Summary
Mutations in peripheral myelin protein 22 (PMP22) can result in the common peripheral neuropathy, Charcot-Marie-Tooth disease (CMTD). The Leu16Pro mutation in PMP22 results in misassembly of the protein, which causes the Trembler-J (TrJ) disease phenotype. Here we elucidate the structural defects present in a partially folded state of TrJ PMP22 that are decisive in promoting CMTD-causing misfolding. In this state transmembrane helices 2-4 (TM2-4) form a molten-globular bundle while transmembrane helix 1 (TM1) is dissociated from this bundle. The TrJ mutation was seen to profoundly disrupt the TM1 helix, resulting in increased backbone dynamics and changes in the tertiary interactions of TM1 with the PMP22 TM2-4 core in the folded state. Consequently, TM1 undergoes enhanced dissociation from the other transmembrane segments in TrJ PMP22, becoming available for recognition and sequestration by protein folding quality control, leading to loss of function and toxic accumulation of aggregates that results in CMTD.
Introduction
Peripheral myelin protein 22 (PMP22) is a 160-residue integral membrane protein that is abundant in compact myelin of the peripheral nervous system (PNS). PMP22 appears to be multi-functional, playing roles in Schwann cell development and proliferation, as well as in myelin formation and maintenance (Jetten and Suter, 2000). PMP22 is essential to myelin homeostasis as reflected by the fact that either trisomy or missense mutations of its gene result in the most common inherited disease of the peripheral nervous system, Charcot-Marie-Tooth Disease (CMTD) and related de-myelinating neuropathies (Berger et al., 2006).
CMTD results in dysmyelination and slowed nerve conduction velocities (Berger et al., 2006). While trisomy of the PMP22 gene causes the common Type 1A form of CMTD, more rare inherited dominant missense mutations that encode single amino acid changes in the PMP22 protein cause closely related disorders (Naef and Suter, 1999; Sanders et al., 2001). Most disease mutation sites are located in the transmembrane segments. For example, replacement of leucine-16 with proline in near the middle of TM1 causes both severe CMTD in humans (sometimes classified as Déjerine-Sottas Syndrome) and the Trembler-J (TrJ) phenotype of peripheral neuropathy in mice. (Jetten and Suter, 2000; Suter et al., 1992; Valentijn et al., 1992)
Wild type (WT) PMP22 is believed to fold with only marginal efficiency in vivo, even under healthy conditions. Approximately 80% of nascent WT is folding-defective and targeted for degradation by the protein folding quality control system of the endoplasmic reticulum (ER) (Pareek et al., 1997). As is also the case for most other CMTD mutant forms of PMP22, the TrJ mutation is believed to reduce this normally modest folding efficiency to near zero (Berger et al., 2006; Naef and Suter, 1999; Sanders et al., 2001; Tobler et al., 1999). The resulting complete loss of native PMP22 function is compounded by the fact that the misfolded TrJ mutant forms toxic aggregates rather than being correctly degraded (D'Urso et al., 1998; Fortun et al., 2006; Fortun et al., 2005; Notterpek et al., 1999), resulting in CMTD.
In previous work it was established that both wild type and TrJ PMP22 are mostly-helical proteins and also that the folding stability of the TrJ mutant is reduced relative to the wild type protein in model membranes at room temperature (Mobley et al., 2007; Myers et al., 2008). Here, we examine the structural properties of a partially folded form of PMP22 to reveal that the Leu16Pro mutation that underlies the TrJ disease phenotype profoundly disrupt the proteins 1st transmembrane segment (TM1), destabilizing the protein and making TM1 available for sequestration by the protein folding quality control apparatus of the endoplasmic reticulum.
Results
Structural Properties of Wild Type PMP22
WT and TrJ (L16P) PMP22 were purified (Figure S1) and reconstituted as micellar complexes (Table S1) with tetradecylphosphocholine (TDPC). Structural studies were carried out under these conditions at 45°C because it has previously been shown that both WT and TrJ forms of PMP22 yield NMR spectra of reasonably high quality under these conditions (Mobley et al., 2007). Here, NMR was used to show that both forms of PMP22 form a late stage folding intermediate state under these conditions, in which TM2-4 populate a molten globular structural state, while extracellular loop 1 (ECL1) is largely disordered.
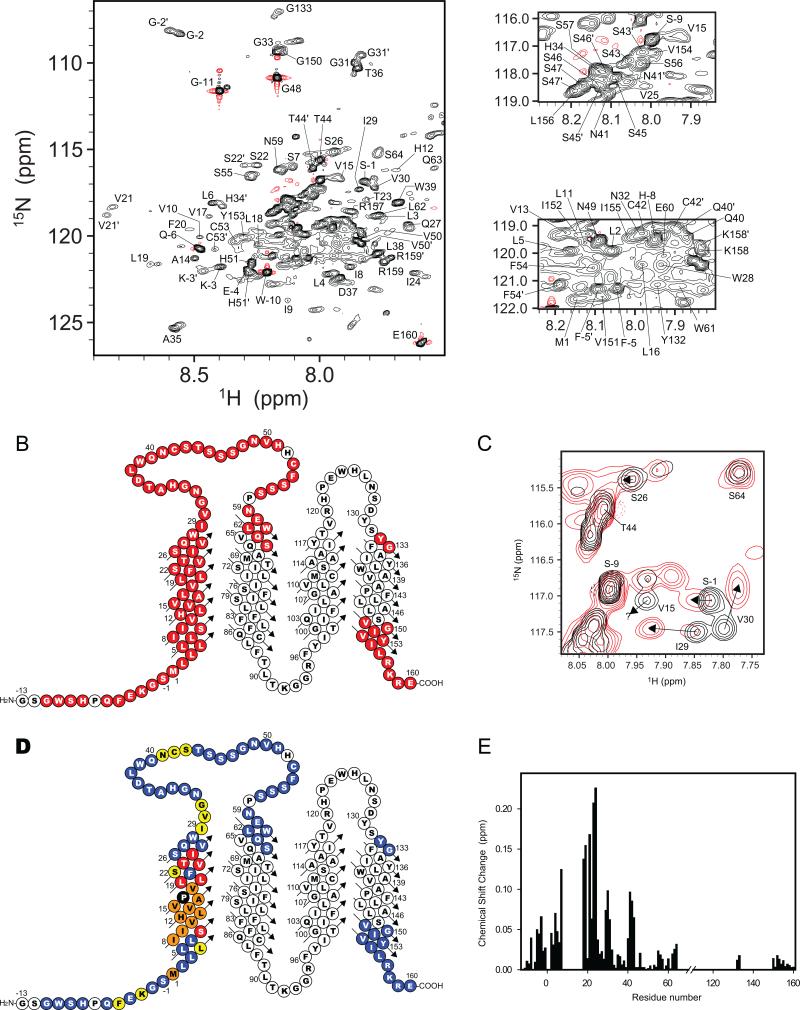
TROSY-based 3-D NMR experiments (Figure S1) were used to assign the backbone amide-1H, 13Cα, 13Cβ , and 15N resonances of WT PMP22. Out of 168 expected backbone amide 1H-15N TROSY resonances from non-proline residues, 85 signals (51%) were assigned (Figures 1A and B). Nearly all expected TROSY resonances from the 1st transmembrane segment (TM1) and 1st extracellular loop (ECL1) were observed and assigned. However, most resonances from TM(2-4) and ECL2 were too broad to detect. Both 1HN-1HN NOE patterns and analysis of the assigned backbone chemical shifts using TALOS+ (Shen et al., 2009) and chemical shift indices (Wishart and Sykes, 1994) revealed that TM1 is a long alpha helix that spans residues 1-30, whereas the first extracellular loop is largely disordered and spans residues 31-57 (Figures 2A and B). Chemical shifts for the few observable resonances from sites located at the beginning of TM2 and at the beginning and ends of TM4 indicate that these transmembrane segments are almost certainly helical as well, which is consistent with far-UV circular dichroism measurements that indicate that WT PMP22 is 63% α-helical (Figure 3A).
Figure 1. NMR spectroscopy of WT and TrJ PMP22.
(A) Panels from an 800 MHz 1H,15N-TROSY NMR spectrum of WT PMP22 showing assigned backbone amide resonances. As for all NMR results presented in this work this spectrum was acquired using labeled (i.e., U-2H,13C,15N) PMP22 in TDPC micelles, pH 5.0 at 45°C. The black and red lines indicate positive and negative contours, respectively. Triple resonance spectra used to assign the resonances are shown in Figure S1. (B) Topology plot showing the approximate sequence spans of WT PMP22's 4 transmembrane helices. Red residues yielded observable TROSY peaks that were assigned for WT. White residues either failed to yield observable amide peaks or, less frequently, were observed but could not be assigned. (C) Superimposed 800 MHz 1H-15N TROSY sub-spectra for TrJ PMP22 (red) and WT PMP22 (black). Residues such as I29 and V30 displayed large chemical shift differences between WT and TrJ as shown by arrows, while residues such as V15 were absent in the spectrum form the mutant because of line-broadening. (D) Topological mapping of the residues in TrJ PMP22 affected by the L16P mutation. The L16P mutation site is highlighted by the black-filled circle. Residues with the chemical shift differences between WT and TrJ of more than 0.1 ppm, 0.05-0.1 ppm, and less than 0.05 ppm are colored red, yellow, and cyan, respectively. Residues with signals that disappeared in the mutant's spectrum are colored orange. (E) Differences in 1H-15N TROSY chemical shifts between TrJ and WT PMP22. The longitudinal axis represents the weighted differences in chemical shifts Δδ (= sqrt[(ΔδH)2 + (ΔδN × 0.18)2]) where ΔδH and ΔδN represent the differences in chemical shift for the amide 1H and 15N, respectively, between WT and TrJ forms of PMP22. Corresponding data for 13Cα is shown in Figure S3.
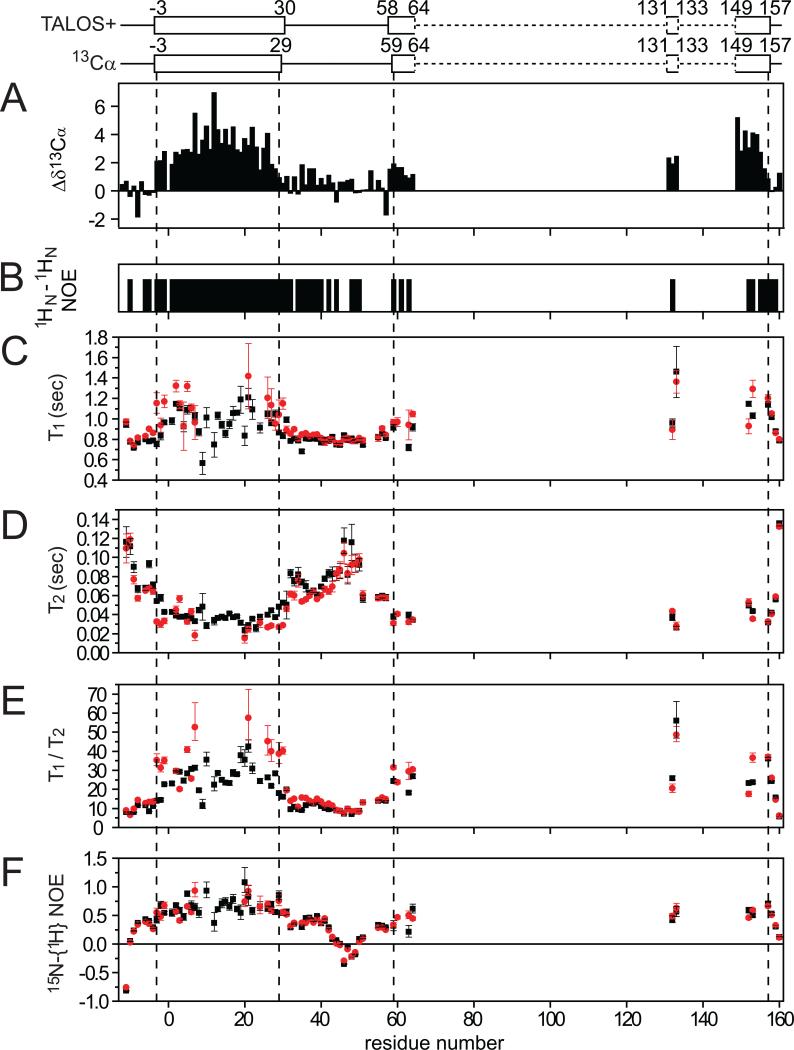
Figure 2. Secondary structural analysis for WT PMP22 (A-B) and NMR relaxation measurements for both WT and TrJ PMP22 (C-F).
Panel A shows the deviation of assigned 13Cα shifts from random coil values, while panel B shows 1HN-1HN NOEs. The diagrams above panel A show the locations of helices that are indicated by either TALOS+ analysis (Shen et al., 2009) of the backbone/Cβ chemical shifts and by chemical shift index analysis based on 13Cα chemical shifts (Wishart and Sykes, 1994). Dashed lines indicated regions where secondary structure cannot be determined because peaks were missing or unassigned. Panels C-F provide the 600 MHz backbone amide 15N NMR relaxation times measured for both wild type PMP22 (black boxes) and the TrJ mutant (red circles). The effective rotational correlation times and translational diffusion coefficients are shown in Table S1.
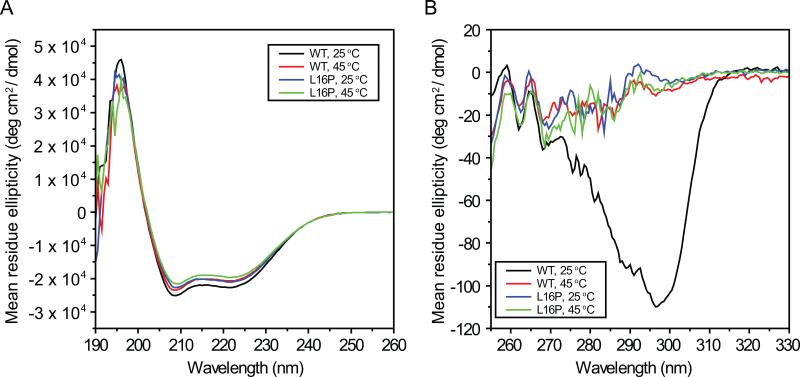
Figure 3. Near- and Far-UV CD spectra of WT AND L16P (TrJ) PMP22 in TDPC micelles at 25 and 45°C.
Samples contained 50 mM acetate (pH 5). Samples for far-UV CD (Panel A) contained 10 uM (WT) or 11 uM (L16P) PMP22, 0.5 mM DTT, and 0.2% TDPC in 0.1 cm cuvettes. Samples for near-UV CD (Panel B) contained 37 uM (WT, 25°C), 32 uM (WT, 45°C) or 48 uM (L16P, both temperatures) PMP22, 1 mM DTT, 200mM NaCl, and 1% TDPC in 1.0 cm cuvettes. From the far-UV CD data, the α-helical content of PMP22 as calculated to be: 58 ± 1% (WT, 25°C), 56 ± 1% (TrJ, 25°C), 63 ± 1% (WT, 45°C), and 61 ± 1% (TrJ, 45°C). The errors represent the standard deviation calculated from three different experimental data. The 25°C spectra closely resemble those reported for PMP22 in a different type of micelles (dodecylphosphocholine micelles) in (Mobley et al., 2007).
The disappearance of nearly all TROSY resonances from TM(2-4) could result either from an excessively large aggregate molecular weight or from intermediate time scale exchange between multiple conformations. NMR-based translational diffusion coefficient measurements for the PMP22/TDPC micellar complex were performed to determine its aggregate molecular weight. The translational diffusion coefficient (Dt) obtained for PMP22 in TDPC micelles was 7.7 (±0.1) × 10-11 m2 sec-1, which corresponds to an aggregate molecular weight of 58 kDa (Table S1). This aggregate molecular weight is further supported by NMR relaxation measurements (see below) and is reasonable since the size of protein-free TDPC micelles has been reported to be 47 kDa (Strop and Brunger, 2005). An apparent molecular weight of 58 kDa is consistent with monomeric WT PMP22 inhabiting micelles containing, on the average, 102 TDPC molecules per micelle. It is inconsistent with micelles containing dimeric PMP22 because only 51 detergent molecules would be incorporated in the complex. 58 kDa is well within the molecular weight range for recording high quality TROSY NMR spectra (Fernandez and Wider, 2003). It can therefore be concluded that the peaks from TM(2-4) are severely broadened not by slow molecular tumbling but instead by intermediate time scale exchange processes, which are characteristic of molten globular helical bundles (Redfield, 2004). This interpretation is supported by fact that WT PMP22's near-UV circular dichroism spectrum at 45°C (Figure 3B) is flat in the 260-310 nm range, as expected for a molten globular protein.
The fact that TM1 and ECL1 yield easily detected TROSY NMR peaks indicates that these structural elements populate states in which they are dissociated from the molten globular domain. This is supported by NMR relaxation measurements that show that most sites in TM1 have 1H-15N NOEs well under 1.0 (Figure 2F). Moreover, the effective rotational correlation time (16 ± 2 nsec) calculated for TM1 from averaged backbone amide 15N T1 and T2 (Figures 2C-E) is smaller by 20% than expected based on the overall size of the PMP22/TDPC mixed micelles estimated from the translational diffusion coefficient, as expected if TM1 spends at least part of its time dissociated from TM(2-4). For ECL1 the mobility is even higher, consistent with this loop being largely disordered.
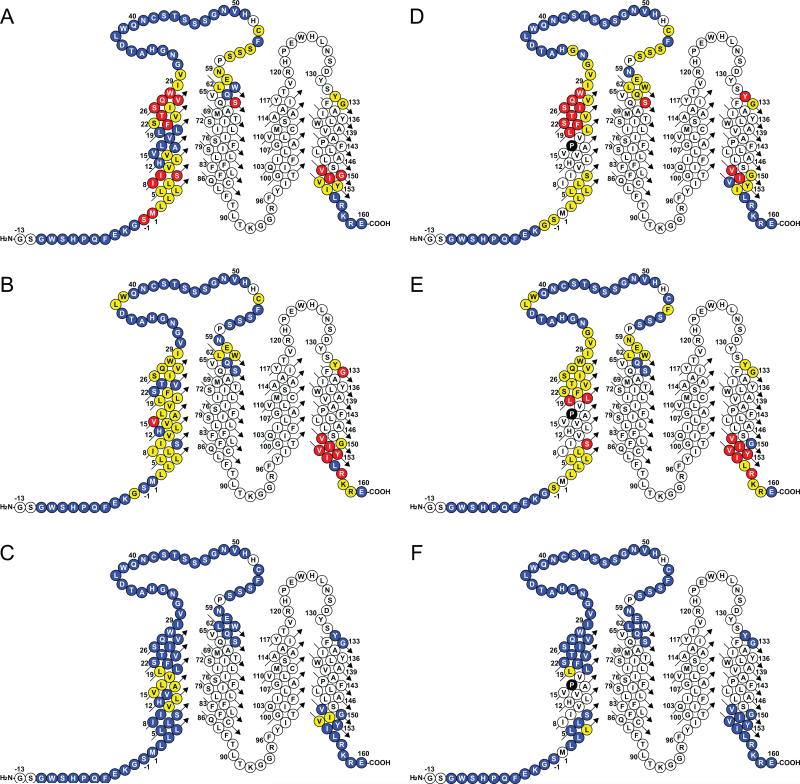
It is clear that at 45°C TM1 of WT PMP22 significantly populates a state in which it is dissociated from tertiary structural interactions with TM(2-4). This led to the question of whether it also populates a state in which it interacts with TM(2-4). This was probed by conducting saturation transfer difference (STD) NMR experiments in which different protons of the TDPC detergent were selectively irradiated while monitoring the TROSY peaks from the protein. Reductions in the intensities of PMP22 TROSY peaks are indicative of direct detergent-residue interactions between the associated protein site and the irradiated moiety of the detergent. These measurements confirmed both the facts that TM1 is a transmembrane segment and that ECL1 is largely water-exposed (Figures 4 and S2). Moreover, the STD data for TM1 exhibited a periodicity that indicates that TM1 is interacting with TM(2-4), but that the interactive faces of the C terminus and center of TM1 are offset by roughly 90 degrees with respect to the long axis of an ideal helix (Figures S2D and E). This likely indicates that the 30 residue TM1 is both tilted and is bent or curved. The fact that protection is for no site complete (Figure 4) lends further support to the conclusion that TM1 also populates a state where it is fully dissociated from interacting with TM(2-4).
Figure 4. Results of STD-NMR studies in which the 1H,15N-TROSY peaks of PMP22 were monitored after saturation transfer from specifically-irradiated moieties of the host detergent, TDPC.
Cyan-colored sites exhibited minimal saturation transfer (Isat/Iref >0.9) indicating little or no contact between the indicated site and the relevant moiety of TDPC that was irradiated. Yellow sites exhibited intermediate interactions ((Isat/Iref is 0.8-0.9), whereas sites that exhibited the most intimate interactions (Isat/Iref <0.8) are colored red. Panels A-C and D-F correspond to WT and TrJ PMP22, respectively. Reliable measurements could not be made for uncolored sites. Panels A and D correspond to presaturation of TDPC protons C1H2 (the carbon on the tetradecyl chain that is adjacent to the head group). Panels B and E correspond to presaturation of C4-13H2, while panels C and F correspond to presaturation of the terminal methyl group C14H3 of the tetradecyl chain. The quantitative measurements that these plots summarize are given in detail in Figure S2.
To summarize, at 45°C and in TDPC micelles TM segments 2-4 of WT PMP22 form a molten-globule like helical bundle. TM1 forms a long alpha helix that interacts transiently with the TM(2-4) bundle, but also spends a significant fraction of its time freely dissociated. It is most likely that this structural state represents a late-stage folding intermediate.
Structural Properties of the Leu16Pro Trembler-J CMTD Form of PMP22
The general appearance of the 45°C TROSY NMR spectrum for the TrJ mutant form of PMP22 is similar to that of WT (Mobley et al., 2007). Resonance assignments were completed for the observed peaks, which indicate that TrJ resembles the wild type protein in that, while a majority of peaks from TM1 and ELC1 are observed, most peaks from TM(2-4) and ECL2 are severely broadened (Figures 1C and D). Lack of ordered tertiary structure is confirmed by the fact that the near-UV CD spectrum does not deviate from the baseline (Figure 3B). This indicates that the overall structures of TrJ and wild type PMP22 at 45°C in TDPC micelles are similar: both forms populate what appears to be a late state folding intermediate. The Leu16Pro CMTD mutation clearly does not induce a catastrophic change in the overall structure of TrJ PMP22 relative to WT. Nevertheless closer examination of the NMR data from TrJ PMP22 does reveal significant local changes in protein structure and dynamics for the mutant.
Whereas all resonances from TM1 are observable in the TROSY spectrum of WT PMP22, for the TrJ/L16P mutant the peaks from residues 8-17 in TM1 are absent as a result of severe linebroadening. Moreover, the chemical shifts of the observable resonances in TM1 are significantly perturbed relative to WT shifts (Figures 1E and S3). These observations indicate that the introduction of a proline at site 16 does more than just induce a kink in the helix. On the other hand far-UV CD indicates no significant difference in overall α-helical content between WT and TrJ (Figure 3A), suggesting that most sites in residues 8-17 remain helical. These observation can be reconciled if the helix in the neighborhood of the mutation site is destabilized such that it undergoes intermediate timescale exchange between a predominant helical conformation and a less populated non-helical structure, resulting in extensive amide peak exchange broadening. This interpretation is supported both by limited proteolysis data (below) and by 15N relaxation rate measurements that show the helical ends of TM1 in the TrJ mutants are significantly more mobile than these same segments in WT (Figure 2E). This indicates that local destabilization of the helices immediately adjacent to the Leu16Pro mutation site results in formation of a flexible hinge that enables wagging motions of the flanking TM1 helical segments.
As observed for WT, TM1 in the TrJ mutant form of PMP22 clearly populates a state in which it is dissociated from the molten globular TM(2-4) domain. However, STD-NMR experiments also confirmed that TM1 transiently interacts with TM(2-4), as was seen for WT. For each of the helical segments flanking the hinge associated with the Leu16Pro mutation it is seen (Figures 4 and S2) that there is a helical face that contains sites that are partially protected from saturation transfer from detergent protons. However, the patterns of site protection from STD are significantly different for the mutant than for wild type. In particular, at the C-terminal end the protected face of the helix appears to have rotated by at least 45° around the helical axis in the TrJ mutant relative to wild type (Figures S2D and G).
In summary, while the overall structure and dynamics of TrJ PMP22 at 45 °C is not grossly different from the WT protein, the L16P mutation results in formation of a hinge in the middle of TM1, leading to wagging motions of the helices on each side of the hinge and altered tertiary structural interactions of TM1 with TM(2-4).
Limited Proteolysis of Wild Type and TrJ PMP22
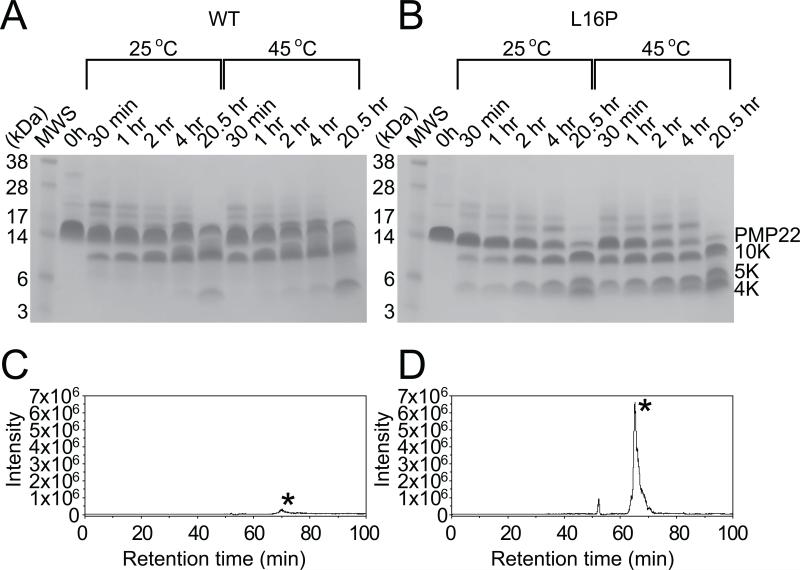
WT and TrJ PMP22 were subjected to limited proteolysis using pepsin under the same sample conditions as used for the NMR experiments. It can be seen (Figures 5A and B) that TrJ PMP22 is more rapidly degraded than WT. This is consistent with the notion that TrJ is less stable than WT PMP22 and is therefore more generally accessible to proteolytic digestion. It is also seen that the pattern of bands observed for cleavage of wild type is different than for TrJ. In order to more quantitatively analyze the time course of cleavage we subjected each proteolytic digest of the time course to separation by liquid chromatography followed by on-line electrospray tandem mass spectrometry. These studies revealed the 5 kDa band that is generated at short times for TrJ (Figure 5B), but only in limited amounts for WT PMP22 (Figure 5A), is predominately the N-terminus of the protein up through residue 38 (Figures 5C and D) located in ECL1. More rapid generation of this 5 kDa fragment for TrJ than the WT PMP22 was confirmed by multiple LC-MS/MS analyses of pepsin digests for 30 min. at 45 °C. Analysis of pepsin digests for 30 min. at 25 °C gave a similar pattern. These results indicate that the structural defects associated with TM1 in the L16P TrJ mutant result in ECL1 becoming more accessible for proteolytic cleavage by pepsin. The increased accessibility of residue 38 likely reflects the enhanced dissociation of TM1 from the TM(2-4) bundle in TrJ relative to WT, which is likely accompanied by an increase in disorder and extension of ECL1, reflected by enhanced cleavage at position 38 in the center of the ECL1 loop. The results of the proteolytic time course experiments are fully consistent with the NMR experimental conclusions of this work and previous evidence (Mobley et al., 2007; Myers et al., 2008) that TrJ PMP22 is conformationally less stable than WT.
Figure 5. Limited Proteolysis of PMP22.
(A) and (B): SDS-PAGE gels giving the time course of limited proteolysis of WT (A) and TrJ (B) PMP22. TDPC micellar solutions of WT (A) or TrJ (B) PMP22 were exposed to pepsin for the amounts of time specified and then subjected to SDS-PAGE followed by coomassie staining (see Methods for additional details). Time courses were run at both 25 and 45°C. (C) AND (D): LC-MS analysis of limited proteolytic digest of TrJ (D) and WT PMP22 (C). Selected ion chromatograms for the molecular ion ([M+5H]5+ at m/z 1097.5 for TrJ (D) and m/z 1100.5 for WT (C)) corresponding to the N-terminus up through residue 38 (marked by asterisk) generated after pepsin digestion of 0.6 μg of PMP 22 for 30 minutes at 45°C. The tandem mass spectra identifying the sequence of these peptides are presented in Figure S4.
Discussion
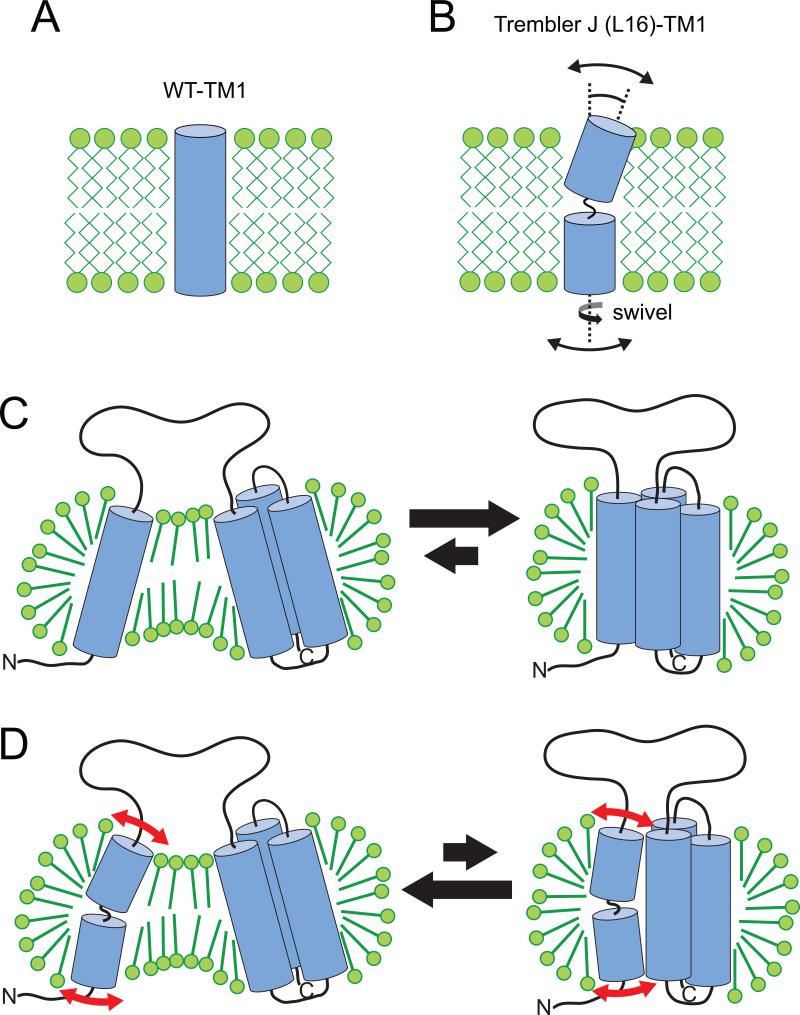
The results of this study define the nature of the structural perturbations in PMP22 that lead to the TrJ phenotype of CMTD. Previous studies established that under micellar conditions similar to those in this study but at lower temperature (25°C), wild type PMP22 exhibits a more ordered tertiary structure in alkylphosphocholine micelles than TrJ as judged by near-UV CD spectroscopy and by the fact that a partially reversible sigmoidal unfolding curve is evident for WT but not for TrJ when a denaturant is titrated into PMP22 solutions (Mobley et al., 2007; Myers et al., 2008). A more detailed structural comparision of the WT and disease mutant forms of the protein could not be conducted at 25°C using NMR spectroscopy because of very poor spectral quality for both forms of that protein at that temperature. Here we conducted structural studies at 45°C, a temperature at which PMP22's NMR spectrum is more optimal. At this higher temperature it was observed that both WT and TrJ PMP22 adopt dynamic partially-folded structures that possibly resemble late-stage states in the folding pathway. For both WT and TrJ, transmembrane segments 2-4 exist as a molten globular helical bundle, while the first transmembrane segment populates two states—one in which it is dissociated from interactions with the TM(2-4) bundle and one in which it undergoes transient interactions with TM(2-4). This is summarized in Figure 6. If indeed this conformational ensemble represents a folding intermediate then it suggests that the folding pathway for PMP22 involves an initial coalescence of TM helices 2-4 to form a loosely packed helical bundle. Mature tertiary interactions in this bundle are consolidated only at the completion of folding, which involves the stable packing of TM1 with the pre-formed TM(2-4) bundle.
Figure 6.
Model for how the L16P mutation of TrJ PMP22 results in destabilization of the protein, resulting in enhanced sequestration by calnexin.
Cell biological studies have established that even wild type PMP22 folds in vivo with only marginal (20%) efficiency, with 4 out of 5 newly-synthesized molecules being deemed folding-defective by the quality control machinery of the endoplasmic reticulum, leading to targeting for degradation via the ERAD pathway (Pareek et al., 1997). The low folding efficiency of wild type PMP22 is consistent with the general instability of wild type PMP22 in detergent micelles, as reflected by its marginal thermal stability (evidenced by loss of ordered tertiary structure during the transition from 25 to 45°C).
While the overall structures of the wild type and TrJ mutant forms of PMP22 are similar, the L16P mutation within TM1 of the TrJ CMTD mutant dramatically alters both the conformation and dynamics of TM1, leading to wagging motions of the helices flanking the hinge-like mutation site and to changes in the interface of TM1 that interacts transiently with TM(2-4). This illuminates why the TrJ disease mutant form of PMP22 folds with such a low (near 0%) efficiency under cellular conditions (Colby et al., 2000; D'Urso et al., 1998; Tobler et al., 2002). For proteins that fold in the ER, low folding efficiency is thought to generally correlate well with low conformational stability (Sanders and Myers, 2004; Wiseman et al., 2007). The increased dynamics of TM1 in the TrJ mutant likely contributes an entropic factor that disfavors the folded state relative to the wild type case. The bend in TM1 that is induced by the L16P mutation as well as the accompanying partial rotation of the helix above site 16 (relative to wild type) is also expected to disrupt normal packing of TM1 with the TM(2-4) bundle, further destabilizing the folded state of the TrJ mutant relative to WT, consistent with our observations in both this and previous work.
Recognition and initial sequestration of both misfolded wild type and TrJ mutant forms in the ER appears to be carried out by the membrane-associated lectin chaperone, calnexin (Dickson et al., 2002; Fontanini et al., 2005). It has been shown that in sciatic nerves of the PNS calnexin binds WT PMP22 to form a complex that has a t1/2 of 11 minutes, while for the corresponding complex with the TrJ mutant the complex is even longer-lived, with a t1/2 of >60 minutes (Dickson et al., 2002). For both WT and TrJ, TM1 appears to play a critical role as a structural element that is directly recognized by calnexin and synthetic polypeptides corresponding to TM1 of both WT and TrJ are bound tightly by calnexin (Fontanini et al., 2005). The present work establishes that TM1 is less avidly bundled with TM(2-4) than these latter three TM segments are to each other. It is therefore logical that calnexin assesses the state of folding of PMP22 based on the accessibility of this TM segment. The recognition of dissociated helices within multispan membrane proteins by calnexin appears to be one of the mechanisms used by ER quality control to recognize membrane proteins that are incompletely folded or misfolded (Korkhov et al., 2008; Li et al., 2010; Swanton et al., 2003). Our observation regarding TM1 dissociation may explain why both WT and TrJ PMP22 navigate ER quality control to fold and traffic beyond the ER with only a low efficiency in vivo.
While calnexin recognizes and associates with the TM1 segments of both wild type and TrJ PMP22, kinetically persistent binding of wild type PMP22 also requires N-glycosylation of PMP22 in ECL1 (Fontanini et al., 2005). In contrast, the TrJ L16P mutant is sequestered by calnexin in a manner that is independent of N-glycosylation (Fontanini et al., 2005). This fact along with the observation that the t1/2 of the complex formed between TrJ PMP22 and calnexin is at least 5X longer than for the complex with WT PMP22 suggests the intriguing possibility that calnexin can not only recognize dissociated helices in multispan membrane proteins as a way of identifying incomplete or incorrect folding, but that calnexin may also bind more avidly to helices that are structurally defective, such as in TrJ. It has previously been shown that calnexin can recognize TM segments that have lipid-exposed acidic residues relative to mutants in which the acidic side chain is replaced by a hydrophobic residue (Li et al., 2010). Perhaps calnexin can recognize multiple forms of defects in TM helices.
The conclusions of this paper lead to a satisfying model for how the TrJ L16P mutation results in misfolding that ultimately leads to CMTD. While WT PMP22 is marginally stable under healthy conditions in which its in vivo folding efficiency is ca. 20%, the L16P mutation of TrJ PMP22 alters the structure and dynamics of TM1 such that folding stability is reduced to the point where the protein primarily inhabits a state in which TM1 dissociated from interactions with TM2-4 and is therefore fully available for recognition by calnexin. Local defects in the conformation of TM1 may also enhance the avidity of binding by calnexin to further increase the efficiency by which TrJ PMP22 is targeted to aggresomes. It will be interesting to determine to what degree other CMTD mutations, many of which are not in TM1, lead to alterations in PMP22 structure, stability, and dynamics that resemble the perturbations documented in this work for the TrJ mutant form. Finally, we also note that missense mutations that encode insertions of proline residues into the TM domains are an unusually common class of disease-causing mutation (Partridge et al., 2002; Sanders, 2005). It is possible that the defects in TM1 observed as a result of the L16P mutation in PMP22 may be shared by many other disease-linked mutant membrane proteins.
Experimental Procedures
Expression and Purification of PMP22
Recombinant PMP22 was produced as a fusion protein with an N-terminus comprised of the first 76 residues of the lambda repressor followed by a deca-histidine tag, a thrombin cleavage site, a STREP tag, and then PMP22 (Figure S1A). The STREP tag was used as a spacer between the N-terminus of PMP22 and the protease site-cleavage could not be effected without this spacer. This construct is slightly different from a construct we previously reported (Mobley et al., 2007) in that a thrombin site now replaces a TEV protease site. Protein expression was carried out in RosettaBlue Escherichia coli (EMD Chemicals, Inc.) grown at 20 °C in M9 minimal media with 100 μg/mL ampicillin and 50 μg/mL chloramphenicol and supplemented with a MEM vitamins (Mediatech, Inc.). Isopropylthiogalactoside was used to induce expression. Uniformly 2H, 13C, and 15N-labeled protein was expressed in D2O-based M9 medium containing 1 g/L 15N-ammonium chloride and 3 g/L 13C-glucose. Amino acid selectively-labeled proteins were expressed in M9 media supplemented with a combination of both labeled and unlabeled amino acids as described (Czerski et al., 2000). Cys, His, Lys, Met, Tyr, Val, and Trp were targeted for selective main chain 15N labeling. Following harvesting cells were resuspended in lysis buffer (75 mM Tris-HCl, pH7.5, 300 mM NaCl, 0.2 mM EDTA, 10 μM BHT) at 20 mL per gram of cell pellet. Lysozyme (0.2 mg/mL of total lysis buffer), RNase (0.02 mg/mL), DNase (0.02 mg/mL), and magnesium acetate (5mM) were added, and the solution was rotated at 4 °C for 30 min. The lysate was then sonicated on ice for 10 min. DTT was then added to a concentration of 0.5 mM. To extract the fusion protein, glycerol was added to 15% and Empigen (a detergent, Emp, Fluka) to 3% and the solution was rotated at 4 °C for 45 min. After centrifugation at 20,000 rpm (48384 g) and 4 °C for 20 min, the supernatant was collected. Ni-NTA resin (QIAGEN; 1.2 mL of resin per gram of cell pellet) was added to the lysate and this mixture was rotated at 4 °C for 45 min. Resin was packed into a chromatography column and washed first with 10 column volumes of Emp/A buffer (40 mM HEPES, pH7.5, 300 mM NaCl, 15% glycerol, 3% Empigen, and 0.5 mM DTT) and second with wash buffer (40 mM HEPES, pH 7.8, 300 mM NaCl, 15% glycerol, 1.5% Empigen, 30 mM imidazole, and 0.5 mM DTT) until the measured absorbance at 280 nm indicated complete elution of weakly bound impurities, usually ~10 column volumes. Detergent was then exchanged from Empigen into dodecylphosphocholine (DPC; Anatrace) by passing 10 column volumes of rinse buffer (25 mM sodium phosphate, pH7.2, 0.5% DPC, and 0.5 mM DTT) through the column. The PMP22 fusion protein was eluted from the column using elution buffer (50 mM Tris-HCl, pH 8.0, 250 mM imidazole, 0.5% DPC, and 0.5 mM DTT). The eluted protein concentration was determined by measuring A280 nm using the extinction coefficient of 1.64 mL mg-1 cm-1. The eluate was diluted with a solution containing 0.5% DPC and 0.5 mM DTT to make a solution containing 0.8 mg/mL of the fusion protein. Cleavage of the fusion partner from PMP22 was performed by adding a stock solution containing 50% glycerol, 2mM EDTA, 0.5 mM DTT, and 50 NIH units/mL thrombin (SIGMA; thrombin from human plasma, >2800 NIH units/mg protein). The final reaction buffer typically contained 0.4 mg/mL fusion PMP22, 25 NIH units/mL thrombin, 11 mM Tris (pH 8), 53 mM imidazole, 0.5 mM DTT, 1 mM EDTA, 0.25% DPC, and 25% glycerol. The pH of the solution was then adjusted to 5.0 and cleaved PMP22 was purified by a cation exchange chromatography using an SP-Sepharose cation exchange column (GE healthcare Life Sciences). Following sample loading the column was washed with 20 column volumes of buffer containing 50 mM acetate (pH 5.0), 0.2% TDPC, and 1 mM DTT to exchange detergent from DPC to TDPC. Cleaved PMP22 was then eluted by elevating (as a gradient) the NaCl concentration to 200 mM. Remaining uncleaved fusion protein was then removed from the eluate from the cation exchange column by adding Ni-NTA resin (0.7 mL resin per 20 mL of PMP22 solution) and then gently mixing at 4 °C for 45 min. After removal of the resin, the PMP22 solution was concentrated in a 10 kDa MWCO Amicon Ultra-4 centrifugal filtering device (Millipore). The buffer was then exchanged into 10 mM acetate (pH 5.0), 10 mM DTT, 1mM EDTA, and 5% D2O by multiple centrifugal ultrafiltration exchanges during which extra detergent was not added. This was the final step in preparing the NMR samples used in these studies. The final PMP22 and TDPC concentrations were typically 0.8~1.0 mM and 284~395 mM (11~15%), respectively.
NMR Measurements
All NMR measurements were carried out at 45°C. Data were collected on Bruker Avance 600 and 800 spectrometers equipped with cryogenic triple-resonance probe heads with z-axis pulsed field gradients. Diffusion experiments were carried out on Avance 600 spectrometer with conventional triple-resonance probe with z-axis pulsed field gradient. The spectra were processed using Topspin and analyzed with Sparky (Goddard, T. D. and Kneller, D. G. University of California, San Francisco). Chemical shifts were referenced to DSS as described (Edison et al., 1994).
2-D 1H,15N-TROSY experiments were carried out at 600 and 800 MHz using asensitivity- and gradient-enhanced TROSY pulse sequence (Weigelt, 1998). Spectra are typically acquired with 128 complex points and an acquisition time of 56 msec in the 15N dimension and 1024 complex points and an acquisition time of 80 msec in the observed dimension.
Triple resonance TROSY versions of HNCA, HN(CO)CA, HNCACB, and HN(CO)CACB experiments (Eletsky et al., 2001; Salzmann et al., 1999) were carried out for 2H, 13C, and 15N-labeled WT-PMP22 in TDPC micelles at 800 MHz. For L16P-PMP22, TROSY versions of HNCA and HN(CO)CA were recorded. Triple resonance experiments were acquired with 32 complex points and an acquisition time of 14.1 msec in the 15N dimension, and 512 complex points and an acquisition time of 40 msec in the observed dimension. Experiments with 13Cα and 13Cα/13Cβ evolution were acquired with 32 and 58 complex points and acquisition times of 6.63 and 4.37 msec in the 13C dimension, respectively. A 1H-15N NOESY-TROSY (Chill et al., 2006; Zhu et al., 1999) spectrum was acquired at 800 MHz field with a mixing time of 80 msec. The spectrum was acquired with 128 complex points and an acquisition time of 26.7 msec in the indirect 1H dimension, 32 complex points and an acquisition time of 14.1 msec in the 15N dimension, and 1024 complex points and an acquisition time of 91.8 msec in the 1H observe dimension. An E-BURP pulse was used as the first 1H pulse to selectively excite amide protons in the NOESY experiment as described (Chill et al., 2006).
HSQC-based T1, T2, and 15N-(1H)-NOE experiments were carried out for 15N-labeled PMP22 (WT or L16P) at 600 MHz. The TDPC concentration was 290 and 284 mM for WT and the L16P mutant, respectively. Gradient-enhanced versions of pulse programs with improved sensitivity were used for these experiments (Farrow et al., 1994; Kay et al., 1992). T1 values were determined from a series of spectra with 0.1, 0.2, 0.4, 0.8, 1.2, 1.6, and 2.0 sec relaxation delays. T2 values were obtained from the spectra with 0, 8.6, 17, 35, 69, 138, and 199 msec relaxation delays. For the L16P (TrJ) mutant, T2 data for an additional relaxation delay of 104 msec was also collected. An additional delay of 1.2 s between scans was also employed when measuring 15N T1 and T2 values. 15N-(1H)-NOE values were determined from the intensity ratios between signals in the spectrum acquired using 3 sec proton presaturation relative to the corresponding signals in the reference spectrum acquired without presaturation. All spectra were recorded as 128 (15N) × 1024 (1H) complex points with 32 scans per t1 point for the T1/T2 experiments and 24 scans for the NOE experiment. Spectral widths were 1703 and 9615 Hz in the 15N and 1H dimensions, respectively.
Diffusion experiments were carried out for 15N-labeled PMP22 in 276 mM TDPC micelles at 600 MHz. 2D 1H-15N-DOSY-HSQC (Brand et al., 2007) pulse schemes employing bipolar pulse pair longitudinal eddy-current delays (BPPLED) (Wu et al., 1995) followed by the sensitivity-enhanced HSQC unit (Schleucher et al., 1994) were used to measure the translational diffusion coefficient (Dt) of PMP22. The detergent Dt was measured by 2D-BPPLED. For 2D experiments, a series of 32 diffusion-weighted one-dimensional spectra with 4096 complex points were recorded in a two-dimensional manner, using a gradient pulse length of 3.5 msec, a 110 msec diffusion delay between the first 90 degree pulses of the two BPP units (large delta), and an inter-scan delay of 6 sec. The pulsed field gradient (PFG) strength was varied from 0.76 to 36 G cm-1. PFG strength was obtained by back-calculating the diffusion coefficient of 1% H2O (1.905 × 10-9 m2 sec-1 at 25°C) (Longsworth, 1960) using a solution containing 1% 1H2O, 99% D2O, and 0.1 mg/ml GdCl3. Spectral widths were 1703 and 9615 Hz in 15N and 1H dimensions, respectively. NMR signal integrals from 6.5 or 6.7 to 9 ppm were used for analysis of the 2D diffusion data. Dt was obtained by fitting the data to the theoretical formula as described (Wu et al., 1995).
Saturation transfer difference (STD) NMR experiments were carried out for 2H, 13C, and 15N-labeled PMP22 in TDPC micelles at 600 MHz field. TDPC protons were selectively irradiated in continuous wave mode for 2 sec and the amide proton signals of the protein were detected by the TROSY pulse scheme following the irradiation (Sakakura et al., 2008). The delay for relaxation between scans was set to 2 sec. Selective irradiation was applied at 0.84 ppm for the C(14)H3 of TDPC, 1.25 ppm for the C(4-13)H2, 3.82 ppm for the C(1)H2. TROSY spectra with and without saturation were recorded in interleaved fashion.
The secondary structure was analyzed by calculating deviations of the observed 13Cα chemical shifts from their residue-dependent random coil values (ΔCα) (Wishart and Sykes, 1994). The TALOS+ program (Shen et al., 2009) was also used to assess the secondary structure using 13Cα, 13Cβ, and 15NH chemical shifts as input data.
The secondary chemical shifts of amide protons were calculated by subtracting random coil values from the shifts measured in the TROSY spectrum of 2H, 13C, 15N-labeled sample (Wishart et al., 1991; Zhou et al., 1992). Corrections were made for the 1JNH/2 shifts of the TROSY.
The effective rotational correlation time of TM1 was calculated using the averaged backbone 15N T1/T2 ratio for residues from Lys-3 through Ile29 with the program r2r1_tm: (http://biochemistry.hs.columbia.edu/labs/palmer/software/r2r1_tm.html).
Circular Dichroism Measurements
CD experiments were carried out using a Jasco J-810 instrument with a Peltier temperature control unit. For far-UV, CD samples were added to a 0.1 cm path length quartz cuvette (Hellma) and spectra were acquired from 185 nm to 260 nm at 25 and 45°C with a 1.5 nm bandwidth, averaging for 4 s at each wavelength. Three spectra were averaged to give the final trace, and blank spectra were subtracted. Protein concentrations were 10 and 11 uM for WT and L16P PMP22, respectively. The protein solutions contained 50 mM acetate (pH 5), 0.5 mM DTT, and 0.2% TDPC. The fractional helicity was calculated as described (Rohl and Baldwin, 1997).
For near-UV CD samples were added to a 1.0 cm path length quartz cuvette (Hellma) and spectra were acquired over the range of 255 nm to 330 nm at 25 and 45°C with a 1 nm bandwidth, averaging for 4 s at each wavelength. 3 spectra were averaged to give the final trace, and blank spectra were subtracted. Protein concentrations were 37, 32, and 48 uM for WT (25°C), WT (45°C), and L16P PMP22, respectively. The protein solutions contained 50 mM acetate (pH 5), 1.0 mM DTT, ca 200 mM NaCl. TDPC concentrations were 1.0% (WT, 25°C), 0.9% (WT, 45°C), and 0.9% (L16P).
Limited Proteolytic Cleavage of PMP22
0.6 mg/ml of PMP22 was digested with 0.6 ug/ml of pepsin (Sigma, pepsin A from porcine stomach mucosa, 3100 units / mg). Pepsin cleavage was carried out in a solution containing 50 mM acetate (pH 5), 0.6/0.4% TDPC (for WT/L16P), 0.5 mM DTT. At a series of time points (30 min, 1h, 2h, 4h, and 20.5h), the reactions were terminated by raising the pH with 20 mM NaOH and heating of the samples at 95°C for 1 min. The cleaved protein fragments were separated by SDS-PAGE and the amount of protein fragments loaded on the gel corresponds to 7 μg of PMP22 (Invitrogen, NuPAGE, 10% Bis-Tris gel).
Mass Spectrometry to Analyze Time Course of Proteolysis of PMP22
Direct analysis of peptic digests obtained from digestion of 0.6 μg of PMP 22 was carried out by LC-MS/MS after sample clean-up using a C4 Ziptip (Millipore, Bedford, MA), and elution in 70% acetonitrile (ACN) (0.1% formic acid). The eluate was dried in a speedvac and reconsitituted in 0.1% formic acid and loaded onto a custom packed C8 trap column (5 cm × 100 um, Jupiter C8, 3 um, 100 Å media). The trap column was coupled to a nanoflow analytical column (11 cm × 75 um, Jupiter C8, 3 um, 100 Å media). Liquid chromatography was performed using the following gradient at a flow rate of 0.4 uL/min: 0-2 min: 2% ACN (0.1% formic acid), 2-30 min: 2-35% ACN (0.1% formic acid), 30-75min: 35-90% ACN (0.1%formic acid). The eluate was directly infused into an LTQ Velos ion trap mass spectrometer (ThermoScientific, San Jose, CA), which was operated in data dependent mode for all the analyses with the top 6 most abundant ions in each MS scan selected for fragmentation in the LTQ. Peptides were identified by manual interpretation with the assistance of Proteome Discoverer (Thermoscientific). Selected ion chromatograms (SIC) were plotted for TrJ and WT peptides and the intensities used to quantitate differences in digestion efficiency at specific cleavage sites.
Supplementary Material
Acknowledgements
We thank K. Danielle Kelley for technical assistance, Dr. Wade Van Horn for useful discussion, and Professor Jun Li for helpful comments. This work was supported by a postdoctoral fellowship to MS (long-term research grant) from the Toyobo Biotechnology Foundation and by US NIH grants RO1 GM47485 and R01 NS058815 (to CRS), and R01 EY13462 (to KLS). The authors declare no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The NMR assignments for the wild type and Leu16Pro (Trembler-J) forms of PMP22 have been deposited into the BioMagResBank, www.bmrb.wisc.edu. (codes 17454 for WT and 17455 for L16P)
Supplemental information
Supplemental information includes four figures and a table, and can be found with this article online.
References
- Berger P, Niemann A, Suter U. Schwann cells and the pathogenesis of inherited motor and sensory neuropathies (Charcot-Marie-Tooth disease). Glia. 2006;54:243–257. doi: 10.1002/glia.20386. [DOI] [PubMed] [Google Scholar]
- Brand T, Cabrita EJ, Morris GA, Gunther R, Hofmann HJ, Berger S. Residue-specific NH exchange rates studied by NMR diffusion experiments. Journal of Magnetic Resonance. 2007;187:97–104. doi: 10.1016/j.jmr.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Science. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J, Nicholson R, Dickson KM, Orfali W, Naef R, Suter U, Snipes GJ. PMP22 carrying the trembler or trembler-J mutation is intracellularly retained in myelinating Schwann cells. Neurobiol Dis. 2000;7:561–573. doi: 10.1006/nbdi.2000.0323. [DOI] [PubMed] [Google Scholar]
- Czerski L, Vinogradova O, Sanders CR. NMR-Based amide hydrogen-deuterium exchange measurements for complex membrane proteins: development and critical evaluation. J Magn Reson. 2000;142:111–119. doi: 10.1006/jmre.1999.1920. [DOI] [PubMed] [Google Scholar]
- D'Urso D, Prior R, Greiner-Petter R, Gabreels-Festen AA, Muller HW. Overloaded endoplasmic reticulum-Golgi compartments, a possible pathomechanism of peripheral neuropathies caused by mutations of the peripheral myelin protein PMP22. J Neurosci. 1998;18:731–740. doi: 10.1523/JNEUROSCI.18-02-00731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson KM, Bergeron JJ, Shames I, Colby J, Nguyen DT, Chevet E, Thomas DY, Snipes GJ. Association of calnexin with mutant peripheral myelin protein-22 ex vivo: a basis for “gain-of-function” ER diseases. Proc Natl Acad Sci U S A. 2002;99:9852–9857. doi: 10.1073/pnas.152621799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edison AS, Abildgaard F, Westler WM, Mooberry ES, Markley JL. Practical introduction to theory and implementation of multinuclear, multidimensional nuclear magnetic resonance experiments. Methods Enzymol. 1994;239:3–79. doi: 10.1016/s0076-6879(94)39003-7. [DOI] [PubMed] [Google Scholar]
- Eletsky A, Kienhofer A, Pervushin K. TROSY NMR with partially deuterated proteins. J Biomol NMR. 2001;20:177–180. doi: 10.1023/a:1011265430149. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Wider G. TROSY in NMR studies of the structure and function of large biological macromolecules. Curr Opin Struct Biol. 2003;13:570–580. doi: 10.1016/j.sbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Fontanini A, Chies R, Snapp EL, Ferrarini M, Fabrizi GM, Brancolini C. Glycan-independent role of calnexin in the intracellular retention of Charcot-Marie-tooth 1A Gas3/PMP22 mutants. J Biol Chem. 2005;280:2378–2387. doi: 10.1074/jbc.M405104200. [DOI] [PubMed] [Google Scholar]
- Fortun J, Go JC, Li J, Amici SA, Dunn WA, Jr., Notterpek L. Alterations in degradative pathways and protein aggregation in a neuropathy model based on PMP22 overexpression. Neurobiol Dis. 2006;22:153–164. doi: 10.1016/j.nbd.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Fortun J, Li J, Go J, Fenstermaker A, Fletcher BS, Notterpek L. Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. J Neurochem. 2005;92:1531–1541. doi: 10.1111/j.1471-4159.2004.02987.x. [DOI] [PubMed] [Google Scholar]
- Jetten AM, Suter U. The peripheral myelin protein 22 and epithelial membrane protein family. Prog Nucleic Acid Res Mol Biol. 2000;64:97–129. doi: 10.1016/s0079-6603(00)64003-5. [DOI] [PubMed] [Google Scholar]
- Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. Journal of the American Chemical Society. 1992;114:10663–10665. [Google Scholar]
- Korkhov VM, Milan-Lobo L, Zuber B, Farhan H, Schmid JA, Freissmuth M, Sitte HH. Peptide-based interactions with calnexin target misassembled membrane proteins into endoplasmic reticulum-derived multilamellar bodies. J Mol Biol. 2008;378:337–352. doi: 10.1016/j.jmb.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Su YY, Wang H, Li L, Wang Q, Bao L. Transmembrane segments prevent surface expression of sodium channel Nav1.8 and promote calnexin-dependent channel degradation. J Biol Chem. 2010;285:32977–32987. doi: 10.1074/jbc.M110.143024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longsworth LG. The mutual diffusion of light and heavy water. J Phys Chem. 1960;64:1914–1917. [Google Scholar]
- Mobley CK, Myers JK, Hadziselimovic A, Ellis CD, Sanders CR. Purification and initiation of structural characterization of human peripheral myelin protein 22, an integral membrane protein linked to peripheral neuropathies. Biochemistry. 2007;46:11185–11195. doi: 10.1021/bi700855j. [DOI] [PubMed] [Google Scholar]
- Myers JK, Mobley CK, Sanders CR. The peripheral neuropathy-linked Trembler and Trembler-J mutant forms of peripheral myelin protein 22 are folding-destabilized. Biochemistry. 2008;47:10620–10629. doi: 10.1021/bi801157p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naef R, Suter U. Impaired intracellular trafficking is a common disease mechanism of PMP22 point mutations in peripheral neuropathies. Neurobiol Dis. 1999;6:1–14. doi: 10.1006/nbdi.1998.0227. [DOI] [PubMed] [Google Scholar]
- Notterpek L, Ryan MC, Tobler AR, Shooter EM. PMP22 accumulation in aggresomes: implications for CMT1A pathology. Neurobiol Dis. 1999;6:450–460. doi: 10.1006/nbdi.1999.0274. [DOI] [PubMed] [Google Scholar]
- Pareek S, Notterpek L, Snipes GJ, Naef R, Sossin W, Laliberte J, Iacampo S, Suter U, Shooter EM, Murphy RA. Neurons promote the translocation of peripheral myelin protein 22 into myelin. J Neurosci. 1997;17:7754–7762. doi: 10.1523/JNEUROSCI.17-20-07754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge AW, Therien AG, Deber CM. Polar mutations in membrane proteins as a biophysical basis for disease. Biopolymers. 2002;66:350–358. doi: 10.1002/bip.10313. [DOI] [PubMed] [Google Scholar]
- Redfield C. Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins. Methods. 2004;34:121–132. doi: 10.1016/j.ymeth.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Rohl CA, Baldwin RL. Comparison of NH exchange and circular dichroism as techniques for measuring the parameters of the helix-coil transition in peptides. Biochemistry. 1997;36:8435–8442. doi: 10.1021/bi9706677. [DOI] [PubMed] [Google Scholar]
- Sakakura M, Oo-Puthinan S, Moriyama C, Kimura T, Moriya J, Irimura T, Shimada I. Carbohydrate binding mechanism of the macrophage galactose-type C-type lectin 1 revealed by saturation transfer experiments. J Biol Chem. 2008;283:33665–33673. doi: 10.1074/jbc.M804067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. Journal of the American Chemical Society. 1999;121:844–848. [Google Scholar]
- Sanders CR. Post-Integration Misassembly of Membrane Proteins and Disease. In: Tamm LK, editor. Protein-Lipid Interactions. Wiley-VCH; Berlin: 2005. pp. 81–94. [Google Scholar]
- Sanders CR, Ismail-Beigi F, McEnery MW. Mutations of peripheral myelin protein 22 result in defective trafficking through mechanisms which may be common to diseases involving tetraspan membrane proteins. Biochemistry. 2001;40:9453–9459. doi: 10.1021/bi010894f. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Myers JK. Disease-related misassembly of membrane proteins. Annu Rev Biophys Biomol Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- Schleucher J, Schwendinger M, Sattler M, Schmidt P, Schedletzky O, Glaser SJ, Sorensen OW, Griesinger C. A General Enhancement Scheme in Heteronuclear Multidimensional NMR Employing Pulsed-Field Gradients. Journal of Biomolecular NMR. 1994;4:301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter U, Moskow JJ, Welcher AA, Snipes GJ, Kosaras B, Sidman RL, Buchberg AM, Shooter EM. A leucine-to-proline mutation in the putative first transmembrane domain of the 22-kDa peripheral myelin protein in the trembler-J mouse. Proc Natl Acad Sci U S A. 1992;89:4382–4386. doi: 10.1073/pnas.89.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton E, High S, Woodman P. Role of calnexin in the glycan-independent quality control of proteolipid protein. EMBO J. 2003;22:2948–2958. doi: 10.1093/emboj/cdg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler AR, Liu N, Mueller L, Shooter EM. Differential aggregation of the Trembler and Trembler J mutants of peripheral myelin protein 22. Proc Natl Acad Sci U S A. 2002;99:483–488. doi: 10.1073/pnas.012593399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler AR, Notterpek L, Naef R, Taylor V, Suter U, Shooter EM. Transport of Trembler-J mutant peripheral myelin protein 22 is blocked in the intermediate compartment and affects the transport of the wild-type protein by direct interaction. J Neurosci. 1999;19:2027–2036. doi: 10.1523/JNEUROSCI.19-06-02027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn LJ, Baas F, Wolterman RA, Hoogendijk JE, van den Bosch NH, Zorn I, Gabreels-Festen AW, de Visser M, Bolhuis PA. Identical point mutations of PMP-22 in Trembler-J mouse and Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;2:288–291. doi: 10.1038/ng1292-288. [DOI] [PubMed] [Google Scholar]
- Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. Journal of the American Chemical Society. 1998;120:10778–10779. [Google Scholar]
- Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE. An adaptable standard for protein export from the endoplasmic reticulum. Cell. 2007;131:809–821. doi: 10.1016/j.cell.2007.10.025. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Wu DH, Chen AD, Johnson CS. An improved diffusion-ordered spectroscopy experiment incorporating bipolar-gradient pulses. Journal of Magnetic Resonance Series A. 1995;115:260–264. [Google Scholar]
- Zhou NE, Zhu BY, Sykes BD, Hodges RS. Relationship between amide proton chemical-shifts and hydrogen-bonding in amphipathic alpha-helical peptides. Journal of the American Chemical Society. 1992;114:4320–4326. [Google Scholar]
- Zhu G, Kong XM, Sze KH. Gradient and sensitivity enhancement of 2D TROSY with water flip-back, 3D NOESY-TROSY and TOCSY-TROSY experiments. Journal of Biomolecular NMR. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.