Summary
The Tet family of enzymes (Tet1/2/3) converts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). Mouse embryonic stem cells (mESCs) highly express Tet1 and have an elevated level of 5hmC. Tet1 has been implicated in ESC maintenance and lineage specification in vitro but its precise function in development is not well defined. To establish the role of Tet1 in pluripotency and development we have generated Tet1 mutant mESCs and mice. Tet1−/− ESCs have reduced levels of 5hmC, subtle changes in global gene expression, are pluripotent and support development of live-born mice in tetraploid complementation assay but display skewed differentiation towards trophectoderm in vitro. Tet1 mutant mice are viable, fertile and grossly normal though some mutant mice have a slightly smaller body size at birth. Our data suggest that Tet1 loss leading to a partial reduction in 5hmC levels does not affect pluripotency in ESCs and is compatible with embryonic and postnatal development.
Keywords: Tet1, DNA demethylation, 5-hydroxymethylcytosine, 5hmC, embryonic development, Tet1 knockout mice
Introduction
DNA methylation is a well-defined epigenetic modification that is essential for normal development and regulation of gene expression. Mechanisms of establishment and maintenance of DNA methylation are well characterized. Conversely, little is known about mechanisms that underlie DNA demethylation (Ooi and Bestor, 2008; Wu and Zhang, 2010). Passive demethylation has been proposed as simply the failure of the maintenance methyltransferase (DNMT1) upon DNA replication (Dean et al., 2003). This model however cannot explain examples of replication-independent DNA demethylation most notably observed in the paternal pronucleus in the zygote and during reprogramming of primordial germ cells. A number of models of active demethylation have been proposed, from simple excision of the methyl group to more intricate pathways that involve mechanisms of DNA repair (Wu and Zhang, 2010).
Recently, the Tet (Ten-eleven translocation) family of methyldioxygenases (Tet1, Tet2 and Tet3) has been implicated in DNA demethylation (Ito et al., 2010; Tahiliani et al., 2009). These enzymes have 2-oxoglutarate (2OG)-and Fe- (II) dependent oxygenase activity and catalyze the hydroxylation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). Given that DNMT1 recognizes 5hmC poorly, this modification could lead to passive demethylation (Valinluck and Sowers, 2007). It has also been proposed that 5hmC could be involved in active DNA demethylation whereby it undergoes spontaneous or enzymatic conversion to cytosine, or serves as an intermediate that can be subjected to DNA repair and then replaced with cytosine (Ooi and Bestor, 2008). Indeed, a recent study reports that Tet1 mediated hydroxylation of 5mC promotes active DNA demethylation in the adult brain through a process that requires base excision repair pathway involving AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA-editing enzyme complex) family of cytidine deaminases (Guo et al., 2011).
Though the Tet enzymes are expressed in various tissues, the 5hmC base is particularly abundant in Purkinje neurons (Kriaucionis and Heintz, 2009) and mouse embryonic stem cells (mESCs) (Ito et al., 2010; Tahiliani et al., 2009). Recently it has been shown that Tet1 and 5hmC are enriched at transcriptional start sites of CpG rich promoters and gene bodies in ES cells, where it promotes DNA demethylation and transcription. Interestingly, Tet1 also binds to polycomb target gene promoters and plays a role in repression of polycomb-controlled developmental regulators (Williams et al., 2011; Wu et al., 2011). These findings suggest that Tet1 could have an important functional role in maintaining pluripotency and development. Using small hairpin RNAs (shRNAs) to knockdown Tet1 in mESCs and one-cell stage embryos, one study reported that Tet1 maintains expression of Nanog and is required for mESC self-renewal and inner cell mass specification (Ito et al., 2010). Conversely, another study found that Tet1 and Tet2 are downstream targets of Oct4 but knockdown of neither of these genes affect Nanog expression or the pluripotent state of ES cells (Koh et al., 2011b). However, the latter study reported that Tet1 depletion skews differentiation towards endoderm-mesoderm lineages in a teratoma assay. Together, these findings imply a potential requirement for Tet1 in maintenance of pluripotency and normal embryogenesis and warrant further elucidation of the role of Tet1 in pluripotency and development.
Results
Generation of Tet1 knockout ES cells
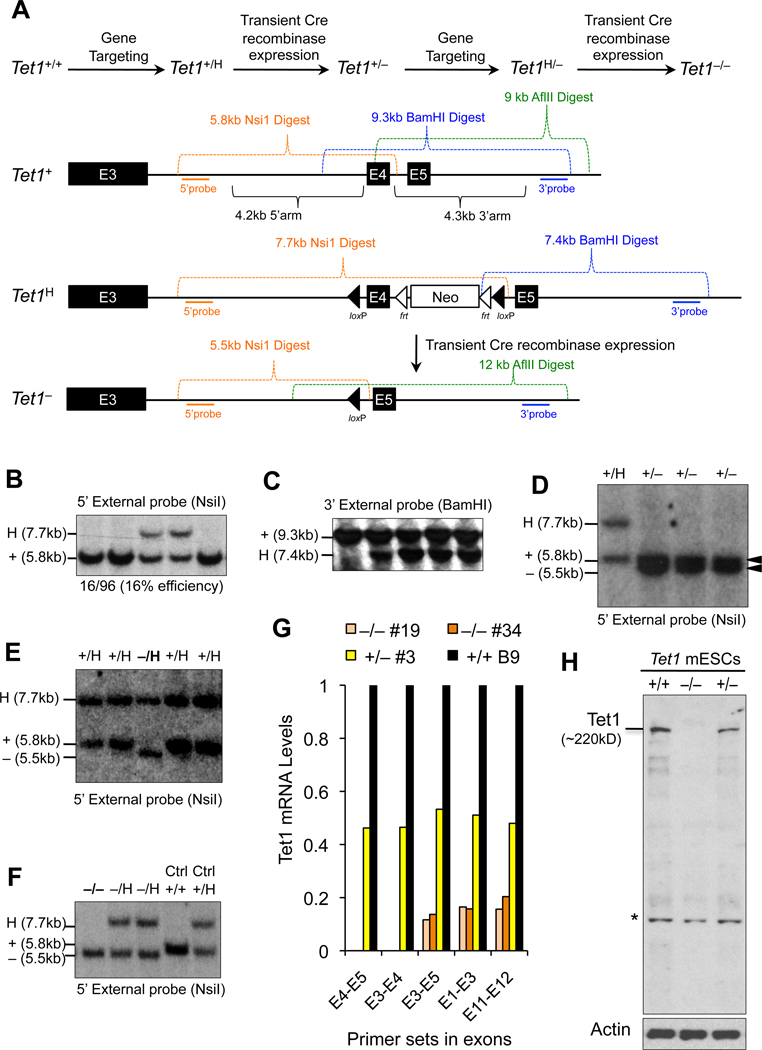
To define the role of Tet1 in maintaining pluripotency and to establish its essential functions during development it is important to utilize a system that stably and completely depletes Tet1 in mESCs. To this end, we applied gene targeting to generate Tet1 knockout mESCs. We flanked exon 4 of Tet1 with loxP sites to generate a conditional 2lox allele (H), which was subsequently excised with transient Cre recombinase expression to produce a Tet1 knockout (−) allele (Figure 1A). Loss of exon 4 in this allele leads to an out of frame fusion of exons 3 and 5 yielding an unstable truncated product lacking the catalytic domain of Tet1. Upon sequential targeting of each allele of Tet1 followed by Cre-mediated excision of exon 4, we obtained Tet1−/− mESCs (Figure 1B–F). We confirmed the absence of transcripts containing exon 4 by RT-qPCR (Figure 1G) and complete depletion of Tet1 protein by western blot (Figure 1H). To measure the presence of any residual transcripts, we used primers spanning the last exons of the gene. As expected for this knockout strategy, we detected some level of residual full-length transcript but none contained exon 4. Moreover, we did not detect any residual low molecular weight truncated proteins in Tet1−/− mESCs by western blot suggesting that indeed Tet1 is completely depleted in these cells.
Figure 1. Generation of Tet1 knockout mouse embryonic stem cells.
(A) Schematic of gene targeting strategy applied to generate Tet1 knockout mouse embryonic stem cells (mESCs). H stands for the properly targeted 2-lox conditional/hypomorphic allele. (B–F) Southern blot confirmation of properly targeted Tet1 mESCs clones. Analysis of Tet1+/H clones (B&C) and Tet1−/H clones (E). Excision of exon 4 by transient Cre recombinase expression to generate Tet1+/− mESCs (D) or Tet1−/− cells (F). Note that the clones shown in these blots are representative clones and do not correspond numerically with each other. (G) Relative Tet1 mRNA levels measured by quantitative RT-PCR in Tet1 mESCs of indicated genotypes using primers in different exons, including the deleted exon 4. Data are normalized to GAPDH. Note that Tet1−/− cells are completely depleted of mRNA containing exon 4. (H) Tet1 protein levels measured by western blot in lysates generated from mESCs of indicated genotypes using anti-Tet1 antibody. Actin is used as a loading control. Note that deletion of exon 4 in Tet1−/− mESCs leads to complete depletion of Tet1 protein. Asterisk indicates nonspecific band.
Tet1 loss in ES cells leads to partial reduction in 5hmC and subtle changes in global DNA methylation and gene expression
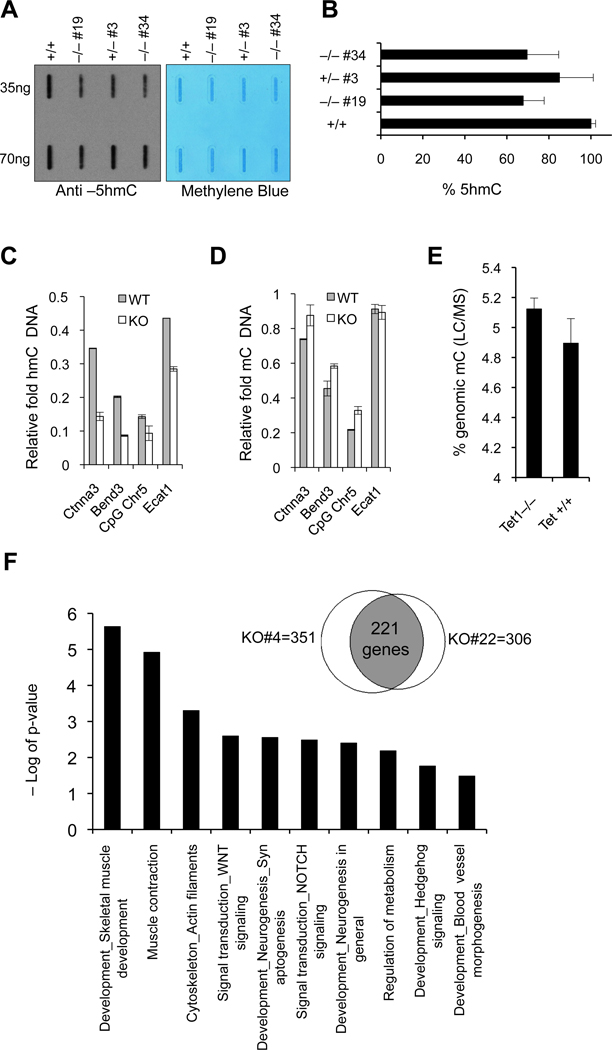
Given the role of Tet1 in converting 5mC to 5hmC, we wanted to determine if loss of Tet1 affects 5hmC levels in Tet1 knockout mESCs. To this end we measured global 5hmC levels in genomic DNA from Tet1+/+, Tet1+/− and Tet1−/− mESCs by a dot blot assay using an antibody against 5hmC. Consistent with previous studies using Tet1 knockdown shRNAs (Ito et al., 2010; Tahiliani et al., 2009), we found that loss of Tet1 did not lead to complete depletion of 5hmC levels but rather to a level reduced by ~35% (Figure 2A&B). To quantify 5hmC levels in a locus specific fashion, we performed glucosylated hydroxymethyl sensitive qPCR (gluc-MSqPCR) using HpaII and MspI restriction enzymes and primers in CpG islands of genes known to contain 5hmC (Ficz et al., 2011) and found that, compared to wild type mES cells, Tet1−/− mESCs had a significant reduction in 5hmC in these CpG islands (Figure 2C), which correlated with a less profound but considerable increase in 5mC content (Figure 2D). To determine how global 5mC levels are affected in the absence of Tet1, we quantified the genomic 5mC content in Tet1+/+ and Tet1−/− mESCs by liquid chromatography/mass spectrometry (LC/MS) (Figure 2E) and found a slight increase of 5mC levels from 4.89% in wild type cells to 5.15% in Tet1−/− cells. These analyses suggest that loss of Tet1 in ES cells leads to a partial reduction in 5hmC levels with a very subtle effect on global DNA methylation. This also suggests that Tet1 is likely not the only enzyme catalyzing 5hmC generation in ES cells and that other Tet family members or pathways participate in this process or compensate for Tet1 loss. Although Tet1 loss did not lead to induction of Tet2 or Tet3 expression in Tet1−/− mESCs, shRNA-mediated knockdown of Tet2 in these cells further reduced 5hmC levels (Figure S1A–D).
Figure 2. Loss of Tet1 leads to partial reduction in 5hmC and subtle changes in global DNA methylation and gene expression profile.
(A) Analysis of 5hmC levels in DNA isolated from Tet1 mESCs of indicated genotypes by dot blot assay using anti-5hmC antibody. Methylene blue staining is used to control for proper transfer. (B) Quantification of intensity of 5hmC signal for each genotype is plotted. Error bars represent SEM. (C&D) Locus specific quantification of 5hmC and 5mC in promoter CpG island regions of indicated genes in Tet1 knockout and wild type mESCs by glucosylation of genomic 5hmC followed by methylation sensitive qPCR (glucMS-qPCR). CpG Chr5 refers to a CpG island in chromosome 5:99466154-99466819. Error bars represent SEM. (E) Global quantification of 5mC in mESCs of indicated genotypes by liquid chromatography/mass spectrometry (LC/MS). Error bars represent SEM. (F) Gene ontology analysis for commonly deregulated genes in two Tet1−/− mESCs compared to wild-type mESCs as determined by microarray gene expression profile analysis. Venn diagram shows in the intersection the number of genes commonly differentially expressed in the knockout cells (versus wild-type cells). Total number of genes differentially regulated between each knockout cell line versus wild-type cell line is also indicated. See also Figure S1 and S2.
To examine whether Tet1 loss and reduction in 5hmC levels influence gene expression of ES cells, we analyzed RNA from two independent Tet1−/− and wild type ES cells by microarray. We found that 221 genes (mostly genes involved in developmental processes) were significantly deregulated by two fold or more in both knockout ES cells compared to wild type mESCs (Figure 2F). While 60% of genes (137 genes) were down regulated, 40% (84 genes) were up regulated (Figure S2) suggesting that Tet1 has both activating and repressive effects on gene expression and is consistent with the current notion on dual roles of Tet1 in transcriptional regulation (Williams et al., 2011; Wu et al., 2011).
Tet1 knockout ES cells are pluripotent and can support development of the embryo proper in a tetraploid complementation assay
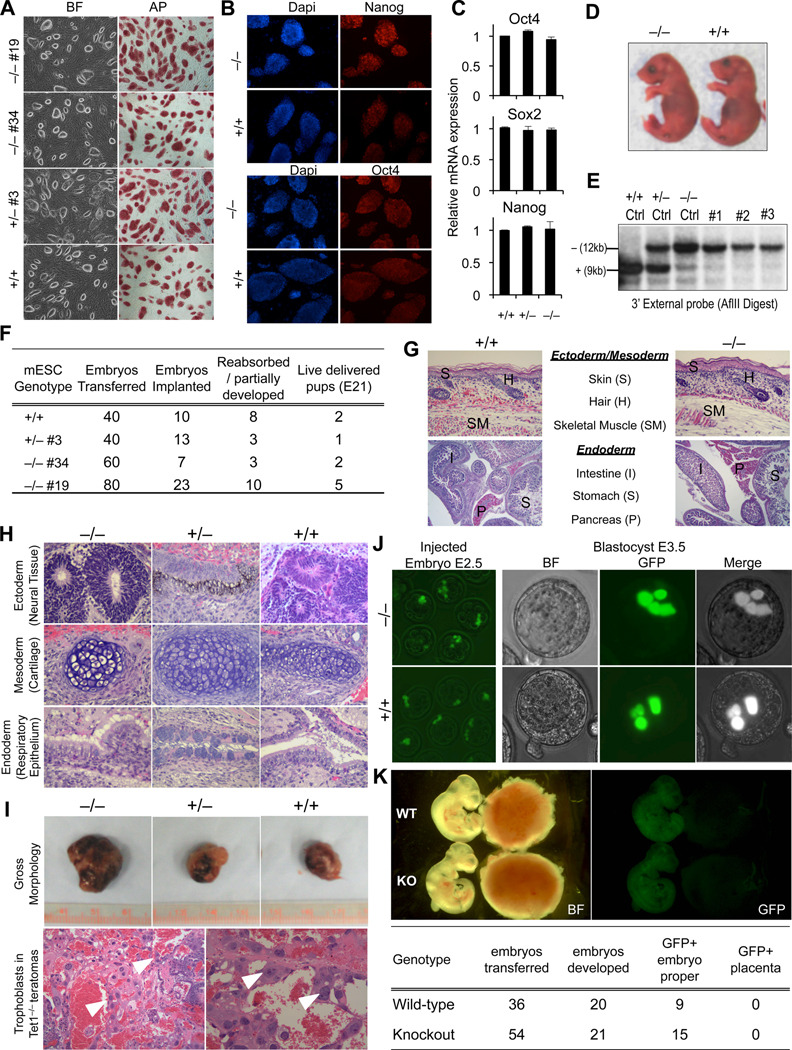
To establish whether depletion of Tet1 influences the pluripotency of ES cells we first examined the morphology and expression of pluripotency markers in Tet1 knockout mESCs cultured either on feeders (Figure 3A) or on gelatin (Figure S3A). Under both conditions and over the course of multiple passages (>15) mutant cells maintained a normal undifferentiated ES cell morphology, stained positive for alkaline phosphatase and expressed the pluripotency markers Oct4, Nanog and Sox2 (Figure 3A–C, Figure S3B). A similar unperturbed ES cell morphology and pluripotency marker expression was observed for Tet1 knockout ES cells subjected to a 60% shRNA-mediated Tet2 knockdown (Figure S1E–G). Tet1 knockout ES cells were capable of forming embryoid bodies (EBs) and could differentiate in vitro to neural progenitor cells (NPs). However, knockout EBs had altered expression of the lineage specification markers Brachyury and Pax6 and the overall yield of EB formation from knockout cells was substantially low due to their increased tendency to attach to the plastic surface and differentiate as early as day 6 of LIF withdrawal (Figure S3C–E) suggesting that Tet1 loss may influence the differentiation potential of ES cells in vitro.
Figure 3. Tet1 knockout ES cells are pluripotent and support post-implantation embryonic development in tetraploid complementation assay.
(A) Alkaline phosphatase staining and morphological appearance of Tet1 mESCs of indicated genotypes on feeders. (B) Immunostaining for pluripotency markers Oct4 and Nanog. (C) Relative expression of core pluripotency network genes Oct4, Sox2 and Nanog measured by quantitative RT-PCR. Data are normalized to GAPDH. Error bars represent SEM. (D–G) Tetraploid (4n) complementation assay for Tet1 knockout mESCs. Gross appearance of pups delivered by c-section at day E21 is shown in D. Confirmation of genotypes of pups from tail DNA by Southern blot is shown in E. Tet1 targeted mESC DNA of indicated genotypes is used as controls for genotyping. Complete data from tetraploid complementation experiments are summarized in E. Histological analysis of various tissues and organs of fixed E21 pups from 4n assay is presented in G. (H) H&E staining of sections of teratomas generated from Tet1 mESCs of indicated genotypes. (I) Gross appearance of teratomas (top) and H&E histological analysis of Tet1 knockout teratomas (bottom). Note the presence of trophoblasts (arrow heads) and blood in knockout teratomas. (J) GFP-labeled Tet1 knockout or wild-type ES cells injected into wild-type 8-cell-stage embryos are traced until the blastocyst stage in a developing embryo. Note that the GFP signal is not seen in the outer layer of the blastocyst, which constitutes the trophectoderm (n=20 for each genotype). (G) Gross and fluorescence images of E10.5 chimeric embryos that were injected at blastocyst stage with GFP-labeled Tet1 knockout or wild-type ES cells. Note that GFP positive cells of both genotypes exclusively contribute to the embryo proper (left) and not the placenta (right). Number of embryos examined in this experiment is presented in the table. See also Figure S3.
To determine if Tet1 knockout mESCs are fully pluripotent and can differentiate to support development of the embryo proper, we performed a tetraploid complementation assay. This is the most stringent test for assessing pluripotency as the cells of the recipient tetraploid (4n) blastocyst can only contribute to the trophectoderm but not the epiblast thus generating embryos that are exclusively derived from the injected diploid mESCs (Eggan et al., 2001; Zhao et al., 2009). We injected two independent Tet1 knockout, one heterozygote and one wild-type mESC clones into B6D2F1 × B6D2F1 4n blastocysts, which were subsequently transferred into pseudo-pregnant female mice. We performed cesarean section to deliver the pups and found that Tet1 knockout, heterozygote and wild-type mESC clones developed into full term live mice with similar efficiencies (Figure 3D–G). Tet1 knockout pups appeared indistinguishable from wild-type pups (Figure 3D) and were able to move and breathe at birth. Attempts at fostering these pups to monitor their postnatal development failed as both wild type and Tet1 knockout mice died within hours of delivery. Low rate of postnatal survival is a common limitation of the tetraploid complementation assay (Eggan et al., 2001). Southern blot analysis of tail DNA obtained from these pups confirmed that they were derived from Tet1 knockout mESCs (Figure 3E). Autopsy and histological analyses failed to reveal any abnormalities in knockout pups (Figure 3G). These findings confirm that Tet1 knockout cells are pluripotent and that loss of Tet1 does not lead to defects that would interfere with normal post-implantation embryonic development to birth.
Consistent with these results, Tet1−/− ES cells were shown to be pluripotent in a teratoma assay forming tumors with differentiated cells derived from the three embryonic germ layers (Figure 3H). However, Tet1 knockout teratomas were large and hemorrhagic which is likely due to excessive trophoblast derived cells that form lumens and promote more blood flow from the host (Figure 3I). Thus, loss of Tet1 seems to skew differentiation towards extraembryonic lineages in the teratoma assay consistent with previous findings with Tet1 knockdown ES cells (Koh et al., 2011a). To determine whether Tet1 loss affects differentiation also in the early embryo, we injected GFP-labeled Tet1 knockout ES cells into 8-cell-stage wild-type (2n) embryos and traced their distribution until the blastocyst stage where we could only locate GFP positive cells in the center inner cell mass region of the blastocyst and not in the outer trophectoderm layer (Figure 3J). When the cells were injected into wild-type blastocysts and implanted into pseudo-pregnant females, the GFP-labeled Tet1 knockout ES cells were exclusively found in the embryo proper and no cells were detected in the placenta (Figure 3K). Thus, the findings from these assays suggest that unlike in the teratoma assay, Tet1 loss seems not to skew differentiation toward trophectoderm in vivo in the context of a developing embryo.
Overtly uncompromised differentiation of Tet1 knockout ES cells is also supported by the 4n complementation assay, which resulted in birth of “all-knockout ESC mice”. However, since some extra-embryonic layers are contributed by the host blastocyst, this assay cannot assess the function of Tet1 in the trophectoderm lineage. Therefore, to definitely establish the function of Tet1 in embryogenesis we generated homozygous mutant mice from intercrossing heterozygous animals.
Tet1 null mice are viable and fertile but vary in body size
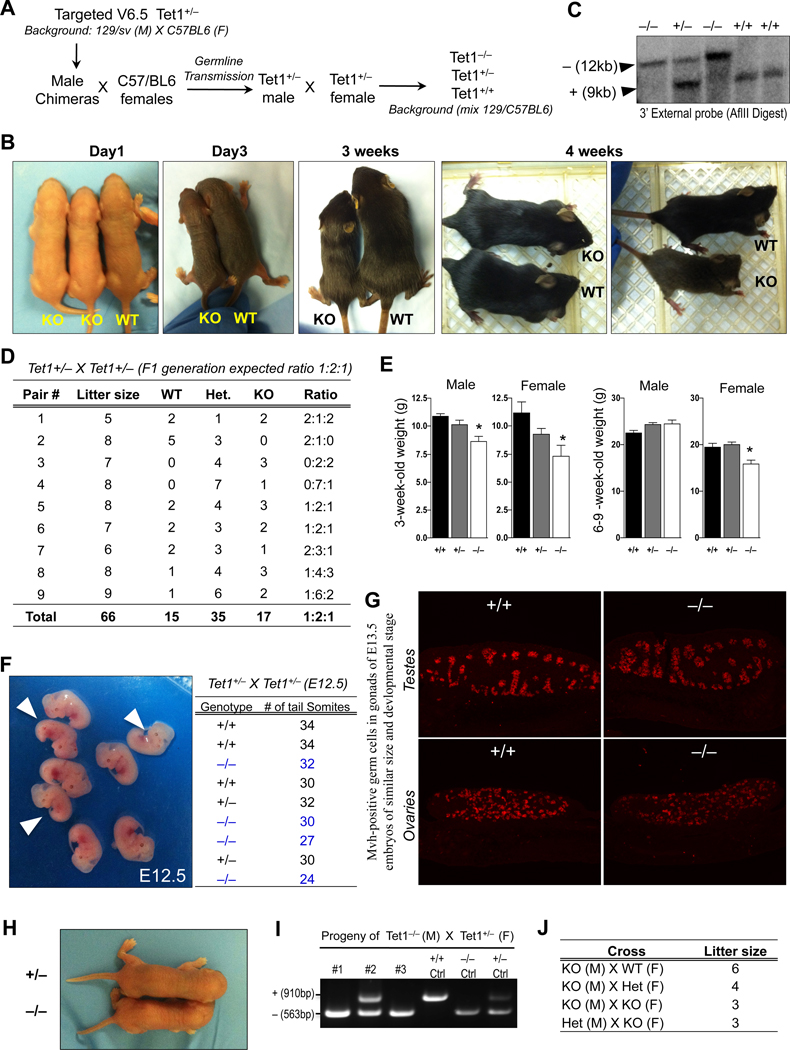
We injected Tet1+/− v6.5 male mES cells into B6D2F1 blastocysts to generate male chimeras, which were bred with C57/BL6 females to obtain heterozygote mice which were grossly normal and fertile (Figure 4A). To generate Tet1 homozygous knockout animals, we intercrossed heterozygote animals at 6–8 week of age. All breeding pairs produced normal sized litters of 5 to 9 pups. However, each litter contained an average of two pups that were considerably smaller in body size than their littermates (Figure 4B). We determined the genotypes of all litters by Southern blot (Figure 4C) and found that wild type, heterozygous and homozygous knockout mice were readily born at the expected 1:2:1 Mendelian ratio suggesting that no Tet1 knockout embryos were lost in utero (Figure 4D). About 75% of the homozygous mutant pups had smaller body size (13/17 pups) at birth. Both male and female mutant mice weighed significantly less than wild type animals at three weeks of age but seemed to gain weight when growing older (Figure 4E). Consistent with the slightly reduced post natal body size, we found that E12.5 knockout embryos from a Tet1+/− X Tet1+/− cross also vary in size ranging between 24 to 32 tail somite pairs compared to heterozygous and wild type embryos that range between 30–34 tail somite pairs at the same embryonic stage (Figure 4F). This suggests that loss of Tet1 during embryogenesis leads to a mild developmental delay, which is partially penetrant producing both, normal sized and smaller embryos.
Figure 4. Tet1 knockout mice are viable but vary in body size.
(A) Schematic of crosses to generate Tet1 knockout mice. (B) Gross appearance of mice of indicated genotypes at various ages. Note that Tet1−/− mice vary in size. (C) Genotype confirmation of mice from Tet1+/− × Tet1+/− cross by southern blot. (D) Table summarizing the litter size and Mendelian ratio of Tet1−/− mice. (E) Mouse body weights at the indicated ages. Error bars represent SD. Asterisks indicate statistically significant than wild type (t-test p-value <0.05). (F) Gross images of E12.5 embryos from Tet1+/− × Tet1+/− cross. Arrowheads point to knockout embryos. Genotype and number of tail somite pairs of embryos is tabulated to the left. (G) Longitudinal sections of E13.5 gonads of indicated genotypes stained with antibody against the germ cell marker Mvh. Note that gonads were harvested from embryos of similar developmental stage and size. (H) Gross images of progeny of Tet1 knockout parents at birth. (J) PCR genotyping confirmation of offspring of Tet1 knockout mice. (K) Table summarizing average litter size from various crosses of 6- to 8-week-old Tet1 knockout mice. See also Figure S4.
With the exception of variability in body size and weight, homozygote animals were grossly normal and appeared healthy. Blood analyses of 4-week-old wild type and knockout mice for CBC and liver enzyme functions showed no major differences except for a slight decrease in the number of neutrophils in knockout animals (Figure S4). Moreover, gonads of both sexes of E13.5 knockout embryos contained mouse vasa homolog (mvh)-positive germ cells (Figure 4G). Mating of homozygous mutant males and females produced viable progeny (Figure 4H–J) though the average litter size (3–6 pups) seemed to be smaller than the average litter size of heterozygous parents (5–9pups). Since Tet1 is expressed in germ cells (Hajkova et al., 2010), the role of Tet1 in gametogenesis and fertility requires further investigation.
Discussion
Ever since the implication of Tet proteins in DNA demethylation and maintenance of pluripotency, research has focused on establishing the biological relevance of these proteins (Ito et al., 2010; Koh et al., 2011b; Tahiliani et al., 2009). Targeted deletion of Tet1 in ES cells and generating Tet1 knockout mice has provided a clean and stable system to study Tet1 function in pluripotency and development. Here, we have provided three lines of evidence to establish that Tet1 is dispensable for ES cell maintenance and its loss compatible with embryonic development and postnatal survival. First, Tet1−/− mESCs remain undifferentiated in vitro and express pluripotency markers Oct4, Sox2 and Nanog and can contribute to tissues of the three embryonic germ layers in teratomas. Second, Tet1 null mESCs can support normal development of the embryo proper in a tetraploid complementation assay suggesting that they are pluripotent and can support post-implantation embryonic development. Third, Tet1 homozygous mutant mice are viable and fertile and born at normal Mendelian ratio, albeit with slight reduction in body size, establishing that loss of Tet1 does not interfere with postnatal survival.
Our findings expand upon the previously published in vitro studies on Tet1 and bring new insights into defining the actual physiological requirements of the gene in pluripotency and development. Previously reported shRNA-based in vitro knockdown studies (Ito et al., 2010; Koh et al., 2011a) together with recent findings on the dual roles of Tet1 in transcriptional regulation and repression of polycomb controlled developmental genes (Williams et al., 2011; Wu et al., 2011), suggested that Tet1 is likely a key player in ES cell maintenance, lineage specification and embryogenesis. One report showed that Tet1 knockdown in mESCs diminished Nanog expression and impaired self-renewal (Ito et al., 2010). We did not observe such a phenotype in Tet1−/− cells and it is possible that ES cell background differences and/or off target effects of shRNAs may explain these differences. Consistent with our findings, another study using a different set of shRNAs to knockdown Tet1 and Tet2 reported that depletion of these proteins does not affect Nanog expression, ES cell self-renewal and pluripotency (Koh et al., 2011b). However, this study found that Tet1 deficiency skews lineage specification toward mesendoderm and trophectoderm at the expense of neuroectoderm in a teratoma assay leading to formation of large hemorrhagic tumors. Although we observed more hemorrhage and an increase in trophoblast-like cells in Tet1 knockout teratomas, we did not observe any overt reduction in formation of neuroectoderm or contribution to placenta upon injection of mutant ESCs into wild-type blastocysts. EBs generated from Tet1 knockout ES cells were capable to form neural progenitors in culture, even though the overall yield of EB formation from knockout cells were substantially lower due to their increased tendency to attach to the plastic surface and differentiate.
The generation of viable mutant mice argues against profound defects in lineage specification due to loss of Tet1. It is possible, therefore, that the teratoma assay or differentiation to EBs uncovered abnormalities that are only detectable in vitro and do not overtly affect embryonic development. We consider two possibilities to explain the seemingly normal in vivo development of mutant embryos and the in vitro differentiation abnormalities of mutant ES cells. (1) Tet1 knockout ES cells exhibit skewed differentiation in non-physiological conditions such as a teratoma or in in vitro assays. (2) Tet1 knockout ES cells have lineage specification defects but any such defects are less pronounced in the context of an embryo compared to a teratoma or EB and are compatible with embryogenesis consistent with the failure of mutant ES cells to contribute to the trophectoderm lineage in vivo.
The generation of viable and fertile Tet1 mutant mice unequivocally indicated that Tet1 deficiency does not prevent embryonic and postnatal development. However, the slight reduction in postnatal body size and weight of Tet1 knockout mice is consistent with Tet1 playing a role during development. Given the evidence that Tet1 regulates gene expression, particularly by repressing polycomb regulated developmental genes (Williams et al., 2011; Wu et al., 2011), it is possible that subtle deregulation of these developmental regulators interfere with timely progression of embryogenesis and trigger a mild developmental delay. Because normal sized knockout mice were generated by tetraploid complementation it is possible that the role of Tet1 is more pronounced in placental function than in embryo development. However, further molecular and histological analysis of embryos at various stages during development is needed to establish if Tet1 loss leads to placental defects.
The three Tet enzymes are differentially regulated during early development. Tet3 is the only Tet enzyme expressed in the oocyte and zygote and is believed to have a role in paternal pronucleus demethylation (Iqbal et al., 2011; Wossidlo et al., 2011). As the zygote develops Tet3 levels diminish at two cell stage and by the blastocyst stage Tet1 and Tet2 are mainly expressed in the inner cell mass (ICM). Both Tet1 and Tet2 are expressed in ES cells with Tet2 expression levels being five fold less than Tet1 (Koh et al., 2011a). We found that loss of Tet1 did not lead to induction of Tet2 and Tet3 mRNA levels in Tet1−/− mESCs. Given that Tet1 knockout ES cells have only a 35% reduction in 5hmC levels, it is likely that expression of Tet2 can compensate for Tet1 loss consistent with the observation that a 60% knockdown of Tet2 in Tet1−/− ES cells further reduced 5hmC levels. Generation of double and triple knockout ES cells and mice for Tet genes will be important for dissecting the developmental roles of the different Tet proteins and 5hmC in vivo.
Embryonic development is a multi-step and complex process. Although we have generated viable Tet1 knockout mice and shown that loss of Tet1 is compatible with pluripotency and development, we cannot exclude subtle defects during development whose effects would manifest later in life, such as neurological and behavioral phenotypes, particularly given the high levels of 5hmC and Tet1 in the adult brain and their potential role in active DNA demethylation (Guo et al., 2011). Moreover, despite the presence of germ cells in knockout gonads, it is important to further study if Tet1 plays any role in gametogenesis, particularly methylation erasure and germ cell maintenance, which requires detailed investigation. Further characterization of Tet1 knockout mice will also allow studying the physiological significance of Tet1 during adult development.
Experimental procedures
Generation and culture of Tet1 knockout mouse embryonic stem cells
Gene targeting procedures for generating Tet1 2-lox conditional/hypomorphic (H) and knockout (−) alleles were as previously described (Dawlaty and van Deursen, 2006). Briefly, Tet1 exon 4 and its flanking 100 base pair regions was amplified from v6.5 mESC DNA and cloned into multiple cloning site 1 (HindIII and KpnI) of pNTKV1901frt-loxP vector. This was followed by cloning 4.2 kb 5’ homology arm and 4.3 kb 3’ homology arm into multiple cloning sites 2 (HpaI) and 3 (SmaI and SacII) of this vector, respectively. Vector was linearized with NotI and electroporated into v6.5 mESCs (mix background: 129/sv(M) X C57BL/6(F)) following standard procedures. Neo resistant clones were picked and screened with 5’ and 3’ external probes and neo sequence internal probe. pPAC–Cre plasmid was electroporated into properly targeted clones to excise exon 4 and generate Tet1 knockout allele. Tet1+/− cells were retargeted to delete the wild-type allele and generate Tet1−/− mESCs. All mESCs in our experiments, unless specified, were cultured on gamma-irradiated DR4 feeders using standard mESC media containing LIF.
Generation of Tet1 Knockout mice
Tet1+/− v6.5 ES line (mix background 129/sv × C57/BL6) was injected into B6D2F1 × B6D2F1 blastocysts and surgically implanted into 2.5 d.p.c. pseudo-pregnant Swiss Webster female mice to generate chimeras following standard procedures. High contribution chimeric animals were bred with C57/BL6 females. Litters with agouti coat color germ line transmission were screened by southern blot and PCR for presence of heterozygote mice. Six week old Tet1+/− animals were intercrossed to generate Tet1−/− mice. Pups were monitored daily for signs of poor health and weighted routinely after birth. Weight calculations and statistical analyses were performed using Prism Graphpad software. For analyses of blood, 400 µl of blood was collected retro-orbitally and analyzed at the Massachusetts Institute of Technology Department of Comparative Medicine Diagnostic and Comparative Pathology lab.
Teratoma and Tetraploid complementation assays
1×106 mESCs were injected subcutaneously into SCID mice (Taconic). 3 weeks after injection mice were euthanized and tumors were removed and fixed in formalin for two days. Then they were imbedded in paraffin, sectioned and stained with hematoxylin and eosin for histological analysis. Tetraploid complementation assay was performed as explained before (Wernig et al., 2007). Briefly, two cell stage B6D2F1 × B6D2F1 embryos were fused by electric pulse. Two hours later, properly fused embryos were separated and cultured in KSOM for two days until they developed into blastocysts. 10–12 mESCs were injected into each blastocyst following standard procedures. 20 injected embryos were surgically transferred into the uterus of 2.5 d.p.c. pseudo-pregnant Swiss Webster female mice. Pregnant mice were sacrificed at day 21 and pups were immediately delivered by cesarean section method.
Gene expression profile and gene ontology analyses
Microarray data were processed and quantile-normalized using Limma package in R (Smyth and Speed, 2003) and (Smyth, 2005). Probe values from the same gene were collapsed into median to get gene expression values. Relative expression was calculated as log2 ratio of gene expression in knockout cells to the wild-type (WT) cells. For downstream analysis, only genes that have a signal of above 5 logs (base 2) in at least one samples and a minimum fold change of 2 in at least one knockout (vs. WT) were retained. Gene ontology analysis was conducted using GeneGO with default parameters. The gene array datasets reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE we just uploaded the files and will get the official accession numbers in 5 days and will include it in the final pdf proof)
Supplementary Material
Acknowledgements
We thank Ruth Flannery for help with animal husbandry, Dongdong Fu for help with histology, J. Kwon and J. Love from Whitehead Genome Technology Core for help with microarray, and F.Soldner, G.Welstead, J. Staerk, K. Saha, B. Carey and L. Medeiros for helpful discussions and critical reading of the manuscript. We are grateful to Dr. R. Bronson for analyzing histology slides and discussion on teratomas and Dr. M. Gehring for help with dot blots. M.M.D is a Damon Runyon Fellow. B.E.P is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. A.W.C. is supported by a Croucher scholarship. D.C.P is an HHMI investigator. R.J. is funded by NIH grants 5-RO1-HDO45022, 5-R37-CA084198, and 5-RO1-CA087869.
References
- Dawlaty MM, van Deursen JM. Gene targeting methods for studying nuclear transport factors in mice. Methods. 2006;39:370–378. doi: 10.1016/j.ymeth.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin Cell Dev Biol. 2003;14:93–100. doi: 10.1016/s1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- Eggan KC, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, Yanagimachi R, Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011a;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 Regulate 5-Hydroxymethylcytosine Production and Cell Lineage Specification in Mouse Embryonic Stem Cells. Stem Cell. 2011b;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SKT, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala HS, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Research. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman RK, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nature Reviews Molecular Cell Biology. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X-y, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo C-l, Ma Q-w, Wang L, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. (Publisher web site, PDF) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.