Abstract
Molecular reductionism has so far failed to deliver the broad-based therapeutic insights that were initially hoped for. This form of reductionism is now being replaced by so-called “systems biology.” This is a nebulously defined approach and/or discipline, with some versions of it relying excessively on hypothesis-neutral approaches and only minimally informed by key physiological concepts such as homeostasis and regulation. In this context, physiology is uniquely positioned to continue to provide impressive levels of both biological and therapeutic insight by using hypothesis-driven “classical” approaches and concepts to help frame what might be described as the “pieces of the puzzle” that emerge from molecular reductionism. The strength of physiology as a “bridge” between reductionism and epidemiology, along with its unparalleled ability to generate therapeutic insights and opportunities justifies increased attention and emphasis on our discipline into the future. Arguments relevant to this set of assertions are advanced and this paper, which was based on the 2011 Adolph Lecture, represents an effort to fill the intellectual void left by reductionism and improve scientific progress.
Keywords: homeostasis, regulation, integrative
this paper reflects ideas that were presented as part of the 2011 Adolph Lecture at the Experimental Biology meeting that was held in Washington, DC. The goal of the talk was to share a physiologist's perspective on what reductionism in general and the “omic” revolution in particular has or has not done for biomedical research and associated therapeutic insights or advances. The main ideas highlighted in the lecture were the following.
Reductionism via various flavors of molecular biology and “omics” has so far failed to deliver its self-promoted revolution in clinical medicine.
Systems biology has a cell-centric focus that is marked by a limited understanding of and application to biology beyond the cell.
The failure of systems biology to recognize and use key concepts from physiology about homeostasis, regulation, redundancy, feedback control, and acclimation/adaptation are major limitations to this poorly defined approach.
While all the attention has been focused on reductionism and more recently systems biology, physiology continues to provide important biomedical insights that lead to therapeutic advances.
As the title demonstrates, my goal in the Adolph Lecture and in this paper was and is to be intentionally provocative and hopefully generate a dialogue with the reductionists. In this context, and because I am “taking sides”, I have adopted what might be called a conversational approach to this paper.
BIOLOGICAL ORTHOPEDIC SURGERY
A key idea or theme that seems to underpin the impetus for reductionism and various flavors of “omics” as applied to biomedical problems might be described as biological orthopedic surgery: “the gene is broken → fix the broken gene → cure the patient.” This thinking clearly seems to explain the enthusiasm about gene therapy that emerged after the discovery of the genetic defect responsible for the most common form of cystic fibrosis and more recently ideas about a limited number of common gene variants explaining the risk for common conditions like atherosclerosis and diabetes (10–12, 43, 51). The line of thinking described above flows from what Denis Noble has critically termed “Neo-Darwinian” thinking about the relationship between genes and phenotype (45, 46). It is exemplified by two quotes, the first from 1989 and second from Francis Collins (the current director of NIH), one of the people involved in the cystic fibrosis gene discovery.
The implications of this research are profound; there will be large spin offs in basic biology, especially cell physiology, but the largest impact will be biomedical (51).
Here we are in 1997, eight years later, and the management of her disease has not changed. . . . .But I will predict that in the course of the next 10 years management of CF will change. . . . .The healthy form of the gene itself may even be used in so-called gene therapy (12).
What is interesting to note is that while gene therapy for cystic fibrosis has failed to materialize in the 20+ years since the gene defect was identified, there are traditional ion channel-based drugs that target the CFTR protein in clinical trials that show promise in cystic fibrosis (18, 66). At one level, the development of these drugs was likely facilitated by the genetic discoveries because they permitted the development of models that advanced the understanding of the biophysics and ultimately pharmacology of the defective channel. However, one is tempted to speculate, for cystic fibrosis and perhaps other diseases, that much faster therapeutic progress might have been made if traditional physiological and pharmacological approaches had been a bigger area of focus. Perhaps the optimism and drive for gene therapy was an example of what might be termed “silver bullet” thinking that I will discuss below.
REDUCTIONISM IS SEDUCTIVE
The type of reductionism that I have termed “biological orthopedic surgery” has a number of attractive features and is at some level very seductive. It is easy to understand, and when it delivers it is associated with a heroic narrative by a lone scientist or team of scientists making a fundamental discovery that solves a problem. This is the sort of silver bullet thinking mentioned above. However, it has been known for some time that both the easy to understand elements and heroic narratives associated with reductionism are mirages. In this context, when the factors that contribute to biomedical breakthroughs were subjected to analysis by Comroe and Drips (13) in the late 1960s and early 1970s via the “retrospectoscope,” biomedical breakthroughs are in fact more nuanced, incremental, and associated with a more serendipitous view of progress vs. the heroic narrative of reductionism.
HEMOGLOBIN IS A SHIFTY MOLECULE
Homeostasis—the ability to regulate key bodily functions within a narrow range in response to either internal (e.g., exercise) or external (e.g., harsh environmental conditions)—is one of the fundamental (perhaps the fundamental) concept in physiology (7). Homeostasis is also subserved by ideas about regulated systems, feedback control, redundant control mechanisms, and adaptation and acclimation over time. These physiological concepts and mechanisms contribute to what might be described as emergent properties, so that the behavior of the system is far more complex and (and likely more robust) than might be predicted on the basis of a single reductionist property (35).
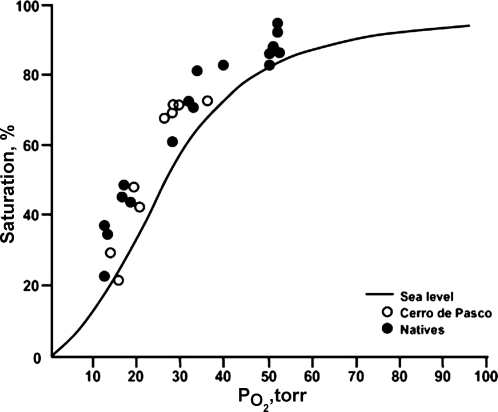
A good, and early, example of this concept comes from the textbook description about the right shift in the oxygen-hemoglobin dissociation curve that occurs at high altitude or during other forms of hypoxia. The standard teaching is that under these conditions there is a rise in 2–3 DPG that allosterically modifies oxygen-hemoglobin dissociation curve and creates a right shift that facilitates the unloading of oxygen at the tissues. However, when measurements of the oxygen-hemoglobin dissociation curve are made in humans who have traveled to high altitude (Fig. 1), under many circumstances there is in fact a net left shift in the oxygen hemoglobin dissociation curve. This left shift is facilitated by the rise in pH and fall in CO2 caused by the hyperventilation driven by systemic hypoxia. Additionally, under some circumstances, it is driven further leftward by a fall in body temperature (68). Furthermore, it is of interest to note that all genetically adapted high altitude animals and the human fetus in the hypoxic intrauterine environment also have left shifted oxygen-hemoglobin dissociation curves, some with P50 values in the teens.
Fig. 1.
Oxygen-hemoglobin dissociation curve demonstrating a left shift among sojourners (○) to high altitude and natives. The left shift in the oxygen-hemoglobin dissociation curve under these circumstances demonstrates that the combined effects of hypocapnia, increased pH, and cold override the simple effects of 2–3 DPG on the oxygen-hemoglobin dissociation curve. These data are an outstanding example of the limits of single mechanism reductionism. They are also consistent with the left shift seen in many genetically adapted animals that are native to high altitude. [Reprinted from Ref. 68, with permission from Elsevier.]
These observations make it seem likely that the main adaptive strategy is to shift the oxygen-hemoglobin dissociation curve to the left to facilitate the “loading” of oxygen at the lung in conditions (altitude) where oxygen availability is limited. This strategy also takes advantage of the fact that the mitochondria in the tissues can work efficiently at very low Po2 values (and that under specific needs such as muscular exercise in hypoxia local increases in [H+] and temperature will reduce the leftward shift in muscle capillaries so that “unloading” of oxygen and tissue O2 levels can be facilitated). It is also of note that the left shift in the oxygen-hemoglobin dissociation curve has been “known” since at least the 1920s. Along these lines, the sequencing of hemoglobin and the understanding of its biophysical properties was one of the earliest triumphs of what has come to be described as molecular biology (55). However, when the interpretation of such discoveries is too narrow, key physiological insights can be missed. The 2–3 DPG story is also an excellent and early example of how physiology trumps reductionist molecular biology as multiple systems and regulatory strategies interact to regulate homeostasis for the whole organism.
PREDICTIVE POWERS OF GENES?
In addition to gene therapy and other molecular treatments for rare diseases, reductionism also made promises about its ability to provide insight about who gets what complex disease like atherosclerosis, diabetes, hypertension, etc. As the quote below demonstrates, this idea became extremely popular after the sequencing of the human genome, and scientific funding agencies like the National Institutes of Health have invested huge sums of money in so-called “genome wide association studies” (GWAS) and other efforts to determine if a few genetic variants are harbingers of future disease in the population as a whole (10, 12, 43).
… because it been known all along that virtually every disease tends to track in families. What has changed is that. . . . .we are now beginning to see possible therapeutic approaches based on gene discoveries that will change the way medicine is practiced (12).
One attractive element of this paradigm was that if a few common variants explained much of the risk for disease like diabetes, then it should be possible to identify those at risk and target them for early intervention. So far, the data from many, if not most or even all of these studies, have been underwhelming (43). First, a large number of variants seem to cause a significant increase in risk, but this increase is small compared with behavioral and environmental factors. An increased risk of several percent seems also likely to fall below what might be described as a phenotypic signal-to-noise ratio. Second, when the gene variants (single nucleotide polymorphisms, SNPs) that have been identified via GWAS or other experimental approaches are tested in large populations, the distribution of risk SNPs is typically strikingly similar in populations with and without disease (50, 63; Fig. 2). Third, when so-called genetic risk scores for disease are compared with predictive algorithms based on traditional risk factors (family history, lifestyle, age, etc.), the genetic risk scores are far less predictive than traditional phenotype-based risk scores. Furthermore, addition of genetic risk elements to phenotypically based scores adds little or no additional predictive power (50, 63). Finally, the idea that identifying prospective genetic risks for complex diseases that include a number of lifestyle and environmental factors (and increasingly even prenatal factors) is fundamentally wishful thinking, because behavioral health issues and culture play such a dominant role in determining who gets what disease when, and it is unclear if people will change their behavior in a positive way if they know prospectively they are at increased risk (24). Paradoxically, perhaps those at reduced genetic risk would pay less attention to behavioral risks.
Fig. 2.
Distribution of so-called high risk genes for cardiovascular disease in women with and without known coronary artery disease. The distribution of risk genes is similar, and construction of a genetic risk score for cardiovascular disease is thus problematic. This is just one example of the limited predictive power of “genomics” as it relates to the ability of relatively common gene variants to predict common diseases. [Borrowed with permission from Ref. 50. Copyright © 2010 American Medical Association. All rights reserved.]
SUCCESS IN PHARMACOGENOMICS AND ANTHROPOLOGY
So far, this paper has offered a sharp critique of the reductionists and taken the position that they over-sold what their technology had to offer on both the individual (gene therapy) basis and also in terms of population risk and intervention. However, there have been some notable successes stemming from application of this technology and two that seem especially worthy of comment. For example, there has been success in so-called pharmacogenomics. It has been well-known for some time that there are “responders” and “non-responders” to many forms of drug therapy. In many cases, this is related to how rapidly drugs are metabolized. In the case of tamoxifen, which had a dramatic effect on the recurrence of breast cancer, individuals with decreased drug metabolism appear to be at increased risk for recurrence. This is especially important for drugs like tamoxifen, which are ingested as pro-drugs with one or more metabolites that are active (56).
Another field where “omic” approaches have yielded dividends is anthropology. Two good examples include discoveries related to the independent development of lactase persistence into adulthood in areas of the world that were early adopters of herding (23, 34). In this context, one can imagine that the ability to digest lactose into adulthood provided the affected individuals a significant survival advantage and thus became the dominant genotype in only a few generations. Another good example that is perhaps counterintuitive relates to the individuals who migrated to the Tibetan plateau. These individuals do not develop chronic mountain sickness even with lifelong living at 3–4,000 m of elevation. These responses contrast to the high altitude natives in the Andes Mountains, who do develop chronic mountain sickness (58, 61, 70). Along these lines, those who migrated to the Tibetan plateau appear to have had selection pressure that favored a less functional variant of the hypoxia-inducible factor that, among other things, prevents them from developing excessive polycythemia, which plays a critical role in chronic mountain sickness.
INTERIM SUMMARY
So far, I have provided a general critique of what might broadly be termed “molecular reductionism”. I have presented evidence that its failure to live up to its self-generated hype is in reality a failure to recognize larger ideas about homeostasis and regulation that are central to physiology. This includes the specific example of the idea of gene therapy for relatively common genetic disorders like cystic fibrosis and also the limited predictive power of gene variants for common diseases. The question now is whether there is some way out of this problem and a better way to use potentially powerful technologies championed by the reductionists in a biomedical context.
IS SYSTEMS BIOLOGY THE ANSWER?
One idea to address the “failure” of molecular reductionism described above is to use a new approach called systems biology. The idea is that if powerful modeling tools and other data analysis techniques could be applied to the data generated via high throughput molecular reductionism, then somehow more meaningful insights would be generated and ultimately exploited for predictive or therapeutic purposes. The rationale for systems biology comes from a sampling of the comments on www.systemsbiology.org web site (34a).
Systems biology is the study of an organism, viewed as an integrated and interacting network of genes, proteins and biochemical reactions which give rise to life. Instead of analyzing individual components or aspects of the organism, such as sugar metabolism or a cell nucleus, systems biologists focus on all the components and the interactions among them, all as part of one system. These interactions are ultimately responsible for an organism's form and functions.
Traditional biology—the kind most of us studied in high school and college, and that many generations of scientists before us have pursued—has focused on identifying individual genes, proteins and cells, and studying their specific functions. But that kind of biology can yield relatively limited insights about the human body.
Biologists, geneticists, and doctors have had limited success in curing complex diseases such as . . . . diabetes because traditional biology generally looks at only a few aspects of an organism at a time.
To a physiologist, there are obvious problems with systems biology. The problems start with the fact that physiology has been attempting for hundreds of years to understand the integrated function of organs and whole organisms that culminated in unifying big ideas about homeostasis and regulation discussed earlier. It is also clear that the type of biology that physiologists have been interested in starting with Harvey and the circulation has been about systems and has used modeling and computational techniques (1, 32, 57). Additionally, at this time the concept of systems biology and how it is defined remains very nebulous (52). Is systems biology a new discipline, an approach, a collection of tools, or merely a new name for integrative physiology generated by individuals who are generally unaware that our field exists (2, 28, 34a, 36, 40, 41, 45, 57)? Clearly physiology has provided and continues to provide insight about human disease, including insight that has led to vast therapeutic advances in recent years (37). Perhaps, the obvious question for the advocates of the cell-centric view of systems biology is did they skip physiology as part of their course work as students?
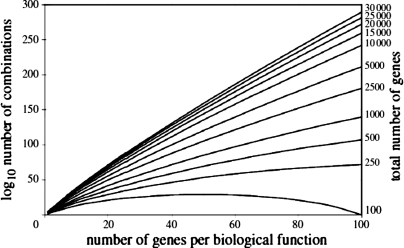
The concerns about systems biology outlined above at some level are about definitions and perhaps intellectual ownership. However, it also seems fair to ask what the long-term outlook for cell-centric systems biology is as an approach to making sense out of the vast amounts of data that can be generated using modern “omic” technology. In this context, there are key intellectual issues related to how data elements are generated, their spatial and temporal relationships, and how many ways they might interact (Fig. 3) that question the very fundamental assumptions about systems biology and its reliance on “bottom up” or “hypothesis neutral” modeling (2, 6, 15, 27, 35, 36, 38, 48, 67). It seems to me that without a narrative approach that includes hypothesis testing and key concepts like homeostasis, systems biology runs the risk of becoming scientific “Abstract Expressionism”. Given the issues discussed earlier with gene therapy and GWAS approaches and the hype that surrounds systems biology, these concerns raise questions about what kind of science and scientific approaches deserve our future attention and funding (2, 24, 35).
Fig. 3.
Simulation of a number of possible combinations of genes gene interactions depending on the number of genes per biological function (x-axis) and the total number of genes in the organism. For biological functions with roughly 50 genes, ∼10150 possible combinations exist for most mammals. This figure shows the immense challenge associated with hypothesis-neutral systems biology and “bottom up” modeling. [Borrowed with permission from Ref. 46.]
REDUCTIONISM STALLS PHYSIOLOGY PROGRESSES
This is not the place for a comprehensive review of the contributions of physiology to biomedical research and therapeutic progress over the last 20–30 years. However, a few highlights that were initially seen as counterintuitive seem warranted. An obvious one is the discovery of EDRF and nitric oxide (25). This observation, which challenged the idea of the endothelium as merely a barrier, led to the discovery of gas-based signaling mechanisms and new therapeutic targets for conditions as diverse as erectile dysfunction and pulmonary hypertension. Would gas-based signaling mechanisms have been discovered by sequencing genes? Physiology has also helped redefine the optimal strategy used during mechanical ventilation in patients with adult respiratory distress syndrome (ARDS; 26). This has led to abandonment of strategies associated with high airway pressures and maintenance of arterial blood gases toward so-called permissive hypercapnia, alternate forms of mechanical ventilation and pressure support. Importantly, these new strategies that emphasize the avoidance of barotrauma have been associated with significant reductions in morbidity and mortality for ARDS. While part of the conventional wisdom now, this strategy was initially seen as counterintuitive.
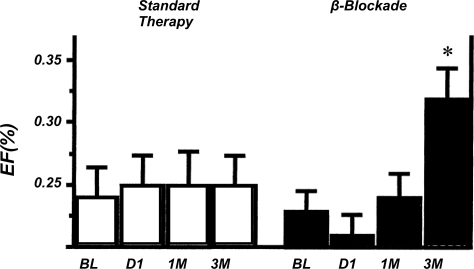
Another example of a counterintuitive physiologically based clinical strategy was the use of beta-blockers in congestive heart failure. For many years these drugs were contraindicated in congestive heart failure (CHF) because it was felt that high sympathetic drive to the heart was required to maintain an adequate cardiac output in CHF. In reality, high sympathetic activity to the heart over time contributed to the progression of the disease and promoted a downward spiral of cardiac remodeling and reduced function (20). Thus the use of beta-blockers along with vasodilator therapy has been revolutionary and can interrupt or slow the downward spiral noted above in patients with congestive heart failure (Fig. 4). Again, the conventional wisdom was turned on its head and provided new insights that ultimately led to improved therapy. In the case of ARDS and congestive heart failure there has also been a two-way street between observations from clinical research conducted “at the bedside” to more fundamental observations in the laboratory.
Fig. 4.
Demonstration that beta-blockade can improve ventricular function (%EF) in humans with congestive heart failure over time. Standard therapy was associated with stable ventricular ejection fraction over 3 mo. By contrast, metoprolol (β-blockade) increased ventricular ejection fraction by >50% over 3 mo (*<0.05 vs. baseline). This finding, while initially counterintuitive, was based on sound physiological reasoning and along with other therapies has improved outcomes for patients with congestive heart failure. [Adapted from Ref. 20, with permission from Wolters Kluwer Health.]
Three other examples of more straight forward physiologically based therapeutic successes in recent years include the long story of improved outcomes for premature infants cared for in the neonatal ICU including altered ventilatory strategies, avoidance of oxygen toxicity, and surfactant therapy (9, 60). These improved outcomes, in the littlest ICU survivors, continue to seem miraculous to individuals who care for these patients and practiced medicine or nursing prior to their use. A second example has been oral rehydration solutions that are life saving in infants and children with diarrheal disease, especially in developing countries where it is a primary and frequent cause of death (8). Finally, in the developed world, where obesity and physical inactivity are leading to a pandemic of type 2 diabetes, physical activity (especially walking training in middle-aged people) has been proven to be highly effective in preventing, limiting, and in some cases reversing type 2 diabetes (16, 29). Each of these therapeutic successes is based on a foundation of physiologically based experimental evidence and insights.
REDUNDANCY, FEEDBACK, AND ACCLIMATION/ADAPTATION
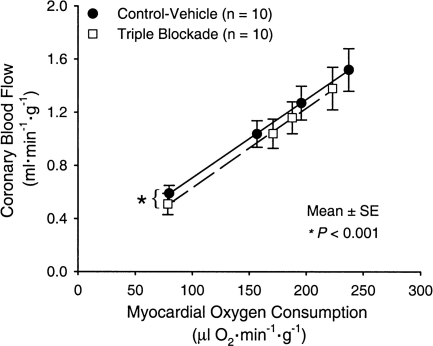
Why has physiology continued to contribute in the era of reductionism? Physiologists are well versed in the overall concept of homeostasis, regulation, feedback, redundancy, and acclimation/adaptation. A classic example of redundancy comes from coronary circulation where coronary vasodilation is tightly linked to myocardial oxygen demand. In this context, a number of vasodilator systems likely contribute to this response. However, pharmacological blockade of one system, or in fact multiple systems, fails to alter this fundamental relationship between coronary vasodilation and myocardial oxygen demand in most species (19, 64; Fig. 5) This suggests that multiple redundant pathways contribute to this critical physiological response so that when one is blocked or absent, oxygen supply to the heart is not threatened when demand rises.
Fig. 5.
Myocardial oxygen demand on the x-axis and coronary blood flow on the y-axis. Note that coronary blood flow rises in proportion to myocardial oxygen demand and that this rise is unaffected by triple inhibition of kATP+ channels, nitric oxide synthase, and adenosine receptor s This is a classic example of the concept of physiological redundancy. This well-known phenomenon may also explain why the absence of many so-called critical genes or proteins has limited impact on overall organ or organism function. This is because so-called redundant systems are able to alter their function and “upregulate” when one or more systems is blocked. [Borrowed with permission from Ref. 64.]
The fundamental relationship between coronary vasodilation and myocardial oxygen demand is also an observation that has had vast therapeutic implications and explains in large part why age specific death rates for cardiovascular disease have fallen dramatically over the last 30–40 years. There are drugs the reduce myocardial oxygen demand, mechanical therapy like stents, bypass surgery that improves myocardial oxygen delivery, and other drugs and lifestyle interventions that can affect both elements of the equation over time (30, 44). This physiological narrative and the progress that has flowed from it is in stark contrast to the relative lack of progress against cancer where there does not seem to be a unifying physiologically based story or model that can be exploited to address the general problem of cancer.
One of the classic feedback control mechanisms in physiology is the arterial baroreflex. While barodenervated animals have relatively normal blood pressure over a given 24 h period, their blood pressure becomes much more variable (14). The relative stability of blood pressure in the long run shows the power of redundant control via renal regulation of arterial pressure. However, for short-term adaptations, essential for things like exercise or changes in posture, feedback control from arterial baroreflexes is essential for normal physiological responses.
An outstanding example of how humans acclimatize and adapt to physiological stress comes from studies that demonstrate that the ability of individuals to exercise in the heat can be remarkably improved by a few weeks of training in the heat (54). This improved exercise tolerance in the heat is associated with expanded plasma volume, increased sweating, and altered thermoregulatory skin blood flow. Another outstanding example is what might be called the adaptability of insulin sensitivity and glucose uptake in skeletal muscle. These variables are extremely sensitive to exercise and changes in daily activity and seem especially relevant in the era of the physical inactivity/obesity pandemic (29, 49, 53, 65).
Ideas about redundancy, feedback control, and acclimation/adaptation are also why physiologists are not that surprised by the ability of various gene knockout animals to survive and thrive (33). At some level this approach is conceptually similar to the classic denervation or high dose pharmacological blockade studies used by physiologists for generations and primarily show the power of the regulatory mechanisms highlighted above to preserve both long term phenotype and homeostasis despite the loss of one or more critical pathways or mechanisms (17). In this context, it is not surprising the yeast can survive without 80% of their genes and the function of these genes only becomes apparent when the organism is stressed (33). Is it too cynical to point out that knockout animals are essentially a “can't lose” experimental approach? If the knockout is lethal or leads to significant phenotypic dysfunction it is essential. If it survives then genetic or other compensatory mechanisms were upregulated to overcome the absence of the essential gene.
Physiology or physiologically based tests can also provide insight into the risk of future disease and/or predictive outcomes. For example, the blood pressure responses to common sympathoexcitatory stress can be used to define those at risk for future hypertension in a way that is potentially much more predictive than any current genetic test. Additionally, tests of autonomic function are strong predictors of outcomes in large populations of humans, and cardiorespiratory fitness is an especially good predictor of all-cause mortality.
TOOLS VS. BIG IDEAS
At some level molecular reductionism and systems biology are at existential cross roads. Are they in fact real disciplines informed by big ideas like homeostasis and regulation, or are they essentially tools and approaches that will facilitate the work of disciplines informed by bigger ideas and more importantly bigger questions and more comprehensive strategies? Based on the concepts and examples highlighted in this paper I would argue that until the vast amounts of data generated by modern “omic” techniques are put in a physiological context it will be an exercise in what Sydney Brenner has deemed “low input, high throughput, no output biology” (6). Along these lines, I want to end on an optimistic note with examples of how physiology is making a difference by applying reductionist tools as part of a more comprehensive approach to important questions. Because the Adolph lecture is sponsored by the Exercise and Environmental Physiology section of the American Physiological Society, relevant examples from related areas will be used. In each case there seems to be an overall hypothesis and a strategy that exploits what might be called responders and non-responders to an intervention.
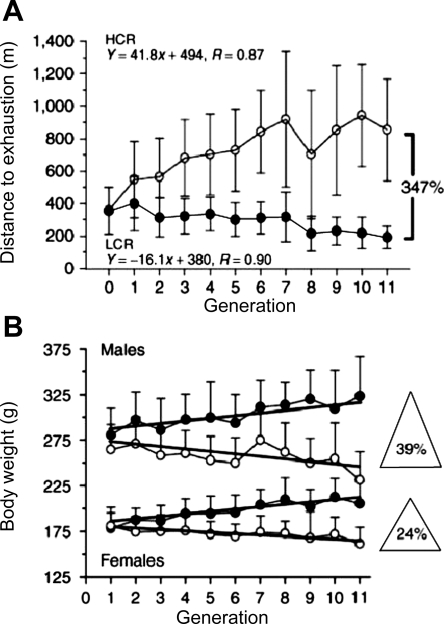
Britton and Koch and colleagues (39, 69) have used selective breeding strategies to develop rats with vastly different inherent aerobic endurance capacities (Fig. 6). These animals have been used in a variety of studies to better understand the gene environment interactions. In many instances the animals selected for low intrinsic aerobic capacity seem to be at increased risk for complex diseases like diabetes, obesity, and heart disease. Additionally, studies using these animals have begun to identify genetic and transcriptional factors and networks that explain in part this increased risk (39).
Fig. 6.
Selective breeding of rats with divergent aerobic capacities. These data show that animals selected for their running capacity diverge dramatically after a few generations and is sustained for many generations. Importantly, at the same time body weight also began to diverge as did a number of risk factors for cardiometabolic disease. Phenotypic studies conducted on these animals in conjunction with more targeted forms of “omic” approaches and other types of molecular reductionism are providing new insights about gene environment interactions. These findings may also have applicability to physically active and inactive humans. The approach of Britton and Koch is a classic example of using reductionist tools in a physiological context to gain new insights with direct applicability to human health and disease. [Reprinted from Ref. 39 with permission from Macmillan Publishers Ltd. Obesity Suppl. copyright 2008.]
Another example of how physiologists are using tools from the “new biology” is the HERITAGE study, which broadly seeks to understand the genetic basis for the differing physiological responses to exercise training in a large number of humans exposed to a standard protocol (3–5). This is an excellent example of how what might be called “high resolution” physiologically based phenotyping in conjunction with genetics. This hypothesis-driven approach also includes uses various “omic” and systems biology approaches and was initiated by physiologists before the terms genomics or systems biology existed. Additionally, like the examples from pharmacogenomics and anthropology discussed earlier, it takes advantage of the fact that there are responders and non-responders in response to a given intervention or environmental stressor.
Finally, my collaborator John Eisenach and I along with our colleagues have performed carefully controlled studies on how common genetic variants in the β2-adrenergic receptor influence a number of physiological responses and how any genotype-based differences might be influenced by dietary sodium (21, 22, 31, 59). These studies were initiated because epidemiological evidence suggested that genetic variation in the β2-adrenergic receptor influenced blood pressure in large populations. In our studies only homozygotes for the genetic variant of interest were recruited in an effort to see the maximum potential physiological effect of the variants. Using this approach, it appears that there are genotype-specific patterns associated with increased cardiac output responses to exercise that may interact with NO-mediated β2-adrenergic receptor peripheral vasodilation. These responses clearly link and mechanistically define how a common gene variant in a key regulatory system can influence a physiological response in humans. They may also provide physiological explanations relevant to the original epidemiological observations on blood pressure and other outcomes, including those in patients with the acute coronary syndrome (42).
SUMMARY
In this paper and in the Adolph Lecture I have highlighted some of the claims associated with molecular reductionism and more recently systems biology. In both cases I have argued that the apparent inability and/or unwillingness of the advocates of these approaches to use key concepts from physiology and ultimately use their tools in a physiological context has limited the contribution of the approaches they advocate. By contrast physiology has continued to use new tools in the service of its big ideas and also continued to provide biomedical insight and therapeutic advances. As the final examples show, it is possible to incorporate reductionist tools in a physiological context to gain broader biomedical insights. Hopefully these insights will fuel the next wave of physiologically inspired therapeutic advances.
GRANTS
My laboratory has been funded continuously by National Institutes of Health since the early 1990s (HL-46493, HL-83947), and our work has also been facilitated by the former Mayo GCRC grant and more recently the CTSA grant (RR-024150). I have also been supported by the American Heart Association, the Mayo Foundation, and the Frank and Shari Caywood Professorship. I would also like to thank the fellows and collaborators who have worked with me, my outstanding technical support staff, and the many subjects who have volunteered for our studies.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author.
ACKNOWLEDGMENTS
The author thanks Drs. Nisha Charkoudian, Doug Seals, and Jerry Dempsey for critical reviews of the manuscript.
REFERENCES
- 1. Auffray C, Noble D. Origins of systems biology in William Harvey's masterpiece on the movement of the heart and the blood in animals. Int J Mol Sci 10: 1658–1669, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beard DA, Kushmerick MJ. Strong inference for systems biology. PLoS Comput Biol 5: 1–10, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, Perusse L, Leon AS, Rao D. Familial aggregation of V̇o2max response to exercise training: results from the HERITAGE Family Study. J Appl Physiol 87: 1003–1008, 1999 [DOI] [PubMed] [Google Scholar]
- 4. Bouchard C, Leon AS, Rao DC, Skinner JS, Wilmore JH, Gagnon J. Aims, design, and measurement protocol. Med Sci Sports Exerc 27: 721–729, 1995 [PubMed] [Google Scholar]
- 5. Bouchard C, Sarzynski MA, Rice TK, Kraus WE, Church TS, Sung YJ, Rao DC, Rankinen T. Genomic predictors of maximal oxygen uptake response to standardized exercise training programs. J Appl Physiol 110: 1160–1170, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brenner S. Sequences and consequences. Phil Trans R Soc B 365: 207–212, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cannon WB. Organization for physiological homeostasis. Physiol Rev 9: 399–431, 1929 [Google Scholar]
- 8. Cheng AC, McDonald JR, Thielman NM. Infectious diarrhea in developed and developing countries. J Clin Gastroenterol 39: 757–773, 2005 [DOI] [PubMed] [Google Scholar]
- 9. Clements JA, Avery ME. Lung surfactant and neonatal respiratory distress syndrome. Am J Respir Crit Care Med 157: S59–S66, 1998 [DOI] [PubMed] [Google Scholar]
- 10. Collins FS. Contemplating the end of the beginning. Genome Res 11: 641–643, 2001 [DOI] [PubMed] [Google Scholar]
- 11. Collins FS. Cystic fibrosis: molecular biology and therapeutic implications. Science 256: 774–779, 1992 [DOI] [PubMed] [Google Scholar]
- 12. Collins FS. The human genome project and the future of medicine. Ann NY Acad Sci USA 882: 42–55, 1999 [DOI] [PubMed] [Google Scholar]
- 13. Comroe JH, Jr, Dripps RD. Ben Franklin and open heart surgery. Circ Res 35: 661–669, 1974 [DOI] [PubMed] [Google Scholar]
- 14. Cowley AW, Jr, Liard JF, Guyton AC. Role of the baroreceptor reflex in daily control of arterial blood pressure and other variables in dogs. Circ Res 32: 564–576, 1973 [DOI] [PubMed] [Google Scholar]
- 15. Csete ME, Doyle JC. Reverse engineering of biological complexity. Science 295: 1664–1669, 2002 [DOI] [PubMed] [Google Scholar]
- 16. Diabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346: 393–403, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Donald DE, Milburn SE, Shepherd JT. Effect of cardiac denervation on the maximal capacity for exercise in the racing greyhound. J Appl Physiol 19: 849–852, 1964 [DOI] [PubMed] [Google Scholar]
- 18. Donaldson SH, Boucher RC. Sodium channels and cystic fibrosis. Chest 132: 1631–1636, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Duncker DJ, Bache RJ. Regulation of coronary blood flow during exercise. Physiol Rev 88: 1009–1086, 2008 [DOI] [PubMed] [Google Scholar]
- 20. Eichhorn EJ, Bristow MR. Medical therapy can improve the biological properties of the chronically failing heart. Circulation 94: 2285–2296, 1996 [DOI] [PubMed] [Google Scholar]
- 21. Eisenach JH, Barnes SA, Pike TL, Sokolnicki LA, Masuki S, Dietz NM, Rehfeldt KH, Turner ST, Joyner MJ. Arg 16/Gly beta-2 adrenergic receptor polymorphism alters the cardiac output response to isometric exercise. J Appl Physiol 99: 1776–1781, 2005 [DOI] [PubMed] [Google Scholar]
- 22. Eisenach JH, McGuire AM, Schwingler RM, Turner ST, Joyner MJ. The Arg16/Gly β-2 adrenergic receptor polymorphism is associated with altered cardiovascular responses to isometric exercise. Physiol Genomics 16: 323–328, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Enattah NS, Jensen TGK, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, El-Shanti H, Seo JK, Alifrangis M, Khalil IF, Natah A, Ali A, Natah S, Comas D, Mehdi SQ, Groop L, Vestergaard EM, Imtiaz F, Rashed MS, Meyer B, Troelsen J, Peltonen L. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet 82: 57–72, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans JP, Meslin EM, Marteau TM, Caulfield T. Genomics. Deflating the genomic bubble. Science 331: 861–862, 2011 [DOI] [PubMed] [Google Scholar]
- 25. Furchgott RF. The discovery of endothelium-derived relaxing factor and its importance in the identification of nitric oxide. JAMA 276: 1186–1188, 1996 [PubMed] [Google Scholar]
- 26. Gillette MA, Hess DR. Ventilator-induced lung injury and the evolution of lung-protective strategies in acute respiratory distress syndrome. Respir Care 46: 130–148, 2001 [PubMed] [Google Scholar]
- 27. Golub TR. Counterpoint: Data first. Nature 464: 679, 2010 [DOI] [PubMed] [Google Scholar]
- 28. Greenhaff PL, Hargreaves M. “Systems biology” in human exercise physiology: is it something different from integrative physiology? J Physiol 589: 1031–1036, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henriksen EJ. Exercise effects of muscle insulin signaling and action. Invited review: effects of acute exercise and exercise training on insulin resistance. J Appl Physiol 93: 788–796, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Heron M, Hoyert DL, Murphy SL, Xu J, Kochanek KD, Tejada-Vera B. Deaths: Final data for 2006. National Vital Statistics Reports; vol 57, no 14 Hyattsville, MD: National Center for Health Statistics, 2009, pp 1–117 [PubMed] [Google Scholar]
- 31. Hesse C, Schroeder DR, Nicholson WT, Hart EC, Curry TB, Penheiter AR, Turner ST, Joyner MJ, Eisenach JH. Beta-2 adrenoceptor gene variation and systemic vasodilatation during ganglionic blockade. J Physiol 588: 2669–2678, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hester RL, Iliescu R, Summers R, Coleman TG. Systems biology and integrative physiological modelling. J Physiol 589: 1053–1060, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St. Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ingram CJE, Mulcare CA, Itan Y, Thomas MG, Swallow DM. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet 124: 579–591, 2009 [DOI] [PubMed] [Google Scholar]
- 34a.Institute for Systems Biology. (Online). http://www.systemsbiology.org [June 22, 2011]
- 35. Joyner MJ, Pedersen BK. Ten questions about systems biology. J Physiol 589: 1017–1030, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joyner MJ. Physiology: alone at the bottom, alone at the top. J Physiol 589: 1005, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joyner MJ. Why physiology matters in medicine. Physiology 26: 72–75, 2011 [DOI] [PubMed] [Google Scholar]
- 38. Kell DB, Oliver SG. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays 26: 99–105, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Koch LG, Britton SL. Development of animal models to test the fundamental basis of gene-environmental interactions. Obesity 16, Suppl 3: S28–S32, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kohl P, Crampin EJ, Quinn TA, Noble D. Systems biology: an approach. Clin Pharmacol Ther 88: 25–33, 2010 [DOI] [PubMed] [Google Scholar]
- 41. Kuster DWD, Merkus D, Van Der Velden J, Verhoeven AJM, Duncker DJ. Integrative Physiology 2.0: integration of systems biology into physiology and its application to cardiovascular homeostasis. J Physiol 589: 1037–1045, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lanfear DE, Jones PJ, Marsh S, Cresci S, McLeod HL, Spertus JA. Beta2-adrenergic receptor genotype and survival among patients receiving beta-blocker therapy after an acute coronary syndrome. JAMA 294: 1526–1533, 2005 [DOI] [PubMed] [Google Scholar]
- 43. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature 461: 747–753, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nelson RR, Gobel FL, Jorgensen CR, Wang K, Wang Y, Taylor HL. Hemodynamic predictors of myocardial oxygen consumption during static and dynamic exercise. Circulation 50: 1179–1189, 1974 [DOI] [PubMed] [Google Scholar]
- 45. Noble D. New-Darwinism, the Modern Synthesis and selfish genes: are they of use in physiology? J Physiol 589: 1007–1015, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Noble D. Differential and integral views of genetic in computational systems biology. Interface Focus 1: 7–15, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noble D. Genes and causation. Phil Trans R Soc A 366: 3001–3015, 2008 [DOI] [PubMed] [Google Scholar]
- 48. Nurse P, Hayles J. The cell in an era of systems biology. Cell 144: 850–854, 2011 [DOI] [PubMed] [Google Scholar]
- 49. Olsen RH, Krogh-Madsen R, Thomsen C, Booth FW, Pedersen BK. Metabolic responses to reduced daily steps in healthy nonexercising men. JAMA 299: 1261–1263, 2008 [DOI] [PubMed] [Google Scholar]
- 50. Paynter NP, Chasman DI, Pare G, Buring JE, Cook NR, Miletich JP, Ridker PM. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA 303: 631–637, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pearson H. One gene, twenty years. Nature 460: 165–169, 2009 [DOI] [PubMed] [Google Scholar]
- 52. Powell A, O'Malley MA, Muller-Wille S, Calvert J, Dupre J. Disciplinary baptisms: a comparison of the naming stores of genetics, molecular biology, genomics, and systems biology. Hist Phil Life Sci 29: 5–32, 2007 [PubMed] [Google Scholar]
- 53. Rogers MA, Yamamoto C, King DS, Hagberg JM, Ehsani AA, Holloszy JO. Improvement in glucose tolerance after 1 wk of exercise in patients with mild NIDDM. Diabetes Care 11: 613–618, 1988 [DOI] [PubMed] [Google Scholar]
- 54. Robinson S, Turrell ES, Belding HS, Horvath SM. Rapid acclimatization to work in hot climates. Am J Physiol 140: 168–176, 1943 [Google Scholar]
- 55. Rossman MG. Chapter 3: Recollection of the events leading to the discovery of the structure of haemoglobin. J Mol Biol 392: 23–32, 2009 [DOI] [PubMed] [Google Scholar]
- 56. Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, Fritz P, Simon W, Suman VJ, Ames MM, Safgren SL, Kuffel MJ, Ulmer HU, Bolander J, Strick R, Beckmann MW, Koelbl H, Weinshilbum RM, Ingle JN, Eichelbaum M, Schwab M, Brauch H. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 302: 1429–1436, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Secomb TW, Pries AR. The microcirculation: physiology at the mesoscale. J Physiol 589: 1047–1042, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, Prchal JT, Ge R. Genetic evidence for high-altitude adaptation in Tibet. Science 329: 72–75, 2010 [DOI] [PubMed] [Google Scholar]
- 59. Snyder EM, Beck KC, Dietz NM, Eisenach JH, Joyner MJ, Turner ST, Johnson BD. Arg16Gly polymorphism of the β2-adrenergic receptor is associated with differences in cardiovascular function at rest and during exercise in humans. J Physiol 571: 121–130, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sol RF. Current trials in the treatment of respiratory failure in preterm infants. Neonatology 95: 368–372, 2009 [DOI] [PubMed] [Google Scholar]
- 61. Storz JF. Genes for high altitudes. Science 329: 40–41, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Talmud PJ, Hingorani AD, Cooper JA, Marmot MG, Brunner EJ, Kumari M, Kivimaki M, Humphries SE. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. Br Med J 340: b4838, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tune JD, Richmond KN, Gorman MW, Feigl EO. K(ATP)(+) channels, nitric oxide, and adenosine are not required for local metabolic coronary vasodilation. Am J Physiol Heart Circ Physiol 280: H868–H875, 2001 [DOI] [PubMed] [Google Scholar]
- 65. van Dieren S, Beulens JW, van der Schouw YT, Grobbee DE, Neal B. The global burden of diabetes and its complications: an emerging pandemic. Eur J Cardiovasc Prev Rehabil 17, Suppl 1: S3–S8, 2010 [DOI] [PubMed] [Google Scholar]
- 66. Verkman AS, Galietta LJV. Chloride channels as drug targets. Nat Rev 8: 153–171, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Weinberg RA. Point: Hypotheses first. Nature 464: 678, 2010 [DOI] [PubMed] [Google Scholar]
- 68. Winslow RM. The role of hemoglobin oxygen affinity in oxygen transport at high altitude. Respir Physiol Neurobiol 158: 121–127, 2007 [DOI] [PubMed] [Google Scholar]
- 69. Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernstrom M, Rezaei K, Lee SJ, Koch LG, Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 307: 418–420, 2005 [DOI] [PubMed] [Google Scholar]
- 70. Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng H, Liu T, He W, Li K, Luo R, Nie X, Wu H, Zhao M, Cao H, Zou J, Shan Y, Li S, Yang Q, Asan Ni P, Tian G, Xu J, Liu X, Jiang T, Wu R, Zhou G, Tang M, Qin J, Wang T, Feng S, Li G, Huasang Luosang J, Wang W, Chen F, Wang Y, Zheng X, Li Z, Bianba Z, Yang G, Wang X, Tang S, Gao G, Chen Y, Luo Z, Gusang L, Cao Z, Zhang Q, Ouyang W, Ren X, Liang H, Zheng H, Huang Y, Li J, Bolud L, Kristiansen K, Li Y, Zhang Y, Zhang X, Li R, Li S, Yang H, Nielsen R, Wang J, Wang J. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]