacter sp. | Acinetobacter calcoaceticus DSM3000 (99%) | X81668.1 |
| UED644 | Bacillus cereus | Bacillus cereus BCHMAC21 (98%) | GU188894.1 |
(−) no matches found in the Bruker Daltonics database.
All isolates showed high sequence similarities to 16S entries in the GeneBank and at Ribosomal Database Project (RDP). URX 291, URX336, URX303, and UED644 have high similarity sequences at species level to Bacillus megaterium (99%), B. subtilis (99%), Bacillus safensis (100%), and B. cereus (98%), respectively. Except for URX303 (B. safensis), all isolates belonging to this genus have already been reported as SHO, and all of them were reported as extracellular producers and had at least alpha-amylase gene cloned and expressed in heterologous system, [21, 25–27].
However, six isolates (URX350, URX356, URX377, URX379, UED487, and UED641) showed similarity sequence to species that, either, have been described recently, therefore have limited information regarding biological properties (URX356 and UED487), or are well known, but have never been associated with amylolytic activity (URX350, URX377, UED641, and URX379).
URX377 and URX379 were both considered as Arthrobacter arilaitensis. To our knowledge this microorganism has never been associated with this enzymatic activity. However, Gratia et al. [28] demonstrated that Arthrobacter psychrolactophilu is an extremophile microbe capable of producing extracellular amylases under low growth temperature, and it is able to clarify wastewater containing starch and protein at 10°C. Other isolates belonging to the Arthrobacter genus have been reported as useful to bioremediation since they can consume p nitrophenol [29].
Isolate UXR356 was identified by 16S sequencing as Solibacillus sylvestris, and no correlation between this species and hydrolytic activity could be found in the literature suggesting that this is novel information. Similarly, UED487 was identified as Isoptericola variabilis, which has not been the target of many reports. However, another species in the genus (Isoptericola jiangsuensis) has been associated with chitin-hydrolyzing activity [30].
UED641 was identified as Acinetobacter calcoaceticus, which has been reported as an exoenzyme producer [31]. However, since this species is considered an opportunistic pathogen and explored under this perspective, it has not been investigated for biotechnological applications.
4. Conclusions
In this paper, we describe 10 amylolytic bacterial strains belonging to very diverse taxonomic groups, identified in a collection of 387 microbes from São Paulo Zoo composting unit, an exotic substrate that has never been prospected before for hydrolytic microorganisms. The increment on amylases activity, observed on the supernatant of five bacterial strains, showed strong induction in response to the combination of starch and temperature (39°C) during a 26-hour period, whereas the other five bacterial isolates did not present such an induction. Initially, we asked the question whether the São Paulo Zoo compost would be a suitable substrate for the discovery of novel hydrolytic microorganisms, producing extracellular robust enzymes for biotechnological application. The data presented demonstrates that not only this is true, but also SHO isolated here shows a distinct pattern of enzyme expression dependent upon substrate induction and high temperature, which are interesting aspects for enzymes aiming biotechnological application.
Acknowledgments
The authors would like to recognize the relevant participation of Dr. Paulo Magalhães Bressan, Dr. João Batista da Cruz, Mr. Carlos Augusto Magalhães, and Mr. João Soares at Fundação Parque Zoológico de São Paulo (FPZSP) for providing all precious information on its composting unit processes and for the compost samples. This work is being conducted under a Partnership Agreement between FPZSP and UNIFESP and was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil, Grants 2009/52030-5 to LJ, 2007/50536-3 to MAV, and Conselho Nacional de Pesquisa (CNPq) postdoctoral fellowship to RFB 2009-8/502889.
References
- 1.Van Der Maarel MJEC, Van Der Veen B, Uitdehaag JCM, Leemhuis H, Dijkhuizen L. Properties and applications of starch-converting enzymes of the α-amylase family. Journal of Biotechnology. 2002;94(2):137–155. doi: 10.1016/s0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 2.Torney F, Moeller L, Scarpa A, Wang K. Genetic engineering approaches to improve bioethanol production from maize. Current Opinion in Biotechnology. 2007;18(3):193–199. doi: 10.1016/j.copbio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Sun H, Zhao P, Ge X, et al. Recent advances in microbial raw starch degrading enzymes. Applied Biochemistry and Biotechnology. 2010;160(4):988–1003. doi: 10.1007/s12010-009-8579-y. [DOI] [PubMed] [Google Scholar]
- 4.Burhan A, Nisa U, Gökhan C, Ömer C, Ashabil A, Osman G. Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochemistry. 2003;38(10):1397–1403. [Google Scholar]
- 5.Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Current Opinion in Biotechnology. 2008;19(3):210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 6.VanFossen AL, Lewis DL, Nichols JD, Kelly RM. Polysaccharide degradation and synthesis by extremely thermophilic anaerobes. Annals of the New York Academy of Sciences. 2008;1125:322–337. doi: 10.1196/annals.1419.017. [DOI] [PubMed] [Google Scholar]
- 7.Schulze KL. Continuous thermophilic composting. Applied Microbiology. 1962;10:108–122. doi: 10.1128/am.10.2.108-122.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waker LP, Nock TD, Gosset JM, VanderGheynst JS. Managing moisture limitations on microbial activity in high solid aerobic decomposting: pilot-scale experimentation. Process Biochemistry. 1999;34:601–612. [Google Scholar]
- 9.Dees PM, Ghiorse WC. Microbial diversity in hot synthetic compost as revealed by PCR-amplified rRNA sequences from cultivated isolates and extracted DNA. FEMS Microbiology Ecology. 2001;35(2):207–216. doi: 10.1111/j.1574-6941.2001.tb00805.x. [DOI] [PubMed] [Google Scholar]
- 10.Higgins CW, Walker LP. Validation of a new model for aerobic organic solids decomposition: simulations with substrate specific kinetics. Process Biochemistry. 2001;36(8-9):875–884. [Google Scholar]
- 11.Niu D, Zuo Z, Shi GY, Wang ZX. High yield recombinant thermostable α-amylase production using an improved Bacillus licheniformis system. Microbial Cell Factories. 2009;8, article no. 58 doi: 10.1186/1475-2859-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitencourt ALV, Vallim MA, Maia D, et al. Core sampling test in large-scale compost cells for microorganism isolation. African Journal of Microbiology Research. 2010;4(15):1631–1634. [Google Scholar]
- 13.Bernfeld P. Enzymes of starch degradation and synthesis. Advances in Enzymology. 1951;12:379–428. doi: 10.1002/9780470122570.ch7. [DOI] [PubMed] [Google Scholar]
- 14.Robyt JF, Bemis S. Use of the AutoAnalyzer for determining the blue value of the amylose-lodine complex and total carbohydrate by phenol-sulfuric acid. Analytical Biochemistry. 1967;19(1):56–60. doi: 10.1016/0003-2697(67)90133-9. [DOI] [PubMed] [Google Scholar]
- 15.Smith BW, Roe JH. A photometric method for the determination of α-amylase in blood and urine, with use of the starch-iodine color. The Journal of Biological Chemistry. 1949;179(1):53–59. [PubMed] [Google Scholar]
- 16.Medda S, Chandra AK. New strains of Bacillus licheniformis and Bacillus coagulans producing thermostable α-amylase active at alkaline pH. Journal of Applied Bacteriology. 1980;48(1):47–58. doi: 10.1111/j.1365-2672.1980.tb05205.x. [DOI] [PubMed] [Google Scholar]
- 17.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry. 1959;31(3):426–428. [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 19.Freiwald A, Sauer S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nature protocols. 2009;4(5):732–742. doi: 10.1038/nprot.2009.37. [DOI] [PubMed] [Google Scholar]
- 20.Ryan SM, Fitzgerald GF, Van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Applied and Environmental Microbiology. 2006;72(8):5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlando AR, Ade P, Di Maggio D. The purification of a novel amylase from Bacillus subtilis and its inhibition by wheat proteins. Biochemical Journal. 1983;209(2):561–564. doi: 10.1042/bj2090561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mabrouk SB, Messaoud EB, Ayadi D, et al. Cloning and sequencing of an original gene encoding a maltogenic amylase from Bacillus sp. US149 strain and characterization of the recombinant activity. Molecular Biotechnology. 2008;38(3):211–219. doi: 10.1007/s12033-007-9017-4. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho NB, De Souza RL, De Castro HF, Zanin GM, Lima ÁS, Soares CMF. Sequential production of amylolytic and lipolytic enzymes by bacterium strain isolated from petroleum contaminated soil. Applied Biochemistry and Biotechnology. 2008;150(1):25–32. doi: 10.1007/s12010-008-8194-3. [DOI] [PubMed] [Google Scholar]
- 24.Kubrak OI, Storey JM, Storey KB, Lushchak VI. Production and properties of α-amylase from Bacillus sp. BKL20. Canadian Journal of Microbiology. 2010;56(4):279–288. doi: 10.1139/w10-014. [DOI] [PubMed] [Google Scholar]
- 25.Nanmori T, Nagai M, Shimizu Y, Shinke R, Mikami B. Cloning of the β-amylase gene from Bacillus cereus and characteristics of the primary structure of the enzyme. Applied and Environmental Microbiology. 1993;59(2):623–627. doi: 10.1128/aem.59.2.623-627.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Lu F, Chen G, et al. High-level expression, purification and characterization of a recombinant medium-temperature α-amylase from Bacillus subtilis. Biotechnology Letters. 2009;32(1):119–124. doi: 10.1007/s10529-009-0112-4. [DOI] [PubMed] [Google Scholar]
- 27.Mabrouk SB, Messaoud EB, Ayadi D, et al. Cloning and sequencing of an original gene encoding a maltogenic amylase from Bacillus sp. US149 strain and characterization of the recombinant activity. Molecular Biotechnology. 2008;38(3):211–219. doi: 10.1007/s12033-007-9017-4. [DOI] [PubMed] [Google Scholar]
- 28.Gratia E, Weekers F, Margesin R, D’Amico S, Thonart P, Feller G. Selection of a cold-adapted bacterium for bioremediation of wastewater at low temperatures. Extremophiles. 2009;13(5):763–768. doi: 10.1007/s00792-009-0264-0. [DOI] [PubMed] [Google Scholar]
- 29.Qiu X, Wu P, Zhang H, Li M, Yan Z. Isolation and characterization of Arthrobacter sp. HY2 capable of degrading a high concentration of p-nitrophenol. Bioresource Technology. 2009;100(21):5243–5248. doi: 10.1016/j.biortech.2009.05.056. [DOI] [PubMed] [Google Scholar]
- 30.Wu Y, Liu F, Liu YC, et al. Identification of chitinases Is-chiA and Is-chiB from Isoptericola jiangsuensis CLG and their characterization. Applied Microbiology and Biotechnology. 2011;89(3):705–713. doi: 10.1007/s00253-010-2917-4. [DOI] [PubMed] [Google Scholar]
- 31.Yavankar S, Pardesi K, Chopade B. Species distribution and physiological characterization of Acinetobacter genospecies from healthy human skin of tribal population in India. Indian Journal of Medical Microbiology. 2007;25(4):336–345. doi: 10.4103/0255-0857.37335. [DOI] [PubMed] [Google Scholar]


