In this issue of the JCI, Wu et al. (1) provide the first direct evidence that brominating species may contribute to the tissue damage that accompanies asthma. The peroxidase of eosinophils generates these reactive species, and the selective recruitment and activation of eosinophils in lung tissue is a hallmark of this inflammatory disease.
Asthma, which is reaching epidemic proportions in the developed world, is characterized by narrowed airways that are hyperresponsive to stimulation (reviewed in refs. 2 and 3). Its clinical hallmarks are paroxysms of dyspnea, cough, and wheezing. The chronic airway narrowing reflects inflammation, extracellular matrix deposition, and hypersecretion of mucus. Allergen exposure and other nonspecific stimuli trigger the constriction of smooth muscle cells in the bronchiolar wall, resulting in acute airway constriction and onset of symptoms.
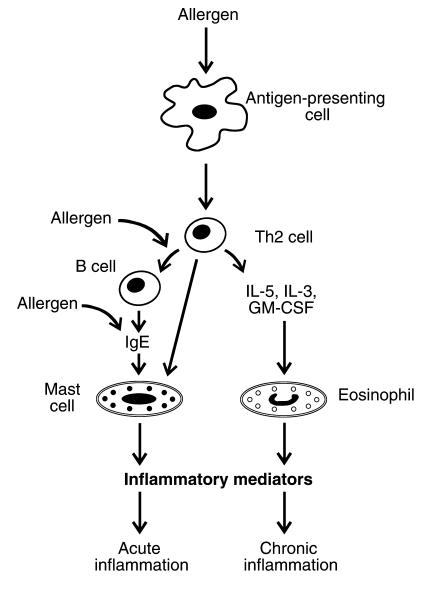
Allergens appear to lack intrinsic toxicity, but they become deleterious when they induce an IgE response (Figure 1) after contacting mucosal surfaces. Antigen-presenting cells then stimulate Th2 cells, a subtype of CD4+ T-helper cells, to form proinflammatory cytokines. In turn, the cytokines prompt B cells to generate allergen-specific IgE. Cytokines also trigger the release of eosinophils from bone marrow and recruit them into lung tissue. In asthmatic individuals, allergen exposure promotes the cross-linking of the IgE to its receptor on mast cells, resulting in mast-cell degranulation and the early phase of bronchoconstriction. The late-phase inflammatory response of asthma involves recruitment and activation of additional cells, including eosinophils, basophils, neutrophils, and macrophages.
Figure 1.
Proposed mechanisms for the pathogenesis of inflammation in asthma. Whereas mast cells are directly stimulated by allergen-specific IgEs and provoke the early phase of asthmatic bronchoconstriction, eosinophils activate later inflammatory responses.
Eosinophils and the eosinophil peroxidase.
Under normal conditions, eosinophils are a minor component of the circulating phagocytic white blood cell population (4). However, in asthmatic individuals and individuals infected with parasites, levels of both IgE and eosinophils are elevated in blood, suggesting that the allergic response originally evolved to play a role in host-defense mechanisms.
Inflammatory mediators generated by activated eosinophils have long been thought to be a key component of the late phase of the allergic response (4, 5). One potential mechanism involves the production of oxidizing intermediates. Activated eosinophils generate such intermediates by first producing superoxide, using a membrane-associated NADPH oxidase (6). Superoxide then dismutates to hydrogen peroxide (H2O2), which is the oxidizing substrate for eosinophil peroxidase, a heme protein secreted by eosinophils (7). This peroxidase resides in cytoplasmic granules, where it contributes to the eosin staining that is the histochemical hallmark of eosinophils. The reactive intermediates generated by this enzyme may account for much of the eosinophil-mediated damage to allergic tissues. Similarly, a functionally and genetically distinct peroxidase, myeloperoxidase, which is produced by neutrophils, monocytes, and macrophages, generates oxidants that are implicated in oxidative reactions in the human artery wall (8, 9).
At plasma concentrations of halide ion (Cl– ∼100 mM, Br– 20–100 μM, I– < 1 μM), the major product of eosinophil peroxidase is hypobromous acid (HOBr) (10): Br– + H2O2 + H+ → HOBr + H2O. In vitro, HOBr reacts with primary amines to form bromamines, and it converts tyrosine to 3-bromotyrosine (11, 12). Eosinophil peroxidase also can use substrates other than Br– in vitro, including thiocyanate and nitrite (13, 14). Certain bodily fluids can contain high concentrations of these substrates, suggesting that this chemistry may be physiologically relevant.
Seeking traces of reactive metabolites.
The oxidative pathways that damage biomolecules in vivo are difficult to identify because the toxic intermediates are short-lived and difficult to measure directly. One solution is to analyze normal and pathological tissues for stable end-products of oxidative reactions that have been identified through in vitro studies (15). For example, unnatural but stable isomers of tyrosine — o-tyrosine and m-tyrosine — result from post-translational modifications of protein-bound phenylalanine residues by hydroxyl radical. Reaction of tyrosine with hypochlorous acid, on the other hand, yields 3-chlorotyrosine, and 3-nitrotyrosine is generated when reactive nitrogen intermediates oxidize tyrosine. Because these abnormal amino acid products are stable to acid hydrolysis, they are potentially useful markers for protein oxidation in vivo (15, 16).
Wu et al. looked for stable markers to determine whether brominating species generated by eosinophil peroxidase are associated with airway hyperreactivity in asthma (1, 12). In vitro studies demonstrated that 3-bromotyrosine represents a specific marker of protein oxidation by eosinophil peroxidase. Moreover, analysis of endobronchial biopsy specimens revealed that lung segments of allergen-challenged asthmatic patients, but not healthy control subjects, exhibited intense eosinophil infiltration. Isotope dilution gas chromatography/mass spectrometric analysis — a sensitive and specific analytical technique — found little difference in baseline levels of 3-bromotyrosine in bronchoalveolar lavage proteins from control and mildly allergic asthmatic individuals. In striking contrast, there was a tenfold increase in the 3-bromotyrosine content of bronchoalveolar lavage proteins when asthmatic patients were challenged with allergen, and this increase was not seen in control subjects. These observations provide the first evidence that reactive brominating intermediates generated by eosinophils modify proteins in vivo.
Bromination of tyrosine by eosinophil peroxidase represents the third example of a halogenation reaction executed by peroxidases in humans. Thyroid hormone synthesis has long been known to involve iodination of tyrosine residues by thyroid peroxidase in a reaction that requires hydrogen peroxide and iodide ion (17). Recent studies have shown that hypochlorous acid generated by myeloperoxidase chlorinates tyrosine residues in vitro (8, 18), and elevated levels of that enzyme and 3-chlorotyrosine have been detected in human atherosclerotic lesions (9, 19), implicating hypochlorous acid generation by myeloperoxidase in protein halogenation in vivo.
The detection of 3-bromotyrosine in bronchoalveolar lavage proteins raises the exciting possibility that oxidants generated by eosinophils are important in the pathogenesis of inflammation in asthma and other allergic disorders. These findings may be of broad significance because eosinophils are implicated in the tissue damage of dermatitis, vasculitis, and the hypereosinophilia syndromes (2, 4). However, detection of amino acid markers cannot establish the physiological relevance of oxidation chemistry. Human studies and the use of animal models that overexpress or lack eosinophil peroxidase will be required. Genetic studies already have implicated several candidate genes in allergic disease (20). If future studies reveal that eosinophil peroxidase–dependent reaction pathways also are important contributors to airway oxidative damage, it might be possible to develop specific eosinophil peroxidase inhibitors for managing asthma and its complications.
References
- 1.Wu W, et al. Eosinophils generate brominating oxidants in allergen-induced asthma. J Clin Invest. 2000;105:1455–1463. doi: 10.1172/JCI9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402(Suppl.):B2–B4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 3.Bochner BS, Undem BJ, Lichtenstein LM. Immunological aspects of allergic asthma. Annu Rev Immunol. 1994;12:295–335. doi: 10.1146/annurev.iy.12.040194.001455. [DOI] [PubMed] [Google Scholar]
- 4.Weller PF. Eosinophils: structure and functions. Curr Opin Immunol. 1994;6:85–90. doi: 10.1016/0952-7915(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 5.Barnes PJ, Chung KF, Page CP. Inflammatory mediators of asthma: an update. Pharmacol Rev. 1998;50:515–596. [PubMed] [Google Scholar]
- 6.Klebanoff SJ, Locksley RM, Jong EC, Rosen H. Oxidative response of phagocytes to parasite invasion. Ciba Found Symp. 1983;99:92–112. doi: 10.1002/9780470720806.ch6. [DOI] [PubMed] [Google Scholar]
- 7.Jong EC, Henderson WR, Klebanoff SJ. Bactericidal activity of eosinophil peroxidase. J Immunol. 1980;124:1378–1382. [PubMed] [Google Scholar]
- 8.Hazen SL, Hsu FF, Mueller DM, Crowley JR, Heinecke JW. Human neutrophils employ chloride gas as an oxidant during phagocytosis. J Clin Invest. 1996;98:1283–1289. doi: 10.1172/JCI118914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazen SL, Heinecke JW. 3-Chlorotyrosine, a specific marker of myeloperoxidase-catalyzed oxidation, is markedly elevated in low density lipoprotein isolated from human atherosclerotic intima. J Clin Invest. 1997;99:2075–2081. doi: 10.1172/JCI119379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss SJ, Test ST, Eckmann CM, Ross D, Regiania S. Brominating oxidants generated by human eosinophils. Science. 1986;234:200–203. doi: 10.1126/science.3018933. [DOI] [PubMed] [Google Scholar]
- 11.Thomas EL, Bozeman PM, Jefferson MM, King CC. Oxidation of bromide by the human leukocyte enzymes myeloperoxidase and eosinophil peroxidase. Formation of bromamines. J Biol Chem. 1995;270:2906–2913. doi: 10.1074/jbc.270.7.2906. [DOI] [PubMed] [Google Scholar]
- 12.Wu W, Chen Y, d’Avignon A, Hazen SL. 3-Bromotyrosine and 3,5-dibromotyrosine are major products of protein oxidation by eosinophil peroxidase. Potential markers for eosinophil-dependent tissue injury in vivo. Biochemistry. 1999;38:3538–3548. doi: 10.1021/bi982401l. [DOI] [PubMed] [Google Scholar]
- 13.Slungaard A, Mahoney JR., Jr Thiocyanate is the major substrate for eosinophil peroxidase in physiologic fluids. Implications for cytotoxicity. J Biol Chem. 1991;266:4903–4910. [PubMed] [Google Scholar]
- 14.Wu W, Chen Y, Hazen SL. Eosinophil peroxidase nitrates protein tyrosyl residues. Implications for oxidative damage by nitrating intermediates in eosinophilic inflammatory disorders. J Biol Chem. 1999;274:25933–25944. doi: 10.1074/jbc.274.36.25933. [DOI] [PubMed] [Google Scholar]
- 15.Heinecke JW. Mass spectrometric quantification of amino acid oxidation products in proteins: insights into pathways that promote LDL oxidation in the human artery wall. FASEB J. 1999;13:1113–1120. doi: 10.1096/fasebj.13.10.1113. [DOI] [PubMed] [Google Scholar]
- 16.Heinecke JW, et al. Detecting oxidative modification of biomolecules with isotope dilution mass spectrometry: sensitive and quantitative assays for oxidized amino acids in proteins and tissues. Methods Enzymol. 1999;300:124–144. doi: 10.1016/s0076-6879(99)00121-4. [DOI] [PubMed] [Google Scholar]
- 17.Ekholm R. Biosynthesis of thyroid hormones. Int Rev Cytol. 1990;120:243–288. doi: 10.1016/s0074-7696(08)61602-2. [DOI] [PubMed] [Google Scholar]
- 18.Domigan NM, Charlton TS, Duncan MW, Winterbourn CC, Kettle AJ. Chlorination of tyrosyl residues in peptides by myeloperoxidase and human neutrophils. J Biol Chem. 1995;270:16542–16548. doi: 10.1074/jbc.270.28.16542. [DOI] [PubMed] [Google Scholar]
- 19.Daugherty A, Rateri DL, Dunn J, Heinecke JW. Myeloperoxidase, a catalyst for lipoprotein oxidation, is expressed in human atherosclerotic lesions. J Clin Invest. 1994;94:437–444. doi: 10.1172/JCI117342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broide DH, Hoffman H, Sriramarao P. Genes that regulate eosinophilic inflammation. Am J Hum Genet. 1999;65:302–307. doi: 10.1086/302520. [DOI] [PMC free article] [PubMed] [Google Scholar]