Abstract
Ligament heals in a synchronized and complex series of events. The remodeling process may last months or years. Experimental evidence suggests the damaged ligament does not recover its normal functional properties. Specific mechanisms to prevent scar formation and to regenerate the original mechanical function remain elusive but likely involve regulation of creeping substitution. Creeping substitution creates a larger hypercellular, hypervascular, and disorganized granulation tissue mass that results in an inefficient and nonregenerative wound healing process for the ligament. Control of creeping substitution may limit the extent of this tissue compromise and reduce the time necessary for healing. The objective of this study is to better understand the mechanism behind scar formation by identifying the extracellular matrix factors and other unique genes of interest differentially expressed during rat ligament healing via microarray. For this study, rat medial collateral ligaments were either surgically transected or left intact. Ligaments were collected at day 3 or 7 postinjury and used for microarray, quantitative PCR, and/or immunohistochemistry. Results were compared with the normal intact ligament. We demonstrate that early ligament healing is characterized by the modulation of several inflammatory and extracellular matrix factors during the first week of injury. Specifically, a number of matrix metalloproteinases and collagens are differentially and significantly expressed during early ligament healing. Additionally, we demonstrate the modulation of three novel genes, periostin, collagen-triple helix repeat containing-1, and serine protease 35 in our ligament healing model. Together, control of granulation tissue creeping substitution and subsequent downstream scar formation is likely to involve these factors.
Keywords: collagen, matrix metalloproteinase, periostin
ligament healing involves a complex, coordinated series of events that results in a ligament that is more scar-like in character than the native tissue. The repair process may extend from months to years. Experimental evidence suggests the injured ligament incompletely recovers its original mechanical properties (25, 28). In an effort to develop an efficient repair process, researchers have tested a number of treatments including tissue engineering approaches, nonsteroidal anti-inflammatory drugs, and ultrasonic or electrical stimulation (28, 35). All show promise, but an incomplete understanding of the healing process makes optimal treatment regimes elusive. With tissue engineering and regenerative medicine, it is essential to understand the normal healing process in a ligament, thereby providing a basis to formulate and evaluate innovative new treatments.
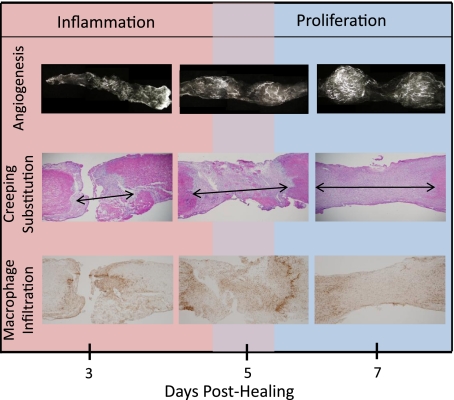
Previous work from our laboratory (8) established a highly active cellular and vascular scenario within the first week after ligament injury. Specifically, macrophage infiltration crested between days 3–5 while blood vessel accumulation peaked between days 7–11 postinjury (Fig. 1). Creeping substitution of damaged tissue into normal uninjured ligament, resulting in the expansion of the original wound size, was also evident during this time (8). The reduction of creeping substitution during inflammation and early proliferation may limit downstream scar formation, but the genetic players remain largely undefined.
Fig. 1.
Typical angiogenic, granulation tissue and macrophage response of a wounded medial collateral ligament (MCL) at 3, 5, and 7 days postinjury. Microangiography of the healing MCL demonstrates low blood vessel infiltration 3 days postinjury (top). By day 5 and 7, blood vessels are prominent. Granulation tissue forms by day 3 and continues to expand within the ligament beyond day 7 (i.e., creeping substitution; middle). Macrophages also appear early during healing and peak around day 5 and 7 (bottom).
Matrix metalloproteinases (MMP) regulate normal tissue remodeling of the extracellular matrix (ECM) by degrading collagen and regulating cell recruitment into tissues. Neutrophils, macrophages, and fibroblasts release various MMPs, modulating inflammation and tissue degradation. During pathologic inflammatory conditions, excessive proteolytic activity is produced to facilitate tissue injury; rheumatoid arthritis and osteoarthritis are associated with increased MMP activity and subsequent collagen and proteoglycan degradation. Control of creeping substitution and subsequent scar formation may therefore modify collagen degradation by the MMPs. Therefore, the objective of this study is to better delineate the genetic factors involved during early ligament healing via microarray analysis. Specifically, this study focuses on identification of MMPs and collagens during early healing.
MATERIALS AND METHODS
Animal model.
This study was approved by the University of Wisconsin Institutional Animal Use and Care Committee. Forty-two skeletally mature Wistar rats were used as an animal model for ligament healing. Animals were randomly placed in one of three groups (n = 14 animals/group): intact control, day 3 postinjury, or day 7 postinjury. Rats were anesthetized via isofluorane. A surgically transected, rather than torn, medial collateral ligament (MCL) was used as an experimental model to create a uniform defect for healing. Rats were subjected to bilateral MCL transection using sterile techniques (n = 37 animals). A 1-cm skin incision was made over the medial aspect of each stifle, and the subcutaneous tissue was dissected to expose the sartorius muscle and underlying MCL. The midpoint of the MCL was completely transected. Severed ends were not sutured and allowed to retract. The muscular, subcutaneous, and subdermal tissue layers were each closed with 4–0 Dexon suture. The remaining 14 animals underwent no transection and served as age-matched intact controls. All animals were allowed unrestricted cage movement immediately after surgery. At day 3 or 7 postinjury (n = 14 animals/day), the MCLs were carefully dissected, measured, weighed, and immediately placed in liquid nitrogen. MCLs from each time point were divided into three pools (3 MCLs/pool) and used for microarray analysis. The contralateral MCLs were used for quantitative PCR (qPCR). MCLs from the remaining five animals per day were snap frozen in optimal cutting temperature media and used for immunohistochemistry.
RNA isolation for microarray and qPCR analysis.
Total RNA was isolated from the MCLs using methods similar to Reno et al. (56), combining the TRIzol (Invitrogen, Carlsbad, CA) method with column fractionalization steps of the RNeasy total RNA kit (Qiagen, Valencia, CA). Tissue was homogenized using a PowerGen 500 homogenizer (Fisher Scientific, Pittsburgh, PA). Subsequent to RNA isolation and yield and purity of RNA were quantified by nanodrop spectrophotometric measurement at 260 nm (Nanodrop Technologies, Wilmington, DE).
Purification and preparation of RNA for affymetrix GeneChip analysis.
Nucleic acid qc, labeling, and hybridizations were performed by the Gene Expression Center, a core research facility within the University of Wisconsin Biotechnology Center. The quality and quantity of each RNA sample were analyzed on the Agilent 2100 BioAnalyzer using the RNA6000 NanoChip and the NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE), respectively. Total RNA was converted to double-stranded cDNA using an oligo dT primer containing the T7 RNA polymerase promoter (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-T24-3′). Briefly, first-strand synthesis was performed by incubating 3–7 μg of total RNA with 100 pmol of T7-d(T)24 primer (University of Wisconsin-Madison DNA Synthesis Facility), 1× first-stand buffer (Invitrogen), 10 mM DTT (Invitrogen), 200 U SuperScript II RT (Invitrogen), and 500 μM dNTPs (GE Healthcare Life Sciences) for 1 h at 37°C. Second-strand synthesis was accomplished by incubation with 1× second-strand buffer, 200 μM dNTPs, 10 U DNA ligase (Invitrogen), 40 U DNA polymerase I (Invitrogen), and 2 U RNaseH (Invitrogen) for 2 h at 16°C followed by a 5-min incubation with 10 U T4 polymerase (Invitrogen). Double-stranded cDNA was purified using the GeneChip Sample Cleanup Module kit (Affymetrix, Santa Clara, CA). A biotin-labeled in vitro transcription reaction was performed using the cDNA template and the Affymetrix 3′-amplification IVT labeling kit. Briefly, double-stranded cDNA was incubated with 1× IVT labeling buffer, 1× labeling NTP mix, and 1× T7 labeling enzyme mix for 16 h at 37°C to yield cRNA. cRNA was purified using the GeneChip Sample Cleanup Module. cRNA yield was determined by absorbance at 260 nM. The cRNA was fragmented at 0.5 μg/μl final concentration in 1× fragmentation buffer (40 mM Tris-acetate pH 8.1, 100 mM potassium acetate, and 30 mM magnesium acetate) for 32 min at 94°C. The size range of the cRNA before (0.1 kb and longer) and after (35–200 base fragments) was checked by agarose gel electrophoresis.
Hybridization and washing.
The Affymetrix Rat GeneChips (Affymetrix) contains >31,000 probe sets representing 28,700 rat genes. A total of nine chips were hybridized using three biological triplicates (containing 3 ligaments per sample) at three time points (intact and days 3 and 7). Microarray hybridization was performed by the University of Wisconsin Biotechnology Gene Expression Center (Madison, WI). GeneChips (Affymetrix) were hybridized with biotin labeled cRNA in 300 μl in the presence of 1× hybridization buffer [100 mM MES, 1 M NaCl (Ambion, Austin, TX), 20 mM EDTA (Ambion), and 0.01% Tween-20 (Pierce Chemical)], 50 pM control oligonucleotide B2 (Affymetrix), 0.1 mg/ml herring sperm DNA (Promega, Madison, WI), 0.5 mg/ml acetylated BSA (Invitrogen), and 1× eukaryotic hybridization controls (bioB, bioC, bioD, and cre at 1.5, 5, 25, and 100 pM, respectively; Affymetrix) for 16 h at 45°C on a rotisserie at 60 rpm. Before application to the GeneChip, samples were heated at 95°C for 5 min, followed by incubation at 45°C for 5 min and spun at 14,000 g for 5 min. Following hybridization, the labeled samples were removed from the GeneChip, stored in the appropriate vial at −20°C, and immediately filled with nonstringent buffer A that contains 6× SSPE (Ambion), 0.01% Tween 20. All GeneChips were postprocessed using the automated Affymetrix GeneChip Fluidics Station 450.
The postprocessing protocol of the Rat 2.0 Genome GeneChip included the following: one wash of 10 cycles with 2 mixes/cycle of nonstringent buffer A at 25°C; one wash of 4 cycles comprised of 15 mixes/cycle with stringent buffer B (100 mM MES, 0.1 M NaCl, and 0.01% Tween 20) at 50°C; staining of the probe array for 10 min at 25°C in streptavidin-phycoerythrin (SAPE) solution [1× MES stain buffer (100 mM MES, 1 M NaCl, 0.05% Tween 20), 2 mg/ml acetylated BSA, and 10 μg/ml SAPE (Molecular Probes)]; poststaining consisting of 10 wash cycles of four mixes/cycle with nonstringent buffer A at 25°C; a second staining of the probe array for 10 min in antibody solution [1× MES stain buffer, 2 mg/ml acetylated BSA, 0.1 mg/ml Normal Goat IgG (Sigma-Aldrich, St. Louis, MO), 3 μg/ml biotinylated antibody (Vector Laboratories)]; a third stain of the probe array for 10 min in SAPE solution at 25°C; and a final wash of 15 cycles of 4 mixes/cycle with nonstringent buffer A at 30°C.
Probe array scan and data acquisition.
To quantify the fluorescent signal from each feature on the GeneChip, all GeneChips were scanned at a wavelength of 570 nm, using the GC3000 G7 scanner. Fluorescent signals corresponding to hybridization intensities were analyzed with the Affymetrix GCOS software.
Microarray analysis.
Quantification of GeneChips was performed by Affymetrix GeneChip Operating Software (AGCC version 2.0). Microarray data were log transformed [log (base 2)], and the log ratio between the intact ligament with day 3 or day 7 was calculated and expressed as fold change. A gene was considered up or downregulated if the day 3 or day 7 samples were at least twofold different than the intact controls. Additionally, data were analyzed using the two-sample Welch t-test to determine significance per gene. To minimize the false discovery rate, the Storey q-value method was performed, and significance was based on q < 0.01. Any q value ≤0.01 per gene was considered significant. Paired t-tests were used to evaluate differences between intact and day 3, intact and day 7, or day 3 and day 7. A critical value of 0.05 was considered as the criterion to select a significant fold change in gene expression between days. Data are presented as fold change of the means of day 3 and day 7 from the intact ligament values (fold change ± SD or fold change ± SE). To validate the expression patterns of microarray analysis with qPCR, the qPCR data was first normalized to the housekeeping gene β-actin. A fold change was then determined between the intact and day 3 samples or the intact and day 7 samples. Paired t-tests were performed to determine qPCR significance between days.
Functional annotation.
Microarray data were functionally grouped using the Database for Annotation, Visualization, and Integrated Discovery (D.A.V.I.D.; http://david.abcc.ncifcrf.gov), integrating functional genomic annotations with intuitive graphical summaries. With the use of the Affymetrix probes as identifiers, the samples were subjected to functional annotation clustering based on function and gene ontology. The categories of interest were further based on the enrichment score of the gene group. The enrichment score, determined by the minus log transformation of the geometric mean P values from all enriched annotation terms, ranks the biological significance of a gene cluster based on the scores of all the annotation terms.
RT and qPCR.
RNA was DNAase treated and reverse transcribed into cDNA using the Superscript III first strand kit (Invitrogen). RNA was added to 50 ng/μl random hexamer, annealing buffer, and RNAse/DNase free water, followed by 2× first-strand reaction mix and RNase enzyme mix. Reverse transcribed samples were diluted 2.5 ng/μl.
To validate microarray genes of interest, quantitative PCR was performed on ligament samples. Primers for collagen type I (Col1a1, Col1a2), type III (Col3a1), type V (Col4a3), type VI (Col6a1, Col6a3), type XI (Col11a1), type XXVII (Col27a1), collagen triple helix repeat containing-1 (Ctrhc1), periostin (Postn), serine protease 35 (Prss35), Mmp3, Mmp11, Mmp12, Mmp13, Mmp14, Mmp19, Timp1, and β-actin primers were utilized (Table 1). Primer sets were either obtained from published reports (11, 18, 26, 33, 36, 64, 66, 74, 77) or designed from sequences available from Genbank from Blast (Basic Local Alignment Search Tool; http://www.ncbi.nlm.nih.gov:80/BLAST).
Table 1.
List of primer sequences for quantitative PCR, the annealing temperature for each primer, and where the primers were obtained
| Target Gene | Sequence | AT | Source |
|---|---|---|---|
| β-Actin | FP: 5′-ATCGCTGACAGGATGCAGAAG-3′ | 58 | Zhu and Altmann (77) |
| RP: 5′-TCAGGAGGAGCAATGATCTTGA-3′ | |||
| Collagen I | FP: 5′-CCCAAGGAAAAGAAGCACGTC-3′ | 55 | Li et al. (26) |
| RP: 5′-AGGTCAGCTGGATAGCGACATC-3′ | |||
| Collagen I | FP: 5′-TACAACGCAGAAGGGGTGTC-3′ | 55 | Tashiro et al. (66) |
| RP: 5′-CCTCAGCAACAAGTTCGACG-3′ | |||
| Collagen III | FP: 5′-ATGGTGGCTTTCAGTTCAGC-3 | 55 | Tashiro et al. (66) |
| RP: 5′-TGTCTTGCTCCATTCACCAG-3′ | |||
| Collagen V | FP: 5′-AGGGACCAACTGGGAAGAGT-3′ | 55 | Li et al. (26) |
| RP: 5′-AAAGTCAGAGGCAGCCAGAT-3′ | |||
| Collagen VI | FP: 5′-AACGTGTGGCTAACCTGGAG-3′ | 53 | Designed |
| RP: 5′-TATTATCAAGCTGCTCCGCC-3′ | |||
| Collagen VI | FP: 5′-TGCTCACCTGAGGCGCTTGC-3′ | 61 | Designed |
| RP: 5′-CCGCCTGCCTCCGTGAAGTG-3′ | |||
| Collagen XI | FP: 5′-TCCGCCTGGAGCTTCCGGTT-3′ | 61 | Designed |
| RP: 5′-GACCCTGAGGTCCGACCGGG-3′ | |||
| Collagen XXVII | FP: 5′-GTCAGATTGGAGCAGGGGTA-3′ | 55 | Designed |
| RP: 5′-ATGCTCTGGATGAGGTTGCT-3′ | |||
| Cthrc1 | FP: 5′-TTCAGGACCTCTTCCCATTG-3′ | 54 | Designed |
| RP: 5′-CACAGAGTCCTTCCACGGAG-3′ | |||
| Mmp3 | FP: 5′-CTCTCTCAAGATGATGTAGATGGTATTC-3′ | 53 | Maeda et al. (33) |
| RP: 5′-AGCTACACATTGGTAAGGTCTCAG-3′ | |||
| Mmp11 | FP: 5′-GGAGACTATTGGCGTTTCCA-3′ | 53 | Designed |
| RP: 5′-AGTAGGCATAGCCCTCAGCA-3′ | |||
| Mmp12 | FP: 5′-CTGGGCAACTGGACACCT-3′ | 60 | Wasserman et al. (74) |
| RP: 5′-CTACATCCGCACGCTTCA-3′ | |||
| Mmp13 | FP: 5′-GCATGAAAACTGTGGGGAGT-3′ | 58 | Fang et al. (11) |
| RP: 5′-AGCTGAAATCTTGCCTTGGA-3′ | |||
| Mmp14 | FP: 5′-GAGGGTCATGAGAAGCAGGC-3′ | 56 | Strup-Perrot et al. (64) |
| RP: 5′-TCAAAGGGTGTGCTGTCGC-3′ | |||
| Mmp19 | FP: 5′-AGGATACTGGCAATGGGATG-3′ | 53 | Designed |
| RP: 5′-TAGCTGCTGAGGGTTGGTCT-3′ | |||
| Timp1 | FP: 5′-GTTCCCTGGCATAATCTGAG-3′ | 52 | Johnson et al. (18) |
| RP: 5′-GGGATGAGGATCTGATCTGT-3′ | |||
| Postn | FP: 5′-AGGAGCCGTGTTTGAGACCAT-3′ | 54 | Hiroi et al. |
| RP: 5′-CGGTGAAAGTGGTTTGCTGTTT-3′ | |||
| Prss35 | FP: 5′-TAGGAGAAGGAGGCAGGTGTATGG-3′ | 53 | Miyakoshi et al. (37) |
| RP: 5′-TAAGCACAGTTGGCAGCGTTTCCG-3′ |
AT, annealing temperature; FP, forward primer; RP, reverse primer.
qPCR was performed using a Bio-Rad thermocycler (Bio-Rad, Hercules, CA). All reactions were carried out using Bio-Rad Sybr Green Supermix (Bio-Rad). Assessment of genes was performed by including negative controls in each qPCR experiment, examining PCR melt curves to ensure specificity, and verifying the primer efficiency was within 90–110% based on the standard curve. The cDNA of the target gene was also normalized to the housekeeping gene β-actin.
Immunohistochemistry.
Immunohistochemistry was performed on frozen sections using mouse monoclonal or rabbit polyclonal antibodies. Sections were fixed with acetone, exposed to 3% hydrogen peroxide to eliminate endogenous peroxidase activity, and blocked using Background Buster (Innovex Biosciences, Richmond, CA). Sections were then incubated for 2 h with rabbit polyclonal periostin (1:1,000; Biovendor, Candler, NC) and mouse monoclonal CTHRC1 (1:500; ab72527; Abcam, Cambridge, MA). After primary antibody exposure, sections were treated with biotin and streptavidin conjugated to horseradish peroxidase using the Stat Q staining kit (Innovex Biosciences). The bound antibody complex was then visualized using DAB. Stained sections were dehydrated, cleared, coverslipped, and viewed under light microscopy. Negative controls omitting the primary antibody were included with each experiment. Positive controls of gut or spleen were also included.
RESULTS
Microarray results.
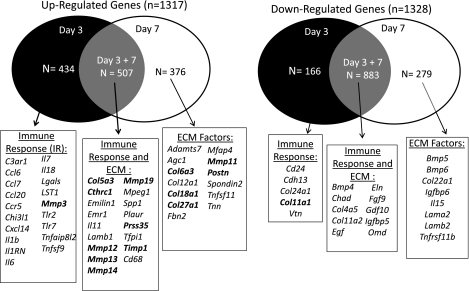
Microarray analysis was conducted to identify differences in the ligament transcriptome between day 3 and day 7 postinjury with the uninjured ligament. A total of 31,000 probe sequences were analyzed. Overall, 2,652 genes from day 3 and/or day 7 were at least twofold greater than the intact controls and achieved a q value of 0.01 or less (Table 2). Of those genes, 2,046 genes were significantly regulated at day 7. Of the day 7 significant targets, 1,161 were downregulated and 883 were upregulated. A total of 1,991 genes were different at day 3 from the intact ligament, with 1,050 upregulated targets and 941 downregulated. Those day 3 and/or day 7 genes significantly different from the intact were then subjected to D.A.V.I.D. and clustered via functionality, resulting in 238 groups. Table 3 lists functional clusters of interest. Seventeen clusters of interest were identified with an enrichment score ranging from 3.15 to 0.15. Functional groups include genes involved in ECM control, metalloenzyme/peptidase/metallopeptidase/proteolysis regulation, immune cell activation/proliferation/differentiation/regulation, proteoglycan/glycoprotein binding, regeneration/limb morphogenesis/wound response, endocytosis/phagocytosis, angiogenesis, cell chemotaxis, and innate response regulation. To further identify relevant gene clusters based on day of injury, genes were selected only if they were significant on one day but not the other and subjected to D.A.V.I.D. analysis. A total of 600 and 655 genes were exclusively regulated at day 3 and day 7, respectively. The remaining 1,391 genes were modulated at both day 3 and 7. The top five functional groups from each day are listed in Tables 4 and 5. Functional annotation grouping via D.A.V.I.D. indicated that many genes are involved in immune regulation at day 3 (Table 4) whereas the day 7 exclusive genes are primarily involved in ECM regulation and angiogenesis (Table 5). Of the 600 genes modulated at day 3, 434 were upregulated and 166 were downregulated compared with the uninjured ligament (Fig. 1). In comparison, 376 genes were upregulated and 279 genes were downregulated in the day 7 group (Fig. 2). Genes of interest are listed along the Venn diagrams and include inflammatory factors, growth factors, and ECM components, including collagens and MMPs.
Table 2.
Number of genes significantly regulated during early wound healing
| Day Postinjury | Total | Upregulated | Downregulated |
|---|---|---|---|
| Day 3 | 1,991 | 941 | 1,050 |
| Day 7 | 2,046 | 883 | 1,161 |
Table 3.
Functional Annotation Clusters of interest obtained from D.A.V.I.D. analysis
| GO Term | Functional Annotation Clustering | ES | Gene Number | P Value |
|---|---|---|---|---|
| Extracellular matrix factors | 3.15 | |||
| GOTERM_CC_FAT | Proteinaceous extracellular matrix | 34 | 8.10E-06 | |
| GOTERM_CC_FAT | Extracellular matrix | 37 | 8.50E-06 | |
| GOTERM_CC_FAT | Extracellular region | 102 | 7.03E-03 | |
| GOTERM_CC_FAT | Extracellular region part | 63 | 1.10E-02 | |
| GOTERM_CC_FAT | Extracellular matrix part | 13 | 3.30E-02 | |
| Immune cell activation/proliferation | 1.59 | |||
| GOTERM_BP_FAT | T-cell activation | 16 | 8.60E-03 | |
| GOTERM_BP_FAT | Lymphocyte proliferation | 8 | 1.20E-02 | |
| GOTERM_BP_FAT | Mononuclear cell proliferation | 8 | 1.40E-02 | |
| GOTERM_BP_FAT | Leukocyte proliferation | 8 | 1.40E-02 | |
| GOTERM_BP_FAT | Leukocyte proliferation | 20 | 1.50E-02 | |
| GOTERM_BP_FAT | Lymphocyte activation | 6 | 4.40E-02 | |
| GOTERM_BP_FAT | T-cell proliferation | 3 | 6.60E-02 | |
| GOTERM_BP_FAT | Activated T-cell proliferation | 4 | 2.00E-01 | |
| GOTERM_BP_FAT | Leukocyte adhesion | |||
| Metalloenzyme regulator activity | 1.56 | |||
| INTERPRO | Netrin domain proteinase inhibitor I35, tissue inhibitor of | 6 | 1.80E-03 | |
| INTERPRO | Metalloproteinase | 3 | 2.60E-02 | |
| PIR_SUPERFAMILY | PIRSF001636:meaalloproteinase inhibitor | 3 | 2.80E-02 | |
| SMART | NTR | 3 | 2.90E-02 | |
| GOTERM_MF_FAT | Metalloenzyme inhibitor activity | 3 | 6.40E-02 | |
| GOTERM_MF_FAT | Metalloendopeptidase inhibitor activity | 3 | 6.40E-02 | |
| GOTERM_MF_FAT | Metalloenzyme regulator activity | 3 | 8.50E-02 | |
| Immune cell activation/differentiaion | 1.39 | |||
| GOTERM_BP_FAT | Leukocyte activation | 26 | 2.30E-03 | |
| GOTERM_BP_FAT | Cell activation | 27 | 5.80E-03 | |
| GOTERM_BP_FAT | T-cell activation | 16 | 8.60E-03 | |
| GOTERM_BP_FAT | Lymphocyte activation | 20 | 1.50E-02 | |
| GOTERM_BP_FAT | T-cell differentiation | 10 | 3.90E-02 | |
| GOTERM_BP_FAT | Immune system development | 26 | 4.70E-02 | |
| GOTERM_BP_FAT | Hemopoietic or lymphoid organ development | 25 | 4.80E-02 | |
| GOTERM_BP_FAT | Hemopoiesis | 23 | 5.00E-02 | |
| GOTERM_BP_FAT | alpha-beta T-cell activation | 5 | 5.40E-02 | |
| GOTERM_BP_FAT | alpha-beta T-cell differentiation | 4 | 9.20E-02 | |
| GOTERM_BP_FAT | Lymphocyte differentiation | 10 | 2.10E-02 | |
| GOTERM_BP_FAT | Leukocyte differentiation | 11 | 3.00E-01 | |
| GOTERM_BP_FAT | Immune response | 27 | 3.40E-01 | |
| Proteoglycan/glycoprotein binding | 1.26 | |||
| GOTERM_MF_FAT | Heparing sulfate proteoglycan binding | 4 | 2.20E-02 | |
| GOTERM_MF_FAT | Proteoglycan binding | 4 | 3.00E-02 | |
| GOTERM_MF_FAT | Glycoprotein binding | 5 | 2.40E-01 | |
| Regeneration | 1.11 | |||
| GOTERM_BP_FAT | Regeneration | 15 | 3.60E-02 | |
| GOTERM_BP_FAT | Developmental growth | 14 | 9.10E-02 | |
| GOTERM_BP_FAT | Tissue regeneration | 7 | 1.00E-01 | |
| GOTERM_BP_FAT | Growth | 20 | 1.10E-01 | |
| Endocytosis/phagocytosis | 0.98 | |||
| GOTERM_BP_FAT | Regulation of endocytosis | 9 | 6.00E-02 | |
| GOTERM_BP_FAT | Positive regulation of endocytosis | 6 | 7.30E-02 | |
| GOTERM_BP_FAT | Regulation of vesicle-mediated transport | 13 | 7.40E-02 | |
| GOTERM_BP_FAT | Regulation of phagocytosis | 4 | 1.40E-01 | |
| GOTERM_BP_FAT | Positive regulation of phagocytosis | 3 | 2.70E-01 | |
| Limb morphogenesis | 0.97 | |||
| GOTERM_BP_FAT | Embryonic limb morphogenesis | 11 | 4.60E-02 | |
| GOTERM_BP_FAT | Embryonic appendage morphogenesis | 11 | 4.60E-02 | |
| GOTERM_BP_FAT | Appendage morphog enesis | 11 | 9.20E-02 | |
| GOTERM_BP_FAT | Limb morphog enesis | 11 | 9.20E-02 | |
| GOTERM_BP_FAT | Appendage development | 11 | 1.20E-01 | |
| GOTERM_BP_FAT | Limb development | 11 | 1.20E-01 | |
| GOTERM_BP_FAT | Embryonic morphogenesis | 19 | 7.10E-01 | |
| Leukocyte/lymphocyte activation | 0.9 | |||
| GOTERM_BP_FAT | Macrophage activation | 4 | 5.30E-02 | |
| GOTERM_BP_FAT | Myeloid leukocyte activation | 7 | 6.90E-02 | |
| GOTERM_BP_FAT | Macrophage activation during immune response | 3 | 8.80E-02 | |
| GOTERM_BP_FAT | Leukocyte activation during immune response | 6 | 9.00E-02 | |
| GOTERM_BP_FAT | Cell activation during immune response | 6 | 9.00E-02 | |
| GOTERM_BP_FAT | T-cell activation during immune response | 3 | 1.90E-01 | |
| GOTERM_BP_FAT | Lymphocyte activation during immune response | 3 | 3.00E-01 | |
| GOTERM_BP_FAT | Myeloid cell activation during immune response | 3 | 3.80E-01 | |
| Peptidase/metallopeptidase activity | 0.86 | |||
| INTERPRO | Peptidase M10A, matrix metallopeptidase | 4 | 3.50E-02 | |
| PIR_SUPERFAMILY | PIRSF001191: peptidase_M10A_matrix | 4 | 4.00E-02 | |
| INTERPRO | Peptidoglycan binding-like | 4 | 5.50E-02 | |
| INTERPRO | Hemopexin/matrixin, repeat | 4 | 7.90E-02 | |
| INTERPRO | Hemopexin/matrixin, repeat | 4 | 7.90E-02 | |
| SMART | HX | 4 | 7.90E-02 | |
| INTERPRO | Peptidase, metallopeptidases | 4 | 9.10E-02 | |
| SMART | ZnMc | 4 | 1.10E-01 | |
| INTERPRO | Peptidase M10A and M12B, matrixin, and damalysin | 4 | 1.40E-01 | |
| INTERPRO | Hemopexin/matrixin, conserved site | 3 | 2.20E-01 | |
| INTERPRO | Peptidase M, neutral zinc metallopeptidases zinc-binding site | 4 | 5.30E-01 | |
| GOTERM_MF_FAT | Metalloendopeptidase activity | 5 | 7.10E-01 | |
| GOTERM_MF_FAT | Metallopeptidase activity | 7 | 8.80E-01 | |
| Angiogenesis | 0.83 | |||
| GOTERM_BP_FAT | Blood vessel morphogenesis | 20 | 5.50E-02 | |
| GOTERM_BP_FAT | Vasculature development | 24 | 5.60E-02 | |
| GOTERM_BP_FAT | Blood vessel development | 23 | 7.30E-02 | |
| GOTERM_BP_FAT | Morphogenesis of a branching structure | 12 | 1.50E-01 | |
| GOTERM_BP_FAT | Angiogenesis | 13 | 1.50E-01 | |
| GOTERM_BP_FAT | Branching morphogenesis of a tube | 9 | 2.20E-01 | |
| GOTERM_BP_FAT | Tube morphogenesis | 14 | 2.40E-01 | |
| GOTERM_BP_FAT | Tube development | 21 | 2.40E-01 | |
| GOTERM_BP_FAT | Epithelium development | 17 | 5.90E-01 | |
| Immune response regulation | 0.62 | |||
| GOTERM_BP_FAT | Positive regulation of defense response | 8 | 9.10E-02 | |
| GOTERM_BP_FAT | Immune response-regulating signal transduction | 7 | 1.50E-01 | |
| GOTERM_BP_FAT | Immune response-regulating cell surface receptor signaling pathway | 6 | 1.70E-01 | |
| GOTERM_BP_FAT | Activation of immune response | 9 | 1.90E-01 | |
| GOTERM_BP_FAT | Immune response-activating signal transduction | 6 | 2.30E-01 | |
| GOTERM_BP_FAT | Immune response-activating cell surface receptor signaling pathway | 5 | 2.90E-01 | |
| GOTERM_BP_FAT | Positive regulation of response to stimulus | 18 | 3.10E-01 | |
| GOTERM_BP_FAT | Antigen receptor-mediated signaling pathway | 4 | 3.80E-01 | |
| GOTERM_BP_FAT | Positive regulation of immune respone | 11 | 3.90E-01 | |
| GOTERM_BP_FAT | T-cell receptor signaling pathway | 3 | 4.60E-01 | |
| Cell chemotaxis | 0.47 | |||
| GOTERM_BP_FAT | Leukocyte chemotaxis | 5 | 2.50E-01 | |
| GOTERM_BP_FAT | Cell chemotaxis | 5 | 3.00E-01 | |
| GOTERM_BP_FAT | Taxis | 8 | 3.10E-01 | |
| GOTERM_BP_FAT | Chemotaxis | 8 | 3.10E-01 | |
| GOTERM_BP_FAT | Leukocyte migration | 6 | 3.20E-01 | |
| GOTERM_BP_FAT | Locomotory behavior | 14 | 6.20E-01 | |
| Innate response regulation | 0.45 | |||
| GOTERM_BP_FAT | Positive regulation of defense response | 8 | 9.10E-02 | |
| GOTERM_BP_FAT | Positive regulation of innate immune response | 3 | 6.20E-01 | |
| GOTERM_BP_FAT | Regulation of innate immune response | 3 | 7.80E-01 | |
| Immune activation | 0.29 | |||
| GOTERM_BP_FAT | Activation of immune response humoral immune response mediated by | 9 | 1.90E-01 | |
| GOTERM_BP_FAT | Circulating immunoglobulin | 4 | 2.00E-01 | |
| GOTERM_BP_FAT | Humoral immune response | 5 | 3.50E-01 | |
| GOTERM_BP_FAT | Immunoglobulin mediated immune response | 5 | 4.20E-01 | |
| GOTERM_BP_FAT | Innate immune response | 7 | 4.40E-01 | |
| GOTERM_BP_FAT | B-cell-mediated immunity | 5 | 4.60E-01 | |
| GOTERM_BP_FAT | Adaptive immune response | 6 | 4.70E-01 | |
| Adaptive immune response based on somateic | ||||
| Recombination of immune receptors built from | ||||
| GOTERM_BP_FAT | Immunoglobulin superfamily domains | 6 | 4.70E-01 | |
| GOTERM_BP_FAT | Protein maturation by peptide bond cleavage | 6 | 5.10E-01 | |
| GOTERM_BP_FAT | Acute inflammatory respone | 7 | 5.90E-01 | |
| GOTERM_BP_FAT | Lymphocyte-mediated immunity | 5 | 6.00E-01 | |
| GOTERM_BP_FAT | Complement activation | 3 | 6.60E-01 | |
| GOTERM_BP_FAT | Immune effecctor process | 8 | 6.60E-01 | |
| Activation of plasma proteins involved in acute | ||||
| GOTERM_BP_FAT | Inflammatory response | 3 | 6.80E-01 | |
| GOTERM_BP_FAT | Leukocyte-mediated immunity | 5 | 7.50E-01 | |
| GOTERM_BP_FAT | Protein processing | 6 | 7.90E-01 | |
| GOTERM_BP_FAT | Protein maturation | 6 | 8.20E-01 | |
| KEGG_PATHWAY | Complement and coagulation cascades | 4 | 8.80E-01 | |
| Wound response | 0.2 | |||
| GOTERM_BP_FAT | Response to wounding | 31 | 4.70E-01 | |
| GOTERM_BP_FAT | Inflammatory response | 14 | 7.30E-01 | |
| GOTERM_BP_FAT | Defense response | 22 | 7.40E-01 | |
| Proteolysis | 0.15 | |||
| GOTERM_MF_FAT | Cysteine-type peptidase activity | 9 | 3.00E-01 | |
| GOTERM_MF_FAT | Endopeptidase activity | 18 | 7.90E-01 | |
| GOTERM_MF_FAT | Peptidase activity peptidase activity, acting on l-amino acid | 25 | 8.90E-01 | |
| GOTERM_MF_FAT | Peptides | 23 | 9.10E-01 | |
| GOTERM_BP_FAT | Proteolysis | 39 | 9.20E-01 |
Significantly differentiated day 3 and day 7 genes were combined and analyzed via Database for Annotation, Visualization, and Integrated Discovery (D.A.V.I.D.) based on gene ontology (GO) terms as identified through biological processes (GOTERM_BP), cellular components (GOTERM_CC), and molecular function (GOTERM_MF), as well as via molecular interactions, predictive protein signatures, biochemical pathways, and protein domains (KEGG Pathway, INTERPRO, SMART, PIR Superfamily). ES, enrichment score.
Table 4.
Functional annotation clustering of the top 5 day 3-specific genes obtained from D.A.V.I.D. analysis
| Annotation Cluster | Functional Annotation Clustering | Gene Numbers |
|---|---|---|
| 1 | Enrichment score: 11.21 | |
| Immune response | 46 | |
| Defense response | 39 | |
| Immune effector process | 21 | |
| Positive regulation of immune system process | 28 | |
| 2 | Enrichment score: 7.83 | |
| Defense response | 39 | |
| Response to wounding | 36 | |
| Inflammatory response | 23 | |
| 3 | Enrichment score: 5.12 | |
| Positive regulation of immune system process | 28 | |
| Immune response-regulating signal transduction | 12 | |
| Positive regulation of immune response | 18 | |
| Positive regulation of response to stimulus | 23 | |
| Immune response-activating signal transduction | 11 | |
| Immune response-regulating cell surface receptor signaling pathway | 10 | |
| Immune response-activating cell surface receptor signaling pathway | 9 | |
| Activation of immune response | 12 | |
| B-cell receptor signaling pathway | 4 | |
| Antigen receptor-mediated signaling pathway | 5 | |
| 4 | Enrichment score: 4.15 | |
| Defense response to bacterium | 12 | |
| Defense response to gram-positive bacteria | 7 | |
| Positive regulation of defense response | 10 | |
| Response to bacterium | 17 | |
| Response to molecule of bacterial origin | 11 | |
| 5 | Enrichment score: 3.71 | |
| Response to organic substance | 57 | |
| Response to estrogen stimulus | 15 | |
| Response to endogenous stimulus | 35 | |
| Response to steroid hormone stimulus | 22 | |
| Response to hormone stimulus | 32 |
Enrichment score ranks the biological significance of a gene cluster. Gene numbers are the total number of genes within each functional subgroup.
Table 5.
Functional annotation clustering of the top 5 day 7-specific genes obtained from D.A.V.I.D. analysis
| Annotation Cluster | Functional Annotation Clustering | Gene Numbers |
|---|---|---|
| 1 | Enrichment score: 5.93 | |
| Cell adhesion | 37 | |
| Biological adhesion | 37 | |
| Cell-cell adhesion | 17 | |
| 2 | Enrichment score: 4.83 | |
| Extracellular region part | 52 | |
| Extracellular matrix | 25 | |
| Proteinaceous extracellular matrix | 20 | |
| Extracellular space | 33 | |
| Extracellular region | 62 | |
| 3 | Enrichment score: 3.52 | |
| Blood vessel morphogenesis | 18 | |
| Blood vessel development | 20 | |
| Vasculature development | 20 | |
| Angiogenesis | 13 | |
| 4 | Enrichment score: 2.39 | |
| Carbohydrate binding | 21 | |
| Polysaccharide binding | 11 | |
| pattern binding | 11 | |
| Glycosaminoglycan binding | 10 | |
| Heparin binding | 7 | |
| 5 | Enrichment score: 2.09 | |
| Fibronectin, type III-like fold | 11 | |
| Fibronectin, type III | 10 | |
| FN3 | 10 | |
| Cytokine receptor activity | 5 |
Enrichment score ranks the biological significance of a gene cluster. Gene numbers are the total number of genes within each functional subgroup.
Fig. 2.
Venn diagrams of specific extracellular matrix (ECM)-related genes up- or downregulated at days 3 and/or day 7 postinjury. Genes of interest involved in specific immune response or ECM regulation at day 3 and 7, respectively, are listed. Genes modulated at both day 3 and 7 are also listed and primarily include factors involved in ECM regulation. Factors analyzed further via quantitative PCR are indicated in bold.
Collagen expression.
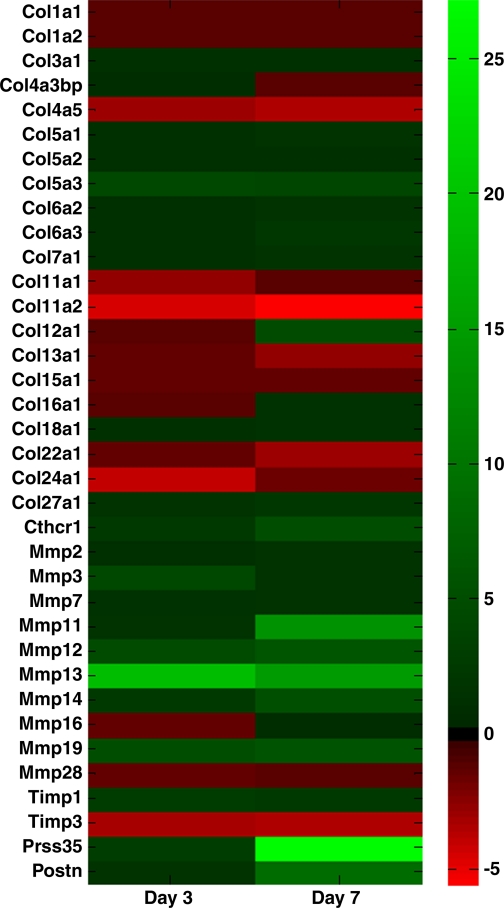
Twenty-one collagen isoforms were detected within the healing ligament via microarray. Ten of the 21 isoforms were significantly changed after injury (Fig. 3). Types I and III collagens did not experience a significant fold change but were the most abundant collagens identified within the ligament regardless of injury. Types IVa3bp, Va1, Va2, VIa2, VIIa1, XVa1, XVIa1, and XVIIIa1 collagens were also constitutively expressed and did not exhibit a significant fold change. In contrast, type IVa5, Va3, VIa3, XIa1, XIa2, XIIa1, XIIIa1, XXIIa1, XXIVa1, and XXVIIa1 were all differentially expressed during healing. At both day 3 and 7, types IVa5 and XIa2 were downregulated while type Va3 was upregulated. The other differentially expressed collagens were only regulated on day 3 (collagen types XIa1 and XXIVa1) or day 7 (collagen types VIa3, XXVIIa1, XIIa1, XIIIa1, and XXIIa1).
Fig. 3.
Heatmap showing changes in genes of interest. Data indicate fold change of day 3 or day 7 genes from the intact normal ligament. Green and red bars indicate an upregulation and downregulation of genes, respectively.
MMP expression.
Ten MMPs were detected in the ligament via microarray. Mmp2 was the primary MMP expressed within the ligament albeit in a constitutive manner (Fig. 3). Mmp3 was the second highest MMP expressed in the intact ligament, followed by Mmp14, 16, 13, 12, 11, 28, 19, and 7. Mmp7, 16, and 28 did not exhibit a twofold difference after injury. In contrast, Mmp3, 11, 12, 13, 14, and 19 all exhibited at least a twofold change between groups. Additionally, Timp1 and Timp3 were significantly up and downregulated at day 3 and day 7 postinjury, respectively.
Validation of selected genes.
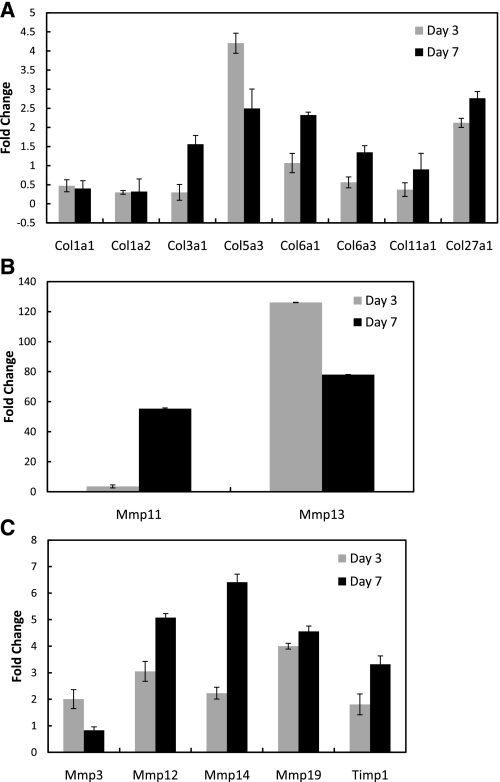
Several of the ECM and collagen genes of interest identified by microarray analysis were further validated via qPCR (Fig. 4). The genes at day 3 and day 7 were first normalized to the housekeeping gene β-actin. The day 3 and day 7 values were then compared (via fold change) to the intact ligament. Similar to the microarray results, type I collagen was not changed over day 3 and day 7 compared with the intact ligament. Type III collagen increased at day 7 compared with day 3 and the intact ligament, but the fold change was under two. Type VI alpha 1 and XI alpha 1 were also increased with injury, but the changes were not over twofold. In contrast, collagen type V alpha 3, VI alpha 1, and XXVII alpha 1 isoforms were significantly increased over the intact control. Expression of type VI alpha 1 and XXVII alpha 1 was highest at day 7 whereas type V alpha 3 was highest at day 3 and then decreased.
Fig. 4.
qPCR results of the genes of interest based on microarray results. A: qPCR analysis of genes found significantly regulated during microarray analysis or typically found in the ligament (as in the case for types I and III collagen). B: Mmps most significantly differentiated (Mmp11 and 13). C: other Mmps and Timp qPCR results. Data are expressed as fold change ± SE. Data were normalized with the housekeeping gene β-actin and compared with the normal intact ligament (and expressed as fold change).
Similar to the microarray results, Mmp3, 11, 12, 13, 14, and 19 all exhibited a twofold increase in gene expression after injury. Mmp11 and 13 demonstrated the greatest change after injury. All Mmps tested were greatest in expression at day 7 excluding Mmp13 and Mmp3, which decreased. Timp1 was also highest at day 7.
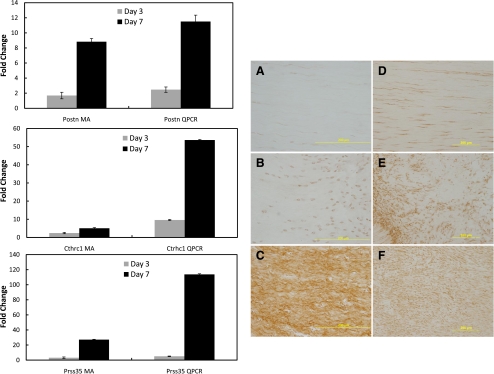
Cthrc1, Prss35, and Postn expression.
Microarray analysis detected three novel genes, Postn, Cthrc1, and Prss35, which were significantly upregulated after injury. Periostin (Postn), collagen triple helix repeat containing-1 (Cthrc1), and serine protease 35 (Prss35) were all significantly increased 7 days postinjury (Fig. 4). qPCR analysis verified these increases. Protein localization of periostin and CTHRC1 was further analyzed via immunohistochemistry. Few regions were positive for either factor in the intact ligament (Fig. 5, A and D). Injury stimulated an increase in both proteins at day 3 (Fig. 5, B and E). Levels were further increased at day 7 (Fig. 5, C and F).
Fig. 5.
Periostin (Postn), collagen triple helix repeat containing 1(Cthrc1), and serine protease 35 (Prss35) expression/localization during healing. qPCR and microarray comparison of Postn (top left), Cthrc1 (middle right), and Prss35 (bottom left) at 3 and 7 days postinjury in relation to the normal intact MCL. Immunohistochemistry (right) of Postn (A, B, and C) and CTHRC1 (D, E, and F) of the intact (A and D), day 3 (B and E), and day 7 (C and F) MCL. Staining for both periostin and CTHRC1 were low in the intact MCL (A and D, respectively). After injury, both periostin and CTHRC1 increased. qPCR and microarray data are expressed as fold change ± SD. qPCR data were normalized with the housekeeping gene β-actin and compared with the normal intact ligament (and expressed as fold change). Microarray data are compared with the normal intact ligament (and expressed as fold change).
DISCUSSION
The aim of this study was to profile transcriptional changes and identify candidate genes to regulate inflammation, ECM turnover, and scar formation during ligament healing. Our previous research (8) demonstrated significant vascular and cellular upregulation within the first week of ligament healing. These results indicated a significant increase in macrophages on days 1–5 and angiogenesis on days 7–11 postinjury. Additionally, the original wound size, based on size of granulation tissue, became increasingly larger (i.e., creeping substitution) at these times. We believe these overlapping immunogenic, cellular, angiogenic, and wound size changes control downstream scar formation. Based on the previous study, days 3 and 7 postinjured ligaments were chosen as time points to determine genetic changes from the intact ligament. Our current results identified a number of Mmps, Timps, and collagens modulated during healing. Specifically, minor collagens such as types XI, XXII, XXIV, and XXVII were identified and differentially expressed within the healing MCL. Unique genes, such as collagen triple helix repeat containing 1, periostin, and serine protease 35 were also identified in our healing model. Of the chosen factors, collagen types Va3, Via2, XIVa1, Cthrc1, Mmp3, Mmp11, Mmp12, Mmp13, Mmp14, and Mmp19, Timp3, Postn, and Prss35 were highly differentiated from the intact ligament, exhibiting a fourfold or greater change. To our knowledge, this is the first study to comprehensively identify and characterize genes involved during early MCL healing. Based on D.A.V.I.D. results, day 3 healing primarily includes genes associated with immune regulation. Our previous study (8) defined the spatial and temporal factors of normal ligament healing and demonstrated a strong upregulation of macrophages, elucidating the enhanced day 3 inflammatory factors in the current study. D.A.V.I.D. analysis further associated the day 7-specific genes to cell adhesion, ECM, and angiogenic functions that coincide with normal granulation tissue formation involving the upregulation of blood vessels, fibroblasts, and collagen turnover. Additional functional group analysis identified MMPs and collagens specific to the ligament. Similar inflammatory factors, MMPs, and collagens were also modulated in the early healing Achilles tendon (17). Recognition of the ligament MMPs and collagens provides a novel report of ligament injury-induced gene expression and sheds light on the possible players involved in the typical functional compromise (creeping substitution and scar formation) associated with normal ligament healing.
Two groups of collagens encoded by 42 genes to generate 29 types currently exist, including the fibril-forming (collagen types I, II, III, V, XI, XXIV, and XXVII) collagens or the nonfibril-forming collagens that are further subgrouped into the fibril associated with interrupted triple helices (FACIT; collagen types IX, XII, XIV, XIX, XX, and XXI), network-forming (collagen types VIII and X), microfibrillar (collagen type VI), anchoring fibril (collagen type VII), transmembrane (collagen types XIII and XVII), basement membrane (collagen type IV), multiplexin (collagen types XV, XVI, and XVIII), or other (type XXII, XXIII, XXV, and XXVI) collagens. In the current study, the fibril-forming (collagen type V, XI, and XXVII), FACIT (type XII collagen), microfibrillar (type VI collagen), transmembrane (type XIII collagen), and basement membrane (type IV collagen) collagens were differentially expressed after injury. The fibril-forming types I and III were the predominant collagens present but were expressed in a constitutive manner.
The fibrillar collagens were the most abundantly expressed in the current MCL model. Types I, III, and V collagens form heterotypic collagen fibrils, the fundamental functional unit of the ligament providing structural support. Type I collagen is the primary ligamentous collagen, consisting of two α1-chains and one α2-chain synthesized in a 2:1 ratio. Type I collagen was constitutively expressed within our healing model, regardless of injury. These results compare to our recent spatial and temporal study describing type I procollagen protein levels in the normal healing ligament (unpublished observations). Type I procollagen did not change the first week of healing but steadily decreased thereafter. These combined results support that type I collagen levels of the intact ligament are not different than the healing ligament within the first week of injury.
Fibrillar type III collagen intersperses within the type I collagen matrix. Type III collagen is commonly used as a marker for scar formation in the ligament. The microarray results showed no significant increase in type III collagen after injury, but the qPCR data indicated a slight increase at day 7. Previous results from our normal healing experiment localizing type III collagen via immunohistochemistry likewise demonstrated no change at day 3 but evidenced an increase at day 7 (unpublished observations). As scar formation progressed, type I collagen decreased and type III collagen increased (unpublished observations).
In contrast to the type I and III collagen microarray results, type V collagen was significantly upregulated at days 3 and 7 postinjury. Collagen type V is a widely distributed, low abundance fibrillar collagen believed to stimulate fibril nucleation and regulate fibril size and organization. Type V collagen mutations are linked to Ehlers-Danlos syndrome, a connective tissue disorder characterized by hyper-expandable skin (61). Levels also increase with hypertrophic scar formation, aging patellar tendons, and degenerating tendons (9, 12). Type V collagen abnormalities are accompanied by unusually large type I collagen diameters and increased fibril disorganization (2). More recently, type V collagen has been identified as an autoantigen of chronic inflammation/fibroproliferation processes and is believed to serve as a candidate marker for tendon pathology (7, 38). Based on these reports and the apparent importance in regulating type I collagen organization, type V collagen may play a critical role in scar formation as well as replacement fibrosis. Type V collagen may also play a fundamental role as an identifier of normal ligament injury.
To our knowledge, this is the first report to identify the fibril-forming types XI, XXIV, and XXVII collagens as well as type XXII collagen within the healing ligament. Type XI collagen is primarily known for its localization in cartilaginous tissues. However, in the current study, we verified the presence and the reduced expression of type XI collagen in the healing ligament. Type XI collagen is assembled as a heterotrimer consisting of α1 (XI)-, α2 (XI)-, and α3 (XI)-chains. Type XI collagen mutants develop Stickler syndrome, a connective tissue disorder resulting in beaded bundles of irregular collagen diameter causing premature arthritis and craniofacial disorders (1, 6, 57). Although its function is largely unknown, these results suggest that type XI collagen functions to regulate fibril diameters. Even less is known about types XXII and XXIV, but both participate in controlling bone and cartilage physiological processes such as collagen fibrillogenesis. Type XXIV is found primarily in the developing eye and skeleton and, to a lesser degree, skin and tendon during periods of accentuated type I collagen fibrillogenesis (20). Unlike types XI, XXII, and XXIV, type XXVII collagen may function in cartilage mineralization, cellular apoptosis, blood vessel invasion, matrix degradation, and type I collagen endochondral bone formation (15). Type XXVIIa1 collagen is primarily, but transiently, expressed in developing cartilage and dermis, the eye, and some epithelial layers during a narrow window of development in several tissues (48). The temporary expression of collagen type XXVII may function in cell signaling or as a supporting environment for invading blood vessels rather than structural stability. Type XXVII collagen upregulation in the current model may also support the transient blood vessel infiltration and matrix turnover during ligament healing. Taken together, these minor collagens may play a role in collagen fibrillogenesis and/or structural support to the MCL.
The fibrillar collagens provide structural support for the matrix, but the nonfibrillar collagens regulate anchoring and organization of the ECM. Type IV collagen is the primary component of basement membranes, forming a collagen, proteoglycan, glycoprotein meshwork. Type IV collagen forms the basement membranes around blood vessels. Angiogenesis increases substantially after injury, peaking around days 7–11 postinjury (8) but collagen type IV expression decreased in the current healing model. Type IV collagen is not required for basement membrane formation but rather for maintenance by mediating cell adhesion to the basement membrane. Injury disrupts normal ligament matrix, and the initial reduction in type IV collagen may permit cell migration to the damaged region, followed by reparative and replacement fibrosis via fibrillar collagen deposition and the eventual deposition of type IV collagen.
The microfibrillar type VI collagen contributes to a network of beaded filaments interwoven with other collagen fibrils in all connective tissues excluding bone. Type VI collagen acts as an adhesive between ECM components and the basement membrane (10). More recently, type VI has been reported to induce myofibroblast differentiation (40). Dermal wound healing studies reported type VI collagen from day 4 to 7 postinjury (59). The current results demonstrated a similar increase in type VI expression at day 7 postinjury, which coincides with myofibroblast upregulation in the healing ligament (unpublished observations). The later increase in type VI collagen may thereby maintain the number of myofibroblasts, which may contribute to creeping substitution and scar formation.
Only one FACIT collagen (type XII collagen) was differentially expressed in our wound healing model. FACIT collagens do not form supramolecular aggregates but link fibrils to other ECM proteins and bind collagen fibrils to regulate fibril growth, spacing, and diameter. Type XII collagen colocalizes with type I collagen and binds other ECM proteins, such as tenascin, to render the tissue more resistant to mechanical stress (42). Type XII collagen is also associated with enhancing the fluidity of collagen fibrils, presumably by reducing the friction between sliding fibrils (42). Additionally, type XII collagen mutants are associated with augmented female anterior cruciate ligament injuries (49). Based on these results, the day 7 increase in type XII collagen likewise provides a structural support between ECM and fibrillar collagens within the MCL.
The transmembrane type XIII collagen exists both as a membrane-bound and soluble molecule. Although its exact role is unknown, it plays an important role in certain adhesive interactions necessary for normal development. Research with type XIII collagen mutants mice developed lymphomas, abnormal basement membranes, and enhanced inflammatory gene expression (68). The reduced expression of type XIII collagen at day 7 in the current study likely reflects the disruption of collagen organization and hence cell-cell adhesion after injury.
Collagen triple helix repeat containing-1 (Cthrc1) is a gene product able to control collagen deposition and modulate transforming growth factor-β (TGF-β; Refs. 23, 51). TGF-β modulates transdifferentiation of fibroblasts to myofibroblasts by promoting expression of smooth muscle actin and subsequent scar formation (29, 34, 46). Cthrc1 overexpression stimulates faster cell migration to the wound and reduces TGF-β signaling, resulting in decreased wound formation, smooth muscle cell proliferation, and collagen deposition (24). Overexpression of Cthrc1 also reduces type Ia1, a2, and type IIIa1 collagen mRNA (24, 51). Based on these results, an increase of CTHRC1 may limit scar formation in the injured ligament by modulating TGF-β signaling. This concept, however, requires additional ligament healing studies using overexpressing and knockout Cthrc1 animals.
The MMPs and tissue inhibitors of MMP (TIMPs) are key enzymes involved in ECM remodeling during ligament healing. MMPs degrade numerous substances in the ECM, including collagen and fibrin, to expedite cell migration, deposit new ECM, and develop new tissue. MMPs also regulate the ECM structure by activating and releasing cytokines and growth factors from the ECM, sending cues to influence cell behavior and action. Four subsets of MMPs exist, including the collagenases, gelatinases, stromelysins, and membrane-type MMPS. In the present study, the collagenase (Mmp13), the stromelysins (Mmp3, Mmp12, and Mmp11), and membrane-bound (Mmp14 and Mmp19) collagen were expressed differentially in the healing ligament. The gelatinase Mmp2 was also expressed strongly in the ligament but in a constitutive manner.
Collagenases are proteases capable of degrading native triple helix fibrillar collagens types I, II, and III. In the current study, only the collagenase Mmp13 significantly increased in expression after injury. This Mmp was also the most significantly influenced by injury. Previous reports (21, 54, 55, 63, 71) demonstrated increased expression of MMP-13 in areas of excessive collagen destruction including osteoarthritis, rheumatoid arthritis, and periodontal inflammation. Message levels also significantly increased in the tendon injury model (5, 65). Cutaneous wound healing in Mmp13 knockout mice resulted in delayed wound closure and reepithelialization, decreased vascularization in granulation tissue, and decreased wound contraction and myofibroblast formation (14). Given these results, MMP-13 appears as the primary MMP involved in fibrillar collagen turnover, cell migration, angiogenesis, and MCL remodeling. Although not tested, MMP-13 may also contribute to the decrease in fibrillar collagens types XI, XXIV, and XXVII. Control of MMP-13-induced collagen degradation may therefore control creeping substitution and subsequent scar formation.
Stromelysins contribute a broad role in tissue development, remodeling, and wound healing. This class is also associated with collagen degradation of types IV, V, IX, and X. The stromelysin MMP-11 is expressed in normal and pathological remodeling processes including inflammation, wound healing, postpartum involution, and tumor proliferation (13, 58, 60, 67, 75). MMP-11 is primarily expressed in fibroblasts and does not degrade the typical ECM substrates (including collagens, gelatins, laminin, and fibronectin) but is involved in tissue remodeling, antiapoptotic events, and inflammation to favor cell survival and tumor progression. Collectively, these results suggest the significant increase in MMP-11 by the day 7 healing ligament may control inflammation and collagen degradation, before angiogenesis and after the inflammatory response. The role of MMP-11 in tumor progression suggests its possible role in modulating ligament creeping substitution.
The current ligament healing model demonstrates an increase in stromelysin MMP-12 starting at day 3 and continuing to day 7. MMP-12 is primarily produced by macrophages and is essential in stimulating macrophage migration. It also functions to digest elastin and other ECM molecules. Expression of MMP-12 is upregulated in several lung diseases (22, 27, 41, 76). A study (32) examining Mmp12 knockout mice reported an increase in Mmp2 and IL-13/IL-4R-induced Mmp13 expression after Mmp12 deficiency. Based on these results, control of MMP-12 may control phenotype-specific macrophage infiltration into the healing ligament and thereby modulate creeping substitution and subsequent scar formation.
MMP-3 degrades a wide range of ECM proteins including types IV, V, XI, and X collagens, proteoglycans, fibronectin, and laminin. Studies (47) with MMP-3 deficient animals suggest this protease functions to detach cells from the underlying basement membrane to initiate cell migration. Additional MMP-3 knockout studies (73) using the lung injury model resulted in reduced lung injury by inhibiting leukocyte recruitment. These studies suggest that the transient increase of MMP-3 in the day 3 ligament recruits cells to the wound site, and promotes inflammation.
Unlike other MMPs, membrane type-MMPs are not secreted into the ECM. Rather, they reside on cell membranes and function by binding to and activating other MMPs. MMP-14 is associated with stimulating MMP-2 and MMP-9 (70). In the current study, Mmp9 was not identified in the healing ligament, whereas Mmp2 was expressed. The increased Mmp14 expression observed at days 3 and 7 within the healing ligament could therefore modulate MMP-2 activity within the healing ligament model. Additionally, MMP-14 is collagenolytic towards type I, II, and III collagens (62, 70) and may likewise mediate collagen breakdown.
Membrane bound MMP-19 functions to degrade ECM molecules including the basement membrane and type IV collagen. This enzyme does not cleave type I collagen. MMP-19 has been localized to vascular smooth muscle cells, endothelial cells, fibroblasts of granulation tissue, monocytes, and macrophages and is upregulated during inflammatory conditions such as arthritis and multiple sclerosis (3, 4, 31, 53). MMP-19-deficient mice demonstrated enhanced tumor invasion, reduced CD8+ T-cell activation and distribution, and a diminished inflammatory reaction determined by lowered cytokine expression, decreased influx of inflammatory cells, lessened cell proliferation, and increased angiogenesis (3, 19). Additional reports described the downregulation of MMP-19 in invasive carcinomas, suggesting its role in tissue homeostasis. Based on these reports, the increase in Mmp19 (likely released by macrophages and fibroblasts) may recruit inflammatory cells and modulate degradation of ECM during inflammation.
TIMPs are natural inhibitors of MMPs. Currently, four members of the TIMP family have been identified and include TIMP-1, TIMP-2, TIMP-3, and TIMP-4. Timp1 was identified in the current healing model and inhibits most MMP activity excluding MMP-14 and MMP-2. Timp3 was also differentially regulated during ligament healing. TIMP-3 is bound to ECM and is not a freely soluble protein but is able to inhibit other molecules outside of MMPs including a disintegrin and metalloproteinase domain (ADAM) and ADAMS with thrombospondin domains (ADAM-TS). Timp2 and Timp4 were not identified in the current healing model.
The interaction between collagens and MMPs during ligament healing is not fully understood but plays a significant role in matrix turnover. In conjunction with previous reports and the current study, types I and III collagens are eventually degraded by collagenases that breakdown the helical structure. In the ligament, MMP-13 primarily controls fibrillar collagen turnover, but membrane-bound MMP-14 may also contribute. Collagen types IV and V are not targeted by collagenases but by gelatinases or stromelysins (30, 69). Gelatinases were not differentially regulated in the current study but the stromelysins were. Thus type IV and V collagens appear to be modulated by MMP-3 and -12 in the ligament model. MMP-3 and -12 also stimulate inflammatory cell migration and detachment while degrading collagen. Type VI collagen is not subject to the same breakdown by the MMPs and is degraded by serine proteases produced primarily by neutrophils and mast cells during inflammation. This suggests type VI collagen levels remain in check in the presence of inflammatory cells but increase as cell number decreases. Later, fibroblast-stimulated MMP-11 may ameliorate the macrophage MMP-induced degradation and initiate collagen deposition. Further study is required to elucidate the functional interactions among the MMPs, TIMPs, CTHRC1, and collagens.
Periostin is a gene of interest based on microarray data and current literature. Prior studies of periodontal ligament (16, 16), remodeling bone (16, 16), carcinomas (50), and myocardial infarcted heart (39, 43, 43) reported an increase in periostin and subsequent increase in deposition and attachment of collagen. Periostin knockout mice exhibit reduced stiffness and diminished collagen formation after myocardial infarction, suggesting improved cardiac function (16, 44, 45). Although these results indicate that periostin participates in collagen synthesis during wound healing, successful ligament healing also requires the spatial and temporal aspects of collagen alignment and production of the appropriate type of collagen. Previous research from our laboratory demonstrates a significant increase in type III collagen and a concomitant decrease in type I procollagen during the first week of healing. These immunohistochemistry results suggest that periostin colocalizes with type III collagen of the healing ligament. Current results indicate that periostin increases during the first week of healing to stimulate fibroblast proliferation and regulate collagen synthesis. Periostin knockout studies are currently underway to determine its role during wound healing.
This study is the first to report the presence of Prss35 within the healing ligament. A paucity of information is known about this extracellular serine protease, but its presence has been reported in the mouse ovary (37, 72). The reported spatial and temporal regulation of PRSS 35 within the mouse ovary suggests it may be involved in tissue remodeling that occurs with ovulation (37, 72). The significant upregulation of Prss35 within the healing MCL at day 7 also suggests its possible contribution to ligament remodeling. More research needs performed to determine the role of this protease.
When interpreting results, several things must be borne in mind. First, the study investigates healing in a rat MCL healing after surgical transection. Second, within this model, protein synthesis and functional expression may not directly correlate with the mRNA levels reported in the study. Third, periostin and CTHRC-1 immunohistochemistry was also performed to verify protein expression of these novel factors. However, many additional factors reported herein would benefit from further protein analysis. In addition, microarray results were not always consistent with qPCR results. Several causes may lead to such discrepancies. For instance, specific gene expression could be defined as absent via microarray but may be detected by the more sensitive qPCR technology. Further, the intrinsic variability in the affinity of labeled samples to the microarray probes could influence the fold change in gene activity between experimental groups. In our study, the entire ligament, rather than the injured region within the ligament, was assayed. The inclusion of the total ligament diluted expression particular genes regulated specifically within the healing region. Lastly, microarray results are influenced by array production, probe labeling, hybridization conditions, and image analysis (52), which may also explain any discrepancies with the qPCR analysis.
Our experiments focused on the first week of ligament healing during inflammation and proliferation, rather than the subsequent remodeling. We believe scar formation is a downstream result of various immune cell-mediated responses to injury. If so, identifying and modulating factors involved in creeping substitution, macrophage infiltration, and angiogenesis would limit scar formation and reduce healing time. A similarly robust analysis of later time points during healing would also supply a wealth of information on the formation and remodeling of the neo-ligament.
In summary, differences in the genetic expression of MMPs, collagens, and the novel genes Postn, Cthrc1, and Prss35 were observed in the early healing ligament. To our knowledge, these results provide the first comprehensive assessment of ECM expression in the MCL and will lead to a better understanding of the molecular mechanisms of creeping substitution and scar formation. Identification of these factors during early healing is novel, but their specific functional roles and interactions require further elucidation.
GRANTS
Financial support was provided by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grants AR049266 and AR059916.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDEMENTS
We acknowledge the petit autem pro rat sacrificium ultimum, the technical and computer support of Sandra Splinter BonDurant and Jean-Yves Sgro, heatmap development of Kayt Frisch, and statistical work of Alejandro Munoz del Rio.
REFERENCES
- 1. Anonymous International nomenclature and classification of the osteochondrodysplasias (1997). International Working Group on Constitutional Diseases of Bone. Am J Med Genet 79: 376–382, 1998 [DOI] [PubMed] [Google Scholar]
- 2. Andrikopoulos K, Liu X, Keene DR, Jaenisch R, Ramirez F. Targeted mutation in the col5a2 gene reveals a regulatory role for type V collagen during matrix assembly. Nat Genet 9: 31–36, 1995 [DOI] [PubMed] [Google Scholar]
- 3. Beck IM, Ruckert R, Brandt K, Mueller MS, Sadowski T, Brauer R, Schirmacher P, Mentlein R, Sedlacek R. MMP19 is essential for T cell development and T cell-mediated cutaneous immune responses. PLos One 3: e2343, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Behera AK, Hildebrand E, Scagliotti J, Steere AC, Hu LT. Induction of host matrix metalloproteinases by Borrelia burgdorferi differs in human and murine lyme arthritis. Infect Immun 73: 126–134, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berglund M, Hart DA, Wiig M. The inflammatory response and hyaluronan synthases in the rabbit flexor tendon and tendon sheath following injury. J Hand Surg Eur Vol 32: 581–587, 2007 [DOI] [PubMed] [Google Scholar]
- 6. Blaschke UK, Eikenberry EF, Hulmes DJ, Galla HJ, Bruckner P. Collagen XI nucleates self-assembly and limits lateral growth of cartilage fibrils. J Biol Chem 275: 10370–10378, 2000 [DOI] [PubMed] [Google Scholar]
- 7. Burlingham WJ, Love RB, Jankowska-Gan E, Haynes LD, Xu Q, Bobadilla JL, Meyer KC, Hayney MS, Braun RK, Greenspan DS, Gopalakrishnan B, Cai J, Brand DD, Yoshida S, Cummings OW, Wilkes DS. IL-17-dependent cellular immunity to collagen type V predisposes to obliterative bronchiolitis in human lung transplants. J Clin Invest 117: 3498–3506, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chamberlain CS, Crowley E, Vanderby R. The spatio-temporal dynamics of ligament healing. Wound Repair Regen 17: 206–215, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dressler MR, Butler DL, Wenstrup R, Awad HA, Smith F, Boivin GP. A potential mechanism for age-related declines in patellar tendon biomechanics. J Orthop Res 20: 1315–1322, 2002 [DOI] [PubMed] [Google Scholar]
- 10. Engvall E, Hessle H, Klier G. Molecular assembly, secretion, and matrix deposition of type VI collagen. J Cell Biol 102: 703–710, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang HL, Lai JJ, Lin WL, Lin WC. A fermented substance from Aspergillus phoenicis reduces liver fibrosis induced by carbon tetrachloride in rats. Biosci Biotechnol Biochem 71: 1154–1161, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Goncalves-Neto J, Witzel SS, Teodoro WR, Carvalho-Junior AE, Fernandes TD, Yoshinari HH. Changes in collagen matrix composition in human posterior tibial tendon dysfunction. Joint Bone Spine 69: 189–194, 2002 [DOI] [PubMed] [Google Scholar]
- 13. Hagglund AC, Ny A, Leonardsson G, Ny T. Regulation and localization of matrix metalloproteinases and tissue inhibitors of metalloproteinases in the mouse ovary during gonadotropin-induced ovulation. Endocrinology 140: 4351–4358, 1999 [DOI] [PubMed] [Google Scholar]
- 14. Hattori N, Mochizuki S, Kishi K, Nakajima T, Takaishi H, D'Armiento J, Okada Y. MMP-13 plays a role in keratinocyte migration, angiogenesis, and contraction in mouse skin wound healing. Am J Pathol 175: 533–546, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hjorten R, Hansen U, Underwood RA, Telfer HE, Fernandes RJ, Krakow D, Sebald E, Wachsmann-Hogiu S, Bruckner P, Jacquet R, Landis WJ, Byers PH, Pace JM. Type XXVII collagen at the transition of cartilage to bone during skeletogenesis. Bone 41: 535–542, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, Toyama Y, Bonewald LF, Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res 14: 1239–1249, 1999 [DOI] [PubMed] [Google Scholar]
- 17. Jelinsky SA, Li L, Ellis D, Archambault J, Li J, St Andre M, Morris C, Seeherman H. Treatment with rhBMP12 or rhBMP13 increase the rate and the quality of rat Achilles tendon repair. J Orthop Res. 2011 Apr 5; doi: 10.1002/jor.21427. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 18. Johnson EC, Jia L, Cepurna WO, Doser TA, Morrison JC. Global changes in optic nerve head gene expression after exposure to elevated intraocular pressure in a rat glaucoma model. Invest Ophthalmol Vis Sci 48: 3161–3177, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jost M, Folgueras AR, Frerart F, Pendas AM, Blacher S, Houard X, Berndt S, Munaut C, Cataldo D, Alvarez J, Melen-Lamalle L, Foidart JM, Lopez-Otin C, Noel A. Earlier onset of tumoral angiogenesis in matrix metalloproteinase-19-deficient mice. Cancer Res 66: 5234–5241, 2006 [DOI] [PubMed] [Google Scholar]
- 20. Koch M, Laub F, Zhou P, Hahn RA, Tanaka S, Burgeson RE, Gerecke DR, Ramirez F, Gordon MK. Collagen XXIV, a vertebrate fibrillar collagen with structural features of invertebrate collagens: selective expression in developing cornea and bone. J Biol Chem 278: 43236–43244, 2003 [DOI] [PubMed] [Google Scholar]
- 21. Konttinen YT, Ainola M, Valleala H, Ma J, Ida H, Mandelin J, Kinne RW, Santavirta S, Sorsa T, Lopez-Otin C, Takagi M. Analysis of 16 different matrix metalloproteinases (MMP-1 to MMP-20) in the synovial membrane: different profiles in trauma and rheumatoid arthritis. Ann Rheum Dis 58: 691–697, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lavigne MC, Thakker P, Gunn J, Wong A, Miyashiro JS, Wasserman AM, Wei SQ, Pelker JW, Kobayashi M, Eppihimer MJ. Human bronchial epithelial cells express and secrete MMP-12. Biochem Biophys Res Commun 324: 534–546, 2004 [DOI] [PubMed] [Google Scholar]
- 23. LeClair R, Lindner V. The role of collagen triple helix repeat containing 1 in injured arteries, collagen expression, and transforming growth factor beta signaling. Trends Cardiovasc Med 17: 202–205, 2007 [DOI] [PubMed] [Google Scholar]
- 24. LeClair RJ, Durmus T, Wang Q, Pyagay P, Terzic A, Lindner V. Cthrc1 is a novel inhibitor of transforming growth factor-beta signaling and neointimal lesion formation. Circ Res 100: 826–833, 2007 [DOI] [PubMed] [Google Scholar]
- 25. Levenson SM, Geever EF, Crowley LV, Oates JF, Berard CW, III, Rosen H. The healing of rat skin wounds. Ann Surg 161: 293–308, 1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem 284: 15676–15684, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lian X, Qin Y, Hossain SA, Yang L, White A, Xu H, Shipley JM, Li T, Senior RM, Du H, Yan C. Overexpression of Stat3C in pulmonary epithelium protects against hyperoxic lung injury. J Immunol 174: 7250–7256, 2005 [DOI] [PubMed] [Google Scholar]
- 28. Lin TW, Cardenas L, Soslowsky LJ. Biomechanics of tendon injury and repair. J Biomech 37: 865–877, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Lindner V. Role of basic fibroblast growth factor and platelet-derived growth factor (B-chain) in neointima formation after arterial injury. Z Kardiol 84, Suppl 4: 137–144, 1995 [PubMed] [Google Scholar]
- 30. Lindsey ML. Novel strategies to delineate matrix metalloproteinase (MMP)-substrate relationships and identify targets to block MMP activity. Mini Rev Med Chem 6: 1243–1248, 2006 [DOI] [PubMed] [Google Scholar]
- 31. Locati M, Deuschle U, Massardi ML, Martinez FO, Sironi M, Sozzani S, Bartfai T, Mantovani A. Analysis of the gene expression profile activated by the CC chemokine ligand 5/RANTES and by lipopolysaccharide in human monocytes. J Immunol 168: 3557–3562, 2002 [DOI] [PubMed] [Google Scholar]
- 32. Madala SK, Pesce JT, Ramalingam TR, Wilson MS, Minnicozzi S, Cheever AW, Thompson RW, Mentink-Kane MM, Wynn TA. Matrix metalloproteinase 12-deficiency augments extracellular matrix degrading metalloproteinases and attenuates IL-13-dependent fibrosis. J Immunol 184: 3955–3963, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maeda E, Shelton JC, Bader DL, Lee DA. Differential regulation of gene expression in isolated tendon fascicles exposed to cyclic tensile strain in vitro. J Appl Physiol 106: 506–512, 2009 [DOI] [PubMed] [Google Scholar]
- 34. Majesky MW, Lindner V, Twardzik DR, Schwartz SM, Reidy MA. Production of transforming growth factor beta 1 during repair of arterial injury. J Clin Invest 88: 904–910, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marsolais D, Cote CH, Frenette J. Neutrophils and macrophages accumulate sequentially following Achilles tendon injury. J Orthop Res 19: 1203–1209, 2001 [DOI] [PubMed] [Google Scholar]
- 36. Miyakoshi K, Murphy MJ, Yeoman RR, Mitra S, Dubay CJ, Hennebold JD. The identification of novel ovarian proteases through the use of genomic and bioinformatic methodologies. Biol Reprod 75: 823–835, 2006 [DOI] [PubMed] [Google Scholar]
- 37. Miyakoshi K, Murphy MJ, Yeoman RR, Mitra S, Dubay CJ, Hennebold JD. The identification of novel ovarian proteases through the use of genomic and bioinformatic methodologies. Biol Reprod 75: 823–835, 2006 [DOI] [PubMed] [Google Scholar]
- 38. Mokone GG, Schwellnus MP, Noakes TD, Collins M. The COL5A1 gene and Achilles tendon pathology. Scand J Med Sci Sports 16: 19–26, 2006 [DOI] [PubMed] [Google Scholar]
- 39. Nakazawa T, Nakajima A, Seki N, Okawa A, Kato M, Moriya H, Amizuka N, Einhorn TA, Yamazaki M. Gene expression of periostin in the early stage of fracture healing detected by cDNA microarray analysis. J Orthop Res 22: 520–525, 2004 [DOI] [PubMed] [Google Scholar]
- 40. Naugle JE, Olson ER, Zhang X, Mase SE, Pilati CF, Maron MB, Folkesson HG, Horne WI, Doane KJ, Meszaros JG. Type VI collagen induces cardiac myofibroblast differentiation: implications for postinfarction remodeling. Am J Physiol Heart Circ Physiol 290: H323–H330, 2006 [DOI] [PubMed] [Google Scholar]
- 41. Nenan S, Planquois JM, Berna P, De Mendez I, Hitier S, Shapiro SD, Boichot E, Lagente V, Bertrand CP. Analysis of the inflammatory response induced by rhMMP-12 catalytic domain instilled in mouse airways. Int Immunopharmacol 5: 511–524, 2005 [DOI] [PubMed] [Google Scholar]
- 42. Nishiyama T, McDonough AM, Bruns RR, Burgeson RE. Type XII and XIV collagens mediate interactions between banded collagen fibers in vitro and may modulate extracellular matrix deformability. J Biol Chem 269: 28193–28199, 1994 [PubMed] [Google Scholar]
- 43. Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Rodriguez R, Trusk T, Potts JD, Goodwin RL, Davis J, Hoffman S, Wen X, Sugi Y, Kern CB, Mjaatvedt CH, Turner DK, Oka T, Conway SJ, Molkentin JD, Forgacs G, Markwald RR. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem 101: 695–711, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Norris RA, Moreno-Rodriguez RA, Sugi Y, Hoffman S, Amos J, Hart MM, Potts JD, Goodwin RL, Markwald RR. Periostin regulates atrioventricular valve maturation. Dev Biol 316: 200–213, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW, Conway SJ, II, Aronow BJ, Robbins J, Molkentin JD. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res 101: 313–321, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84: 767–801, 2004 [DOI] [PubMed] [Google Scholar]
- 47. Pilcher BK, Dumin JA, Sudbeck BD, Krane SM, Welgus HG, Parks WC. The activity of collagenase-1 is required for keratinocyte migration on a type I collagen matrix. J Cell Biol 137: 1445–1457, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Plumb DA, Dhir V, Mironov A, Ferrara L, Poulsom R, Kadler KE, Thornton DJ, Briggs MD, Boot-Handford RP. Collagen XXVII is developmentally regulated and forms thin fibrillar structures distinct from those of classical vertebrate fibrillar collagens. J Biol Chem 282: 12791–12795, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Posthumus M, September AV, O'Cuinneagain D, van der Merwe W, Schwellnus MP, Collins M. The association between the COL12A1 gene and anterior cruciate ligament ruptures. Br J Sports Med. 2010 doi: 10.1136/bjsm.2009.060756. [DOI] [PubMed] [Google Scholar]
- 50. Puglisi F, Puppin C, Pegolo E, Andreetta C, Pascoletti G, D'Aurizio F, Pandolfi M, Fasola G, Piga A, Damante G, Di Loreto C. Expression of periostin in human breast cancer. J Clin Pathol 61: 494–498, 2008 [DOI] [PubMed] [Google Scholar]
- 51. Pyagay P, Heroult M, Wang Q, Lehnert W, Belden J, Liaw L, Friesel RE, Lindner V. Collagen triple helix repeat containing 1, a novel secreted protein in injured and diseased arteries, inhibits collagen expression and promotes cell migration. Circ Res 96: 261–268, 2005 [DOI] [PubMed] [Google Scholar]
- 52. Rajeevan MS, Ranamukhaarachchi DG, Vernon SD, Unger ER. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25: 443–451, 2001 [DOI] [PubMed] [Google Scholar]
- 53. Ramanathan M, Weinstock-Guttman B, Nguyen LT, Badgett D, Miller C, Patrick K, Brownscheidle C, Jacobs L. In vivo gene expression revealed by cDNA arrays: the pattern in relapsing-remitting multiple sclerosis patients compared with normal subjects. J Neuroimmunol 116: 213–219, 2001 [DOI] [PubMed] [Google Scholar]
- 54. Ravanti L, Kahari VM. Matrix metalloproteinases in wound repair (review). Int J Mol Med 6: 391–407, 2000 [PubMed] [Google Scholar]
- 55. Reboul P, Pelletier JP, Tardif G, Cloutier JM, Martel-Pelletier J. The new collagenase, collagenase-3, is expressed and synthesized by human chondrocytes but not by synoviocytes. A role in osteoarthritis. J Clin Invest 97: 2011–2019, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reno C, Marchuk L, Sciore P, Frank CB, Hart DA. Rapid isolation of total RNA from small samples of hypocellular, dense connective tissues. Biotechniques 22: 1082–1086, 1997 [DOI] [PubMed] [Google Scholar]
- 57. Richards AJ, Baguley DM, Yates JR, Lane C, Nicol M, Harper PS, Scott JD, Snead MP. Variation in the vitreous phenotype of Stickler syndrome can be caused by different amino acid substitutions in the X position of the type II collagen Gly-X-Y triple helix. Am J Hum Genet 67: 1083–1094, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rio MC. From a unique cell to metastasis is a long way to go: clues to stromelysin-3 participation. Biochimie 87: 299–306, 2005 [DOI] [PubMed] [Google Scholar]
- 59. Romanos GE, Strub JR. Effect of Tissucol on connective tissue matrix during wound healing: an immunohistochemical study in rat skin. J Biomed Mater Res 39: 462–468, 1998 [DOI] [PubMed] [Google Scholar]
- 60. Rouyer N, Wolf C, Chenard MP, Rio MC, Chambon P, Bellocq JP, Basset P. Stromelysin-3 gene expression in human cancer: an overview. Invasion Metastasis 14: 269–275, 1994 [PubMed] [Google Scholar]
- 61. Schwarze U, Atkinson M, Hoffman GG, Greenspan DS, Byers PH. Null alleles of the COL5A1 gene of type V collagen are a cause of the classical forms of Ehlers-Danlos syndrome (types I and II). Am J Hum Genet 66: 1757–1765, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci USA 93: 3942–3946, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stahle-Backdahl M, Sandstedt B, Bruce K, Lindahl A, Jimenez MG, Vega JA, Lopez-Otin C. Collagenase-3 (MMP-13) is expressed during human fetal ossification and re-expressed in postnatal bone remodeling and in rheumatoid arthritis. Lab Invest 76: 717–728, 1997 [PubMed] [Google Scholar]
- 64. Strup-Perrot C, Vozenin-Brotons MC, Vandamme M, Linard C, Mathe D. Expression of matrix metalloproteinases and tissue inhibitor metalloproteinases increases in X-irradiated rat ileum despite the disappearance of CD8a T cells. World J Gastroenterol 11: 6312–6321, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tang Z, Yang L, Zhang J, Xue R, Wang Y, Chen PC, Sung KL. Coordinated expression of MMPs and TIMPs in rat knee intra-articular tissues after ACL injury. Connect Tissue Res 50: 315–322, 2009 [PubMed] [Google Scholar]
- 66. Tashiro T, Hiraoka H, Ikeda Y, Ohnuki T, Suzuki R, Ochi T, Nakamura K, Fukui N. Effect of GDF-5 on ligament healing. J Orthop Res 24: 71–79, 2006 [DOI] [PubMed] [Google Scholar]
- 67. Thewes M, Worret WI, Engst R, Ring J. Stromelysin-3 (ST-3): immunohistochemical characterization of the matrix metalloproteinase (MMP)-11 in benign and malignant skin tumours and other skin disorders. Clin Exp Dermatol 24: 122–126, 1999 [DOI] [PubMed] [Google Scholar]
- 68. Tuomisto A, Sund M, Tahkola J, Latvanlehto A, Savolainen ER, Autio-Harmainen H, Liakka A, Sormunen R, Vuoristo J, West A, Lahesmaa R, Morse HC, Pihlajaniemi T., III A mutant collagen XIII alters intestinal expression of immune response genes and predisposes transgenic mice to develop B-cell lymphomas. Cancer Res 68: 10324–10332, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tyagi SC. Proteinases and myocardial extracellular matrix turnover. Mol Cell Biochem 168: 1–12, 1997 [DOI] [PubMed] [Google Scholar]
- 70. Ueno H, Nakamura H, Inoue M, Imai K, Noguchi M, Sato H, Seiki M, Okada Y. Expression and tissue localization of membrane-types 1, 2, and 3 matrix metalloproteinases in human invasive breast carcinomas. Cancer Res 57: 2055–2060, 1997 [PubMed] [Google Scholar]
- 71. Uitto VJ, Airola K, Vaalamo M, Johansson N, Putnins EE, Firth JD, Salonen J, Lopez-Otin C, Saarialho-Kere U, Kahari VM. Collagenase-3 (matrix metalloproteinase-13) expression is induced in oral mucosal epithelium during chronic inflammation. Am J Pathol 152: 1489–1499, 1998 [PMC free article] [PubMed] [Google Scholar]
- 72. Wahlberg P, Nylander A, Ahlskog N, Liu K, Ny T. Expression and localization of the serine proteases high-temperature requirement factor A1, serine protease 23, and serine protease 35 in the mouse ovary. Endocrinology 149: 5070–5077, 2008 [DOI] [PubMed] [Google Scholar]
- 73. Warner RL, Beltran L, Younkin EM, Lewis CS, Weiss SJ, Varani J, Johnson KJ. Role of stromelysin 1 and gelatinase B in experimental acute lung injury. Am J Respir Cell Mol Biol 24: 537–544, 2001 [DOI] [PubMed] [Google Scholar]
- 74. Wasserman JK, Zhu X, Schlichter LC. Evolution of the inflammatory response in the brain following intracerebral hemorrhage and effects of delayed minocycline treatment. Brain Res 1180: 140–154, 2007 [DOI] [PubMed] [Google Scholar]
- 75. Wolf C, Chenard MP, Durand de Grossouvre P, Bellocq JP, Chambon P, Basset P. Breast-cancer-associated stromelysin-3 gene is expressed in basal cell carcinoma and during cutaneous wound healing. J Invest Dermatol 99: 870–872, 1992 [DOI] [PubMed] [Google Scholar]
- 76. Xie S, Issa R, Sukkar MB, Oltmanns U, Bhavsar PK, Papi A, Caramori G, Adcock I, Chung KF. Induction and regulation of matrix metalloproteinase-12 in human airway smooth muscle cells. Respir Res 6: 148, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhu LJ, Altmann SW. mRNA and 18S-RNA coapplication-reverse transcription for quantitative gene expression analysis. Anal Biochem 345: 102–109, 2005 [DOI] [PubMed] [Google Scholar]