Abstract
Endothelin-converting enzyme-1 and -2 (ECE-1 and -2) are membrane-bound metalloproteases that can cleave biologically the inactive endothelin-1 (ET-1) precursor to form active ET-1 in vitro. We previously reported developmental defects in specific subsets of neural crest–derived tissues, including branchial arch–derived craniofacial structures, aortic arch arteries, and the cardiac outflow tract in ECE-1 knockout mice. To examine the role of ECE-2 in cardiovascular development, we have now generated a null mutation in ECE-2 by homologous recombination. ECE-2 null mice develop normally, are healthy into adulthood, are fertile in both sexes, and live a normal life span. However, when they are bred into an ECE-1–null background, defects in cardiac outflow structures become more severe than those in ECE-1 single knockout embryos. In addition, ECE-1–/–; ECE-2–/– double null embryos exhibited abnormal atrioventricular valve formation, a phenotype never seen in ECE-1 single knockout embryos. In the developing mouse heart, ECE-2 mRNA is expressed in the endocardial cushion mesenchyme from embyronic day (E) 12.5, in contrast to the endocardial expression of ECE-1. Levels of mature ET-1 and ET-2 in whole ECE-1–/–; ECE-2–/– embryos at E12.5 do not differ appreciably from those of ECE-1–/– embryos. The significant residual ET-1/ET-2 in the ECE-1–/–; ECE-2–/– embryos indicates that proteases distinct from ECE-1 and ECE-2 can carry out ET-1 activation in vivo.
Introduction
Endothelins (ETs) are a family of small peptides composed of three structurally related isoforms called ET-1, ET-2, and ET-3 (1, 2). ETs act on two subtypes of G protein–coupled heptahelical receptors, the ET A receptor (ETA) and the ET B receptor (ETB) (3, 4). ETA binds ET-1 and ET-2 but not ET-3 at physiological conditions, whereas ETB can bind all three ligands with equal high affinity. Recent gene-inactivation studies of the components of the ET pathway have revealed unexpected roles of these peptides in mammalian development. Mice deficient for ET-1 (5, 6) or ETA (7) show defects in development of subsets of cephalic and cardiac neural crest derivatives, including branchial arch–derived craniofacial tissues, and great vessel and cardiac outflow structures. Mice deficient for ET-3 (8) or ETB (9) show spotted coat color and aganglionic megacolon resulting from the defects in the development of neural crest–derived melanocytes and enteric neurons, respectively.
The biosynthesis of ETs requires two-step proteolytic processing for the final production of biologically active peptides. Approximately 200-residue preproendothelins are first cleaved by a furin-like processing protease(s) into biologically inactive intermediates called big ETs. Big ETs are then proteolytically activated via cleavage at the common Trp21 residue by the highly specific ET-converting enzymes (ECEs). Two isoenzymes of ECE (ECE-1 and ECE-2) have so far been identified (10, 11). Both are type II integral membrane proteins, belonging to a family of membrane-bound metalloproteases. They have a short cytoplasmic tail in the NH2-terminus, followed by a membrane-spanning region and a large extracellular domain containing a zinc-binding motif essential for enzymatic activity. Both ECE-1 and ECE-2 catalyze the conversion of big ET-1 most efficiently among the three big ETs in vitro, with a neutral and acidic optimum pH of 6.8 and 5.6 for ECE-1 and ECE-2, respectively. Recent characterization of ECE-1 deficient mice showed various defects in specific subsets of cephalic, cardiac, vagal, and trunk neural crest derivatives, reproducing the combined phenotype of both ET-1/ETA– and ET-3/ETB–deficient mice (12, 13). This established that ECE-1 was a bona fide converting enzyme for both big ET-1 and big ET-3 in vivo. As expected, tissue ET-3 levels measured in whole-embryo extracts from ECE-1–/– mutants were markedly reduced compared with those of wild-type embryos. However, significant amount of mature ET-1/ET-2 remained in these embryos, indicating that there is another protease(s) responsible for the production of mature ET-1/ET-2.
Cardiac abnormalities observed in ECE-1–/– mice include defects in patterning of the great vessels and formation of the outflow tracts. Common great vessel malformations include interruption of the aortic arch between the left common carotid artery and left subclavian artery, as well as absence of the right subclavian artery. Among outflow tract abnormalities, a perimembranous interventricular septal defect (VSD) is observed in all of the ECE-1–/– embryos. In addition, an overriding aorta is commonly seen, followed by double outlet right ventricle (DORV). Persistent truncus arteriosus (PTA) is seen only in a very small number of cases.
In the present study, we examined the possibility that big ET-1 is an in vivo substrate for ECE-2 and studied the possible role of ECE-2 in embryonic development. ECE-2 mRNA was most abundantly expressed in the adult brain followed by adrenal glands, ovary, and uterus. In the developing heart, ECE-2 mRNA was exclusively expressed in the mesenchyme of endocardial cushions. We found that ECE-2–/– knockout mice were healthy into adulthood and fertile, showing no detectable defects in embryonic development. However, when the null ECE-2 alleles were introduced into an ECE-1 null background, the cardiac defects became more severe than those of seen in ECE-1–/– single mutants. This indicates that there is a functional redundancy between ECE-1 and ECE-2 in embryonic development.
Methods
Mice.
All animal experiment procedures were reviewed and approved by the Institutional Animal Care and Research Advisory Committee of the University of Texas Southwestern Medical Center. Mice deficient for ECE-1 were generated by homologous recombination as described previously (12), maintained on a C57BL6/J-129/SvEv hybrid genetic background, and housed in a conventional animal colony with a 14-hour–light/10-hour–dark cycle.
Gene targeting.
To isolate the mouse ECE-2 genomic fragment, we screened a λFIX II mouse 129/Sv genomic DNA library (Stratagene Inc., La Jolla, California, USA) with a 110-bp ECE-2 cDNA probe described previously (11). Two phage clones encompassing a 14-kb DNA fragment that contains the catalytic domain of ECE-2 were isolated. To construct the targeting vector, we used a universal neo-TK template plasmid vector described previously (9). A 3′ homologous 2.0-kb BamHI-SacI fragment and a 5′ homologous 6.0-kb SalI fragment were cloned into the unique BamHI and XhoI sites of the targeting vector, respectively. The exons encoding the zinc-binding catalytic domain of ECE-2 were replaced with a neor cassette (see Figure 3a). SM-1 ES cells derived from a 129/SvEv mouse strain were cultured on an irradiated LIF-producing STO feeder layer, transfected with linearized targeting vector, and doubly selected in G418 and FIAU as described previously (9). Surviving clones were individually picked, and homologous recombination was confirmed by PCR with primers in the 3′ portion of the neor gene and ECE-2 genomic region external to the short arm (see Figure 3a and data not shown). Recombinant embryonic stem (ES) cell clones were injected into the blastocysts obtained from C57BL6/J females, and resulting chimeric males were mated with C57BL6/J females to obtain F1 mice carrying the targeted allele.
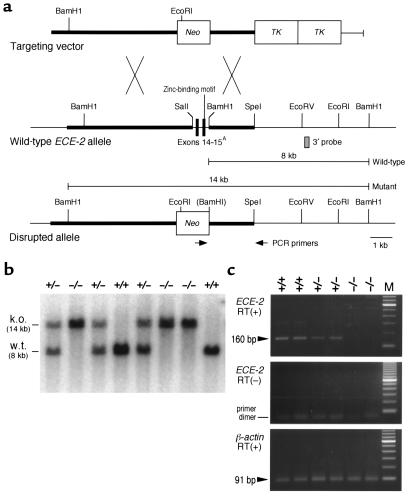
Figure 3.
Generation of ECE-2–/– mice by homologous recombination. (a) Targeting strategy. The exon encoding the zinc-binding domain of ECE-2 is replaced with a neor cassette driven by the RNA polymerase II promoter. Two tandem repeats of thymidine kinase (TK) are used for the negative selection with FIAU (see Methods). PCR primers in the neor gene and 3′ region external to the short arm of targeting vector are shown by arrows. AExon numbers are indicated according to the equivalent exons of the human ECE-1 gene (32); other ECE-2 exons are not shown. (b) Southern blot analysis of tail DNA from the offspring of ECE-2+/– intercrosses. DNA was digested with BamHI and hybridized with a 3′ probe (indicated by the gray box in panel a). k.o., targeted allele; w.t., wild-type allele. (c) RT-PCR of the ECE-2 transcript using total RNA extracted from the brain of ECE-2+/+, ECE-2+/–, and ECE-2–/– mice. Primers amplify the exon encoding the essential zinc-binding domain of ECE-2. The upper panel shows that a 160-bp transcript fragment is detected in wild-type and ECE-2+/– mice, but is absent in ECE-2–/– mice. The middle panel shows that no transcripts are detected without RT incubation. The lower panel shows detection of β-actin transcripts, indicating intact RNA preparations in all lanes.
Genotyping.
Genomic DNA was prepared from tail biopsies at postnatal day 21 or from embryo yolk sac and were subjected to PCR and/or Southern blot analysis. Oligonucleotide primers used for the detection of the wild-type ECE-2 allele were as follows: upstream, 5′-GCCATCTTACAGTAGAGGAG-3′ and downstream, 5′-CTAGAATGGGCCCCTACCTT-3′. Primers used for the detection of the mutated allele were: upstream, 5′-GGATGCGGTGGGCTCTATGGCTTCTGA-3′ and downstream, 5′-ACCATGCAGACCCAACTATGCTTCT-3′. The reaction was cycled 30 times (30 seconds at 94°C, 1 minute at 60°C, and 3 minutes at 72°C), which amplified an approximately 800-bp fragment of the portion of ECE-2 gene disrupted in the mutant allele (for detection of wild-type allele), and a 2.3-kb fragment including the neor gene and the short arm of the targeting vector (for the detection of mutant allele) (see Figure 3a). For Southern blot analysis, 10 μg of genomic DNA was digested with BamHI and probed with the 3′ external probe or was digested with EcoRI and hybridized with a neor probe (see Figure 3a).
Northern blot analysis.
RNA was extracted from adult mouse tissues using RNA STAT-60 (TEL-TEST “B” Inc., Friendswood, Texas, USA) as instructed by the manufacturer. Total RNA (15 μg) was denatured in 1 M deionized glyoxal/50% dimethyl sulfoxide/10 mM phosphate buffer (pH 7.0) and separated in a 1.2% agarose gel. RNA-transferred membranes were prehybridized and hybridized as described previously (14). For probes, partial cDNAs for mouse ECE-1 and ECE-2 were obtained by screening a mouse brain cDNA library using a 110-bp RT-PCR product and a 92-bp RT-PCR product as a probe, respectively (11). A 1.1-kb EcoRI fragment containing the 5′ coding region of ECE-1 cDNA and a 1.2-kb EcoRI-PstI fragment containing the 5′ coding region of ECE-2 cDNA were random primed 32P-labeled and used as probes.
RT-PCR analysis.
Total RNA was extracted from the adult brain of ECE-2+/+, ECE-2+/–, and ECE-2–/– mice and from the heart of embryonic day (E) 12.5 and E13.5 embryos. First-strand cDNA was synthesized from 1 μg of total RNA using SuperScript II RT (Life Technologies Inc., Grand Island, New York, USA) with oligo(dT) primers, according to manufacturer’s recommendations. For DNase I treatment of the samples, each RNA was treated with 10 U RNase-free DNase I for 15 minutes at room temperature before the first-strand synthesis. PCR was then performed on cDNA samples with specific primers 5′-CCCGTGAACGCTTACTACCTT-3′ and 5′-GGTCATCAAAGGCATGTGTCA-3′, which amplified a 160-bp of ECE-2 cDNA and a 280-bp of ECE-2 genomic DNA. Thirty cycles (30 seconds at 94°C, 1 minute at 60°C, and 3 minutes at 72°C) were used for the RT-PCR of ECE-2 cDNA. As an internal control, primers 5′-GGATCCGGTCGTACCACAGGCATTGTGATG-3′ and 5′-GAATTCGGAGAGCATAGCCCTCGTAGATGG-3′ were used to amplify a 91-bp fragment of the mouse β-actin cDNA with the same condition as just described.
Generation of embryos double homozygous for ECE-1 and ECE-2 mutated alleles.
Homozygous ECE-2–/– males on a C57BL6/J-129/SvEv hybrid background were mated to ECE-1+/– heterozygous females. Resulting double heterozygous females were mated again to ECE-2–/– homozygous males to produce ECE-1+/–; ECE-2–/– mice. These mice were then intercrossed to obtain embryos that were ECE-2–/– and wild-type, heterozygous or homozygous for the disrupted ECE-1 allele. The morning that the vaginal plug was found was counted as E0.5, and pregnant females were sacrificed by inhalation of CO2 on the designated day of pregnancy. ECE-1 heterozygous crosses were also performed to obtain embryos that were wild type, ECE-1+/–, or ECE-1–/–. ECE-1 heterozygous crosses were also performed to obtain embryos that were wildtype, ECE-1+/–, or ECE-1–/–. Wildtype littermates were used as controls throughout the present study.
Determination of tissue ET peptides.
E12.5 whole embryos were homogenized immediately after dissection by Polytron (Brinkmann, Littau, Switzerland) (20,000 rpm) in 20 × volume of 1 M acetic acid containing 0.01 mM pepstatin A for 30 seconds, and the homogenates were immediately placed in a boiling water bath for 10 minutes. After centrifugation at 15,000 g, supernatant was concentrated with Sep-pak C18 columns (Varian, Harbor City, California, USA) as described previously (12). Immunoreactive mature ET-1/ET-2 level was determined by a sandwich-type enzyme immunoassay (EIA) (15) with synthetic human ET-1 as standard in triplicate wells. This mature ET-1 EIA fully cross-reacts with mouse ET-1 and ET-2, but not detectably with ET-3 or any of the big ETs.
Histological analysis.
For histological analyses of the heart, embryos were fixed in 10% neutral buffered formalin and embedded in paraffin. Continuous 4-μm serial sections of whole embryos were made in transverse and sagittal directions and stained with hematoxylin and eosin.
Sectional in situ hybridization.
Sectional in situ hybridizations for ECE-2 mRNA was performed as described previously (7, 13). To generate riboprobes, a 448-bp XbaI-XhoI fragment of the mouse ECE-2 cDNA was transcribed in both sense and antisense directions and used in parallel.
Results
Tissue distribution of ECE-2 mRNA in adult mice.
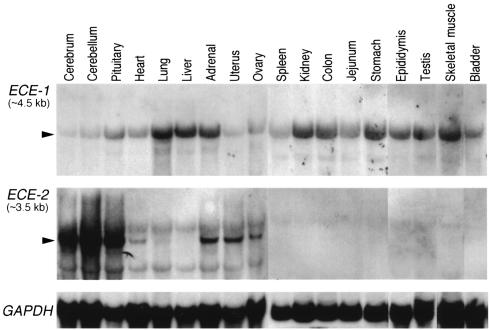
We examined the expression of ECE-2 mRNA in various adult mouse tissues by Northern blot analysis and compared it with ECE-1 mRNA expression (Figure 1). ECE-2 mRNA was expressed predominantly in the central nervous system, including cerebrum, cerebellum, and pituitary, followed by adrenal glands, ovary, and uterus. Notably, ECE-2 expression was also observed in small amounts in the heart. ECE-1 mRNA was strongly expressed in various tissues, including lung, liver, adrenal glands, kidney, digestive tracts, epididymis, testis, and skeletal muscles. Although the expression of ECE-1 mRNA was observed in almost all tissues examined, ECE-2 expression showed a highly restricted pattern of distribution. In the adult brain where abundant message was detected by Northern analysis, ECE-2 mRNA was expressed in the heterogeneous populations of neurons in large areas encompassing thalamus, hypothalamus, amygdala, dentate gyrus, and CA3 (data not shown).
Figure 1.
Northern blot analysis of ECE-1 mRNA (upper panel) and ECE-2 mRNA (middle panel) in adult mouse tissues. Rehybridization with GAPDH probe is shown as an internal control for the amounts of RNA loaded (lower panel). Arrowheads show specific signals for ECE-1 and ECE-2 mRNA.
Expression of ECE-2 mRNA in the embryonic brain and the endocardial cushion.
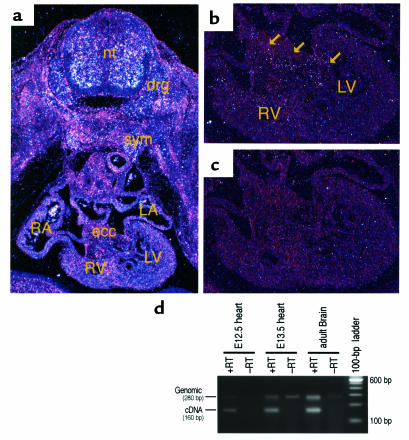
In E10.5 embryos, weak and diffuse expression of ECE-2 mRNA was detected in the mesenchyme of embryos and parts of neural tube (data not shown). The overall expression of ECE-2 mRNA remained weak until E12.5, except that layers of undifferentiated motor neurons were positive for ECE-2 expression (data not shown). At E13.5, clustered signals were detected in the anterior part of the neural tube, dorsal root ganglia, and bilateral sympathetic trunk (Figure 2a). In the developing heart, ECE-2 mRNA was detected in the endocardial cushion (Figure 2b). The endocardium and myocardium proper of the heart were negative for the expression of ECE-2 mRNA. In addition, peribronchial mesenchyme and limb mesenchyme expressed low levels of ECE-2 mRNA (data not shown). RT-PCR analysis confirmed that the ECE-2 transcript was detectable in E12.5 and E13.5 mouse embryonic heart (Figure 2d).
Figure 2.
Detection of ECE-2 transcripts in wild-type mouse embryos by in situ hybridization analysis (a, b, and c) and RT-PCR (d). (a) Transverse section of E13.5 wild-type mouse embryos showing strong expression of ECE-2 in the anterior part of the neural tube (nt), dorsal root ganglia (drg), and bilateral sympathetic trunk (sym). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. (b) Higher magnification of a, showing the heart. Note that ECE-2 expression (arrows) is detected in the mesenchyme of the endocardial cushion (ecc). (c) The sense probe did not show any positive signal. (d) ECE-2 expression is detectable by RT-PCR in E12.5 and E13.5 mouse hearts.
Generation of ECE-2 null mice.
ECE-2 is a membrane-bound metalloprotease and shares 59% amino acid identity with ECE-1 (11). A zinc-binding catalytic domain in the COOH-terminus is essential for the enzymatic activity for both ECE-1 and ECE-2. It is shown in ECE-1 null embryos that the disruption of this zinc-binding domain can completely abolish the enzymatic activity (12). To inactivate the mouse ECE-2 gene in vivo, the exon encoding the analogous zinc-binding motif was replaced with a neor cassette by homologous recombination in ES cells (Figure 3a). Eight recombinant ES cell clones were established, and three clones were injected into blastocysts obtained from C57BL6/J females. Three chimeric males from two independent ES cell clones gave germ line transmission. Heterozygous F1 mice were healthy and fertile and were intercrossed to obtain homozygous ECE-2–/– mice (Figure 3b). Genotyping of postnatal day 21 mice showed a mendelian inheritance, suggesting that there was no embryonic or neonatal lethality in ECE-2–/– mice. Both female and male ECE-2–/– mice were fertile and healthy and indistinguishable from wild-type littermates in their growth rates. Life span of ECE-2–/– mice was indistinguishable from that of wild-type and ECE-2+/– mice. There were no gross neurological or behavioral abnormalities that suggested any defects in the central nervous system. Histological examinations of various tissues including brain, pituitary glands, spinal cord, heart, lung, liver, gall bladder, stomach, duodenum, jejunum, ileum, colon, pancreas, spleen, kidney, adrenal glands, ovary, uterus, testis, bladder, aorta, skeletal muscle, and adipose tissue did not reveal any abnormalities in ECE-2–/– mice. RT-PCR was performed using total RNA extracted from the brain of adult ECE-2+/+, ECE-2+/–, and ECE-2–/– mice. A 160-bp band corresponding to reverse transcribed ECE-2 cDNA was detected in wild-type and ECE-2+/– mice but not in ECE-2–/– mice (Figure 3c). Without RT, no signal was detected in all lanes. In contrast, 91 bp of β-actin cDNA fragment was detected in every genotype (Figure 3c), and it was not detected without the RT reaction (data not shown). These data confirmed that both of ECE-2 alleles were disrupted in ECE-2–/– mice.
Generation of ECE-1–/–; ECE-2–/– embryos.
To examine the possible functional redundancy between ECE-1 and ECE-2, we introduced the ECE-2–/– mutation into the ECE-1-null background. ECE-1+/–; ECE-2–/– mice were generated by first crossing ECE-1+/– mice to ECE-2–/– mice. The resultant ECE-1+/–; ECE-2–/– mice were healthy and fertile and were indistinguishable from ECE-1+/– mice. These mice were then intercrossed to obtain ECE-1–/–; ECE-2–/– embryos. Because ECE-1–/– embryos showed midgestational embryonic lethality with incomplete penetrance (12), we first examined whether the null mutation of ECE-2 affected the embryonic lethality in ECE-1–/– embryos. ECE-1+/–; ECE-2–/– mice were intercrossed, and pregnant females were sacrificed by cesarean section between E16.0 and E20.0. Genotyping of resulting embryos revealed that alive ECE-1–/–; ECE-2–/– embryos consisted of only eight (4.9%) of 162 embryos examined. The ratio of double homozygous embryos that survived until birth tended to be less than that of single ECE-1–/– embryos, 16 (7.1%) of 225 embryos, although there was no statistically significant difference between the two. Gross examinations of near-term or E20.0 ECE-1–/–; ECE-2–/– embryos revealed hypoplastic pinnae and mandible, and shrunken anterior neck, which was identical to ECE-1–/– single knockout embryos (12). There were no additional gross abnormalities that were unique to ECE-1–/–; ECE-2–/– embryos.
ET peptide levels in E12.5 double homozygous embryos.
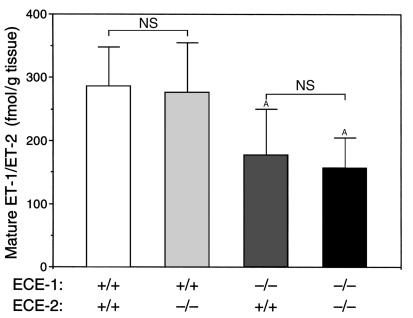
To examine the role of ECE-2 in the proteolytic cleavage of big ET-1 in vivo, mature ET-1 levels were measured by EIA in E12.5 whole-embryo extracts from wild-type, homozygous ECE-1–/–, homozygous ECE-2–/–, and double homozygous ECE-1–/–; ECE-2–/– embryos. Because the number of alive ECE-1–/– and ECE-1–/–; ECE-2–/– embryos decrease after E12.5 owing to embryonic lethality, we collected embryos at E12.5, when the largest numbers of live embryos were available and when ECE-2 is expressed abundantly. Combined mature ET-1/ET-2 levels in wild-type and ECE-2–/– embryos did not differ, whereas ECE-1–/– single mutant embryos showed significantly decreased (but well detectable) levels of mature ET-1/ET-2 compared with those of wild-type and ECE-2–/– embryos. In double homozygous ECE-1–/–; ECE-2–/– embryos, the combined ET-1/ET-2 level did not appreciably differ from that of single homozygous ECE-1–/– mutants (Figure 4). These findings indicated that an absence of ECE-2 did not significantly affect the production of mature ET-1/ET-2 in vivo at E12.5, as judged at the level of whole embryos.
Figure 4.
Tissue ET levels in E12.5 embryos. Combined level of mature ET-1/ET-2 is measured in wild-type (n = 8), ECE-1–/– (n = 5), ECE-2–/– (n = 7), and ECE-1–/–; ECE-2–/– (n = 7) embryos. Values are means ± SD. Statistical analysis by unpaired Student’s t test. AP < 0.05 vs. wild-type. NS, not significant.
Exacerbated cardiac abnormalities in ECE-1–/–; ECE-2–/– embryos.
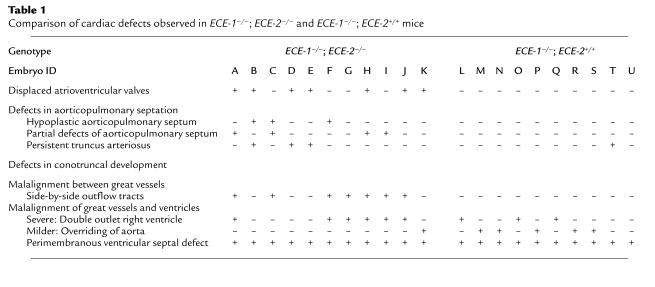
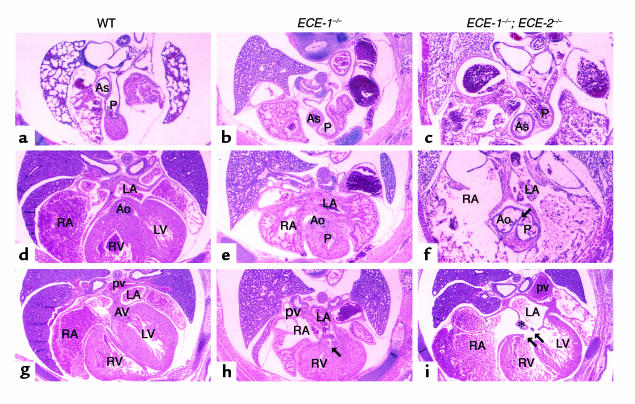
Histological examinations of E16.0-E20.0 double homozygous ECE-1–/–; ECE-2–/– embryos showed defects in multiple neural crest-derived tissues, which were identical to those observed in ECE-1–/– single mutant embryos (12). In addition, ECE-1–/–; ECE-2–/– embryos developed cardiac abnormalities that were more severe than those of ECE-1–/– single mutant embryos (Table 1). In transverse sections of wild-type embryos, the pulmonary trunk (P in Figure 5a) was located more rostral and anterior to the aortic outflow (Ao in Figure 5d; two outflow tracts did not appear in the same transverse plane; also see Figure 7a). In addition, the ascending aorta is seen to the right and posterior to the pulmonary trunk owing to crossing of the two outflow tracts (Figure 5a). In single ECE-1–/– knockout mice, aortic outflow was shifted rostrally and to the right so that pulmonary trunk and ascending aorta appeared in the same plane (Figure 5e; also see Figure 7b). Distance between ascending aorta and pulmonary trunk was shortened compared to wild-type embryos (Figure 5b). However, the spiraling of the aorticopulmonary septum occurred normally so that relative alignment between the two outflow tracts appeared normal. In ECE-1–/–; ECE-2–/– embryos, alignment of the two distinct outflow tracts was more severely impaired. Aortic outflow was located anterior to and right of the pulmonary trunk, and two valves were seen side by side on the transverse section (Figure 5f; also see Figure 7c). The ascending aorta shifted further anterior to the pulmonary trunk as compared with ECE-1–/– embryos (Figure 5c). In addition, the aortic outflow frequently originated from the right ventricle without crossing over the pulmonary outflow (DORV, 6/11). This suggested that the defect was due to the malalignment of the great vessels and ventricles, as well as to malalignment between the great vessels resulting from impaired spiraling of aorticopulmonary septum. At the level of the interventricular septum, a well-developed membranous and muscular septum was seen in wild-type embryos (Figures 5g and 6a). VSD was observed in all the cases of single ECE-1–/– embryos (Figure 5h, arrow; Figure 6b arrowhead) and ECE-1–/–; ECE-2–/– double knockout embryos (Figure 5i, double arrow; Figure 6, c, e, and f, arrowheads). The area of septal defects was larger in ECE-1–/–; ECE-2–/– embryos than that of ECE-1–/– embryos (compare Figure 5h with i).
Table 1.
Comparison of cardiac defects observed in ECE-1–/–; ECE-2–/– and ECE-1–/–; ECE-2+/+ mice
Figure 5.
Transverse sections of the heart of term wild-type (a, d, and g), ECE-1–/– (b, e, and h), and ECE-1–/–; ECE-2–/– (c, f, and i) embryos stained with hematoxylin and eosin. Original magnification is ×10. (a, d, and g) At the level of the pulmonary trunk (P), the ascending aorta (As) is seen as posterior and right to the pulmonary trunk in wild-type embryos. The aortic outflow (Ao) comes from the left ventricle (LV) (d), and the left AV valve opens to the left ventricle (g). (b, e, and h) In ECE-1–/– embryos, relative distance between the ascending aorta and the pulmonary trunk shortens, and the ascending aorta is seen anteriorly (b). Owing to the malalignment of outflow tracts, aortic and pulmonary tracts are seen in the same plane, originating from the right ventricle (RV) (DORV). Two great vessels do cross over (e). A small VSD is indicated by an arrow (h). (c, f, and i) In double homozygous embryos, the ascending aorta is seen further anteriorly, with the pulmonary trunk seen posteriorly to ascending aorta (c). Aortic and pulmonary outflows both originate from the right ventricle (DORV), and the two great vessels do not cross over. Note hypoplasia of the muscular wall of the great vessels (arrow) (f). (i) In a severe case, formation of the endocardial cushion (asterisk) is markedly impaired and the AV valve is not formed. A large VSD is indicated by double arrows. AV, arterioventricular valve; pv, pulmonary vein.
Figure 7.
Schematic representation of the alignment between aortic (A) and pulmonary (P) outflows in a frontal section (upper panel), alignment of A and P valves in a transverse section (middle panel), and alignment between A and P vessels and right (R) and left (L) ventricles (lower panel). (a) In wild-type embryos, aortic and pulmonary outflow tracts spiral around each other. The pulmonary valve locates rostrally (above the plane of the drawing; indicated by dotted lines) and anteriorly to the aortic valve so that two valves do not appear in a same plane. (b) In ECE-1–/– mutant embryos, the alignment between A and P outflows remains in a spiral position, whereas the aortic valve and outflow shift rostrally and to the right so that two valves appear in the same plane. The aorta often overrides the interventricular septum. (c) In ECE-1–/–; ECE-2–/– double mutant embryos, the aortic and pulmonary outflows run side by side in a parallel position, often showing double outlet right ventricle (DORV).
Figure 6.
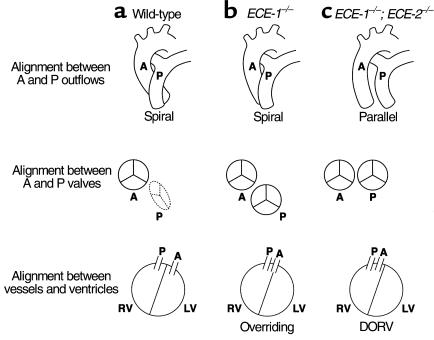
Parasagittal sections of the heart of term wild-type (a), ECE-1–/– (b), and ECE-1–/–; ECE-2–/– (c and d) embryos, and frontal sections of ECE-1–/–; ECE-2–/– embryos (e and f) stained with hematoxylin and eosin. Original magnification is ×11. (a) The interventricular septum (ivs) and aortic outflow (Ao) are seen. (b) Overriding of aorta with a small VSD (arrowhead) is seen in an ECE-1–/– embryo. (c) Persistent truncus arteriosus in an ECE-1–/–; ECE-2–/– embryo. One outflow (arrow) is separated into aortic outflow (Ao) and pulmonary (P) outflows. A large VSD (arrowhead) is shown. Displacement of the left AV valve (asterisk) is seen, opening toward the VSD. (d) Aorticopulmonary window in an ECE-1–/–; ECE-2–/– embryo. Septation of the aortic and pulmonary outflow tracts is incomplete at the base of the outflow tract (arrow). Note that the pulmonary outflow becomes the dorsal aorta (da) via the ductus arteriosus (dc). Pa, pulmonary artery. (e) Pulmonary and aortic outflow tracts originate from the right ventricle (DORV) in an ECE-1–/–; ECE-2–/– embryo. A large conotruncal ridge defect is seen underneath the aortic and pulmonary valves (filled diamond). A large VSD (arrowhead) is also shown. Note that the muscular layers of the pulmonary trunk (P) and aorta (Ao) are hypoplastic, and thin mesenchyme is observed between the two vessels (arrow). (f) DORV in an ECE-1–/–; ECE-2–/– embryo. The conotruncal ridge (filled diamond) is well developed compared with that in e. The arrowhead indicates a VSD.
ECE-1–/–; ECE-2–/– embryos also showed various defects in aorticopulmonary septation (Table 1). The severe form of the aorticopulmonary septum defect, PTA (Figure 6c, arrow), as well as more localized septal defects were observed in ECE-1–/–; ECE-2–/– embryos. These defects appeared as an aorticopulmonary window that showed a communication between aortic outflow and pulmonary outflow just above the semilunar valves (Figure 6d, arrow), or an abnormal opening between the aorta and pulmonary trunk distal to the branching of the pulmonary arteries (data not shown). In both cases, aortic outflow and pulmonary outflow arose from the right ventricle and maintained parallel positions without crossing over. In addition, formation of the distinct demarcation between the opposing surface of the pulmonary trunk and ascending aorta by the muscular layers of the vessels was impaired in some of ECE-1–/–; ECE-2–/– embryos (Figures 5f and 6e, arrow). There were at least 5–7 layers of smooth muscle cells in the septum of wild-type embryos and single ECE-1–/– embryos (data not shown), whereas only 2–3 layers were observed in ECE-1–/–; ECE-2–/– embryos. This resulted in the formation of thinner mesenchymal demarcation between the pulmonary trunk and ascending aorta.
In ECE-1–/–; ECE-2–/– embryos, abnormal formation of the atrioventricular (AV) valves was also observed. AV valves opened toward a ventricular septal defect (Figure 6c, asterisk) but not to the left ventricle. In severe cases, the endocardial cushion was hypoplastic and did not form AV valves at all (Figure 5i, asterisk). In addition, various cardiac abnormalities including atrial septal defect (data not shown) and a large conotruncal ridge defect (Figure 6e, diamond) were seen in some embryos. Collectively, these defects in double homozygous embryos were significantly more severe than those of ECE-1–/– single knockout embryos.
Discussion
Role of ECE-2 in ET-mediated signal transduction pathways.
We have generated a null mutation in the ECE-2 gene in mice and found that there is no detectable developmental defects in these mice. This indicates that ECE-2 is a redundant protease for the activation of big ETs in vivo. However, the phenotype of ECE-1 null embryos worsens on an ECE-2 null background. This genetic interaction of the ECE-1 and ECE-2 genes strongly suggests that ECE-2 also participates in the formation of active ET-1 in vivo, indicating a functional redundancy between ECE-1 and ECE-2. That the ET-1/ET-2 levels in the whole ECE-1–/–; ECE-2–/– embryos do not appreciably differ from those of ECE-1–/– embryos suggests that proteolytic activation of big ET-1 by ECE-2 occurs only locally. Thus, the amount of mature ET-1 cleaved by ECE-2 may not be reflected in the measurements using a whole-embryo extracts. The significant amount of remaining ET-1/ET-2 in ECE-1–/–; ECE-2–/– embryos demonstrates that there is another protease(s), distinct from ECE-1 or ECE-2, that is responsible for the production of mature ET-1 and/or ET-2 in vivo.
Redundant functions of ECE-1 and ECE-2 in the formation of the cardiac outflow structures.
Double homozygous ECE-1–/–; ECE-2–/– embryos show broader and more severe cardiac abnormalities than ECE-1–/– embryos (Table 1 and Figure 7). Defects in aorticopulmonary septation were frequently seen in ECE-1–/–; ECE-2–/– embryos. These abnormalities include total or localized defects of the aorticopulmonary septum, which resulted in PTA or aorticopulmonary window. Defects in spiraling of the conotruncal ridges and aorticopulmonary septum are also a distinct feature of double knockout embryos. These defects resulted in a parallel position of the aortic and pulmonary outflow tracts without crossing over each other, with the two valves seen side by side on the same transverse plane. In addition, abnormal AV valve formation (which is never seen in ECE-1–/– single knockout embryos) was frequently observed in these mutants, indicating that defects extend to the endocardial cushion of the AV canal.
Completion of outflow tract development requires formation of the conotruncal septum which demarcates the right (pulmonary) and left (aortic) ventricular outflow tracts below the semilunar valves, and the formation of the aorticopulmonary septum that separates the two outflow tracts above the valves. Interaction of diverse cell types including the endocardium, myocardium, and mesenchyme of the endocardial cushions, as well as extracardiac cells such as postmigratory neural crest cells, are essential for the morphogenesis of the outflow tracts (16–18). The present studies, along with our recent gene-targeting experiments in the mouse ECE-1 gene, demonstrate that the neural crest is indeed an essential component of cardiac outflow formation and that signals mediated by the ET-1/ECE-1/ETA and ET-1/ECE-2/ETA pathways play critical roles in the development of the outflow tracts of the heart. The cellular mechanism of ET-1/ETA-mediated signaling in outflow tract formation may involve inductive signals mediated by ET-1, originating from the endocardium of the outflow tract, through ETA receptors that are expressed in the neural crest–derived mesenchyme of the outflow tract (13). Interestingly, the endocardium of the outflow tract is thought to have a different origin from cardiogenic mesoderm (19). Endocardium of the outflow tract is derived from embryonic angioblasts that come from head mesoderm, moving through the developing tongue and proximal lower jaw. It is fascinating to hypothesize that an ET-1/ETA-mediated paracrine signaling mechanism functions between the endothelium and mesenchyme of the outflow tracts, both of which are of extracardiac origin.
The role of ECE-2 in cardiac development is further substantiated by its unique localization in the developing heart. Unlike the other ET-related molecules (i.e., ET-1, ECE-1, ETA, and ETB), ECE-2 mRNA is largely absent in the heart until around E12.5, and it seems exclusively expressed in the mesenchyme of endocardial cushions. ET-1 mRNA is predominantly expressed in the endocardium of the outflow tract and the endothelium of branchial arch arteries (13). ETA mRNA is expressed in migratory and postmigratory neural crest cells, i.e., mesenchyme of the branchial arch arteries and outflow tracts, in addition to the myocardium of the heart (7, 12). Expression of ECE-1 mRNA is found in the endothelium and mesenchyme of the branchial arch arteries and in the endocardium overlaying the outflow tracts, atrium, ventricle, and endocardial cushion. Importantly, however, ECE-1 mRNA is not expressed in the mesenchyme of endocardial cushions. ECE-2 may contribute to the local production of ET-1 in endocardial cushions. Thus, when ECE-1 is present, secreted big ET-1 can be converted to mature ET-1 on the surface of ECE-1–expressing cells and mask the phenotype that would result from the absence of ECE-2. Conversely, in the single homozygous ECE-1–/– embryos, the deficits in the generation of mature ET-1 may be partially compensated by ECE-2 in the endocardial cushion mesenchyme. Stochastic variation in the details of phenotypes seen in ECE-1–/–; ECE-2–/– embryos may in part be the result of fluctuation in the amount of mature ET-1 available in the cardiac tissues. The production of ET-1 may be affected by the local production as well as local conversion of maternally derived big ETs by ECE-1 and/or ECE-2 in the developing heart (12).
The role of ECE-2 in the formation of the AV valve is also worth noting. Displacement of the AV valve is seen exclusively in ECE-1–/–; ECE-2–/– double mutants. AV valves are known to be derived from mesenchymal outgrowth of cardiac cushions that reside between the endocardium and myocardium of the primitive heart. Early endocardial cells migrate into cardiac cushions and transform themselves into mesenchyme in the presence of myocardium. Growth factors such as TGF-β2 and BMP-4, as well as extracellular matrix components, are thought to be involved in this process (16). In addition, mice deficient for neuregulins and their receptors show defects in the formation of endocardial cushions, indicating that signaling mediated by neuregulins also play a role in the development of the endocardial cushions (20, 21). Because AV valve abnormalities are rarely seen in neural crest–ablated chick embryos (17), it is likely that ECE-2 is involved in endocardial cushion development that is independent of neural crest cells. It is also possible that ECE-2 functions in endocardial cushions by a cleaving nonET peptide(s) essential for valve formation.
Genes involved in the formation of cardiac outflow structures.
There are a number of genes documented to be involved in the formation of cardiac outflow structures. Mutations in the Pax-3 (Splotch) gene (22), neurofibromatosis type-1 (NF-1) gene (23), Sox-4 gene (24), activin type IIB receptor gene (25), neurotrophin-3 gene (26), TGF-β2 gene (27), Hoxa-3 gene (28), connexin-43 gene (29), and double homozygous mutants for subtypes of RAR genes (30) are all known to cause defects in outflow structures by possibly affecting neural crest development. In particular, cardiac phenotypes seen in TGF-β2 null embryos (27) are similar to those observed in ECE-1–/–; ECE-2–/– homozygous embryos. Because TGF-βs are known potent stimulators of ET-1 production in endothelial cells (1), it is possible that the cardiac defects observed in TGF-β2 null embryos are the consequence of a downregulation of ET-1 mRNA in these mutants. RXRα mutant mice also show defects in aorticopulmonary septation and conotruncal ridges, as well as other defects in AV cushions and the ventricular myocardium. However, the cellular mechanism of these defects seems not to be related to cardiac neural crest development, suggesting that there are several distinct genetic pathways that are involved in the formation of the cardiac outflow structures (31).
In summary, analyses of ECE-1–/–; ECE-2–/– double homozygous embryos demonstrate that ECE-2 contributes to the production of mature ET-1 probably in the endocardial cushion mesenchyme and that loss of ECE-2 in the absence of ECE-1 can cause more severe defects in cardiac outflow structures including the aorticopulmonary septum and the conotruncal ridges, as well as defects in endocardial cushion–derived structures. This strongly indicates that big ET-1 is one of the bona fide substrates for ECE-2 in vivo. Double homozygous mutant embryos can serve as a suitable animal model to analyze the human congenital cardiovascular malformations.
Acknowledgments
We thank E. Hinnant for technical assistance, and H. Yamagishi, A. Arai, and C. Gariepy for helpful discussions. N. Emoto and D.E. Clouthier are former Associates, and M. Yanagisawa is an Investigator, of the Howard Hughes Medical Institute. This work is supported in part by research funds from the Perot Family Foundation, the W.M. Keck Foundation, and the Excellence in Education Fund.
Footnotes
Shin-ichi Takeda’s present address is: First Department of Surgery, Faculty of Medicine, Osaka University, Osaka, Japan.
Noriaki Emoto’s present address is: International Center for Medical Research, Kobe University School of Medicine, Kobe, Japan.
References
- 1.Yanagisawa M, et al. A novel vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411–415. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- 2.Inoue A, et al. The human endothelin family: three structurally and pharmacologically distinct isopeptides predicted by three separate genes. Proc Natl Acad Sci USA. 1989;86:2863–2867. doi: 10.1073/pnas.86.8.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai H, Hori S, Aramori I, Ohkubo H, Nakanishi S. Cloning and expression of a cDNA encoding an endothelin receptor. Nature. 1990;348:730–732. doi: 10.1038/348730a0. [DOI] [PubMed] [Google Scholar]
- 4.Sakurai T, et al. Cloning of a cDNA encoding a non-isopeptide–selective subtype of the endothelin receptor. Nature. 1990;348:732–735. doi: 10.1038/348732a0. [DOI] [PubMed] [Google Scholar]
- 5.Kurihara Y, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- 6.Kurihara Y, et al. Aortic arch malformations and ventricular septal defect in mice deficient in endothelin-1. J Clin Invest. 1995;96:293–300. doi: 10.1172/JCI118033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clouthier DE, et al. Cranial and cardiac neural crest defects in endothelin-A receptor–deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- 8.Baynash AG, et al. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 9.Hosoda K, et al. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 10.Xu D, et al. ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell. 1994;78:473–485. doi: 10.1016/0092-8674(94)90425-1. [DOI] [PubMed] [Google Scholar]
- 11.Emoto N, Yanagisawa M. Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH optimum. J Biol Chem. 1995;270:15262–15268. doi: 10.1074/jbc.270.25.15262. [DOI] [PubMed] [Google Scholar]
- 12.Yanagisawa H, et al. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- 13.Yanagisawa H, et al. Role of endothelin-1/endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J Clin Invest. 1998;102:22–33. doi: 10.1172/JCI2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taurog JD, Maika SD, Simmons WA, Breban M, Hammer RE. Susceptibility to inflammatory disease in HLA-B27 transgenic rat lines correlates with the level of B27 expression. J Immunol. 1993;150:4168–4178. [PubMed] [Google Scholar]
- 15.Suzuki N, et al. A sandwich-type enzyme immunoassay to detect immunoreactive big-endothelin-1 in plasma. J Immunol Methods. 1990;127:165–170. doi: 10.1016/0022-1759(90)90065-4. [DOI] [PubMed] [Google Scholar]
- 16.Eisenberg LM, Markwald RR. Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ Res. 1995;77:1–6. doi: 10.1161/01.res.77.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 18.Noden DM, Poelmann RE, Gittenberger-de Groot AC. Cell origins and tissue boundaries during outflow tract development. Trends Cardiovasc Med. 1995;5:69–75. doi: 10.1016/S1050-1738(99)80002-4. [DOI] [PubMed] [Google Scholar]
- 19.Noden DM. Origins and patterning of avian outflow tract endothelium. Development. 1991;111:867–876. doi: 10.1242/dev.111.4.867. [DOI] [PubMed] [Google Scholar]
- 20.Gassmann M, et al. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee K-F, et al. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 22.Franz T. The Splotch (Sp1H) and Splotch-delayed (Spd) alleles: differential phenotypic effects on neural crest and limb musculature. Anat Embryol (Berl) 1993;187:371–377. doi: 10.1007/BF00185895. [DOI] [PubMed] [Google Scholar]
- 23.Brannan CI, et al. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest–derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 24.Schilham MW, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 25.Oh SP, Li E. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 1997;11:1812–1826. doi: 10.1101/gad.11.14.1812. [DOI] [PubMed] [Google Scholar]
- 26.Donovan MJ, Hahn R, Tessarollo L, Hempstead BL. Identification of an essential nonneuronal function of neurotrophin 3 in mammalian cardiac development. Nat Genet. 1996;14:210–213. doi: 10.1038/ng1096-210. [DOI] [PubMed] [Google Scholar]
- 27.Sanford LP, et al. TGFβ2 knockout mice have multiple developmental defects that are non-overlapping with other TGFβ knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chisaka O, Capecchi MR. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature. 1991;350:473–479. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 29.Huang G, et al. Alteration in connexin 43 gap junction gene dosage impairs conotruncal heart development. Dev Biol. 1998;198:32–44. doi: 10.1006/dbio.1998.8891. [DOI] [PubMed] [Google Scholar]
- 30.Mendelsohn C, et al. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- 31.Gruber PJ, et al. RXRα deficiency confers genetic susceptibility for aortic sac, conotruncal, atrioventricular cushion, and ventricular muscle defects in mice. J Clin Invest. 1996;98:1332–1343. doi: 10.1172/JCI118920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdenaire O, Rohrbacher E, Mattei M-G. Organization of the gene encoding the human endothelin-converting enzyme (ECE-1) J Biol Chem. 1995;270:29794–29798. doi: 10.1074/jbc.270.50.29794. [DOI] [PubMed] [Google Scholar]