Abstract
Tight regulation of TGF-β superfamily signaling is important for normal cellular functions and tissue homeostasis. Since TGF-β superfamily signaling pathways are activated by a short phosphorylation cascade, from receptor phosphorylation to subsequent phosphorylation and activation of downstream signal transducer R-Smads, reversible phosphorylation serves as a critical step to assure the proper TGF-β signaling. This article will review the current progress on the understanding of dynamic phosphorylation in TGF-β signaling and the essential role of protein phosphatases in this process.
Keywords: TGF-β signaling, Smads, phosphorylation, dephosphorylation, protein phosphatases
Introduction
Transforming growth factor-β (TGF-β) superfamily signaling controls numerous cellular responses from cell proliferation, differentiation, extracellular matrix remodeling to embryonic development in species ranging from worms to mammals [1–4]. Consequently, TGF-β signaling plays a key role in the pathogenesis of many diseases, including cancer and autoimmune and fibrotic diseases [5–11]. The activity of TGF-β signaling is tightly regulated via various mechanisms [12]. Accumulating evidence reveals how dephosphorylation on several pathway components can modulate the activation of TGF-β signaling pathway. This underscores the important role of protein phosphatases in controlling TGF-β signaling and maintaining tissue homeostasis.
Overview of the TGF-β superfamily signaling pathway
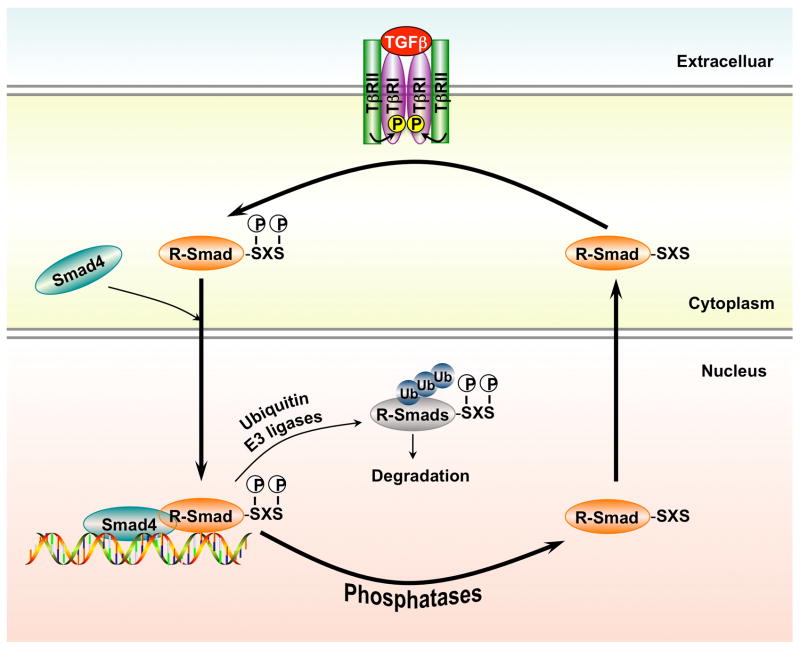
The TGF-β superfamily of cytokines contain two families, the TGF-β/Activin/Nodal sfamily and the bone morphogenetic protein (BMP)/growth and differentiation factor (GDF)/Mullerian inhibiting substance (MIS) family. Upon the ligand binding to receptors at the cell surface, two type I and two type II receptors form a tetrameric complex. The human genome encodes 7 type I receptors (i.e. activin receptor-like kinases 1–7 or ALK1–7) and 5 type II receptors (i.e. TβRII, ActR-II, ActR-IIB, BMPR-II and AMHR-II). Both type I and type II receptors contain an N-terminal extracellular ligand-binding domain, a transmembrane region, and a cytoplasmic serine/threonine kinase domain [13–16]. In the ligand-bound complex of type I and type II receptors, the type II receptor kinase activates the type I receptor kinase through phosphorylation in the GS motif of the type I receptor. The type I receptor then propagates the signal through phosphorylation of the receptor-activated Smads (R-Smads) in the canonical Smad pathway (Figure 1) [17–20].
Figure 1. Canonical TGF-β signaling.
TβRI, type I TGF-β receptor; TβRII, type I TGF-β receptor; R-Smad, receptor-activated receptor.
Smad proteins are the central mediators for TGF-β superfamily signaling [21–25] and are classified into three groups. The first group is the R-Smads, of which Smad1, 5 and 8 are primarily activated by the BMP-specific type I receptors, while Smad2 and Smad3 by the TGF-β subfamily type I receptors. The second group has the common mediator Smad (Co-Smad, e.g. Smad4 in mammals). The third Smad group includes inhibitory Smads (I-Smad, e.g. mammalian Smad6 and Smad7). The R-Smad and Co-Smad proteins contain two conserved structural domains, the N-terminal MH1 domain and the C-terminal MH2 domain. The R-Smads, but not the Co-Smads, contain a characteristic SXS motif at their C termini. Upon ligand stimulation, R-Smad proteins are phosphorylated in the SXS motif by type I receptors, leading to the activation of a series of downstream events. Phosphorylated R-Smads form a heter-oligomeric (often trimeric) complex with Smad4. The Smad complex is imported into the nucleus and regulates the expression of target genes by direct binding to target gene promoter and/or through the interaction with transcriptional cofactors in a cell type-specific manner [26–31]. I-Smads, on the other hand, inhibit TGF-β signaling in a feedback mechanism through inhibition of R-Smad phosphorylation, prevention of complex formation between R-Smad and Co-Smad, or transcriptional repression [32–33].
Besides the canonical Smad-mediated signaling pathway, it has been shown that TGF-β superfamily ligands can also regulate cellular or physiological processes through non-canonical pathways by the activation of other signaling molecules (e.g. MAPK, Akt, Src, mTOR) independent of Smad proteins [34–35], which amplifies the complexity of TGF-β signaling.
Posttranscriptional modulation ofTGF-β superfamily signaling
As TGF-β superfamily signaling regulates a broad range of cellular responses, it is logical that the activity of TGF-β signaling must be tightly controlled. Reduction in the level of ligands, internalization and degradation of the receptors, and induction of I-Smads represent means through which TGF-β signals are modulated. Recently, it is evident that in addition to reversible phosphorylation (to be further discussed in sections below), TGF-β signaling is also regulated by various types of posttranscriptional modifications such as ubiquitination, sumoylation, and acetylation [36].
Smads are regulated by the ubiquitination-proteasome pathway. Smurf (Smad Ubiquitination-related factor) family members, Smurf1, Smurf2 and related NEDD4-2, which share extensive sequence similarity, have been identified as E3 ligases involved in type I receptor and R-Smad ubiquitination and degradation. Smurf1 targets Smad1 and Smad5 for ubiquitination and subsequent proteasome-mediated degradation [37]. Smurf2 is also involved in Smad1 and Smad2 ubiquitination and proteasome-mediated degradation [38–41].
Smad3, a closely-related R-Smad to Smad2, also binds to Smurf2 and NEDD4-2, yet Smad3 is not degraded by these HECT-class E3 ligases [38–40]. Still, Smad3 is regulated by proteasomal degradation and targeted for ubiquitination via different classes of ubiquitin E3 ligase. Smad3 interacts with Rbx1 (also called ROC1 and Hrt1), a RING-containing component of the SCFβTrCP complex (also called SCFFbw1a), which is partly responsible for Smad3 degradation [42]. Additionally, carboxyl terminus of Hsc70-interacting protein (CHIP) serves as a U-box dependent E3 ligase that can directly mediate the ubiquitination and degradation of Smad3 [43] and also Smad1 [44].
Smad4 is central to the canonical TGF-β superfamily signaling. Although it is a fairly stable protein, Smad4 can be targeted by polyubiquitination via various E3 ligases such as Skp2 [45], Jab1 [46] and Smurfs [47]. In addition, Smad4 is monoubiquitinated at Lys507 [48–49] and sumoylated at Lys113 and Lys159 [50–51]. These modifications regulate stability and signaling strength in developmental contexts. The importance of Smad4 regulation by sumoylation and ubiquitination events is further underscored by interplay between phosphorylation and ubiquitin/ubiquitin-like modifications in cancer, where certain somatic mutations on Smad4 switch Smad4 from sumoylation to ubiquitination and ultimately cause rapid degradation of the tumor suppressor [45].
Besides Smads, ubiquitination of activated TGF-β receptors are also mediated by Smurfs and related NEDD4-2, leading to the proteasome-dependent degradation of TGF-β receptors [40, 52–53]. Smad7 plays an important role in this process [54]. Smad7 resides in the nucleus in unstimulated cells and is translocated to the plasma membrane upon ligand stimulation [55–56]. In addition to its ability to directly inhibit R-Smad phosphorylation, Smad7 recruits Smurf1 and Smurf2 to the activated type I TGF-β receptor complex and promote ubiquitination and turnover of the receptors as well as Smad7 and Smurfs themselves. In accordance, Smurf2 also mediates proteasomal degradation of the TGF-β receptors that is induced by inhibition of the molecular chaperone HSP90 [57].
Smurf-mediated ubiquitination of Smad7 is antagonized by acetylation. Smad7 is acetylated in nucleus, as a result of interaction with the transcriptional co-activator p300 [58]. This acetylation blocks ubiquitination on the same residues, resulting in a higher concentration of Smad7 available to bind to receptors and block receptor-R-Smad binding. Conversely, Smad7 can be deacetylated by HDAC1, which decreases the stability of Smad7 by enhancing its ubiquitination [59]. Thus, the activity of Smad7 is regulated by a balance among acetylation, deacetylation, and ubiquitination.
Protein phosphatases
While protein kinases phosphorylate, and in most cases, activate the activities of their substrates, protein phosphatases reverses the actions of protein kinases and thus inactivate the substrates. This reversible phosphorylation provides the balance for proper function of signaling molecules. Sequence analysis predicts that there are ~150 putative protein phosphatases in human genome which can be classified into three groups: 38 tyrosine phosphatases (PTP) family members, ~40 protein serine/threonine phosphatases (PS/TP) members, and around 68 dual specific serine/threonine/tyrosine phosphatases (DUSP) [60–61]. Furthermore, the PS/TP family is classified into three subfamilies by their structurally distinctness: PPM, PPP and FCP/SCP. The PPM (metal ion-dependent protein phosphatases) subfamily consists of PPM1A/PP2Cα, PPM1B/PP2Cβ and other PP2C domain–containing proteins [60–62]. Members of this subfamily are monomeric phosphatases with a single subunit, the catalytic subunit. The PPP subfamily mainly includes protein phosphatase 1 (PP1), protein phosphatase 2A (PP2A), protein phosphatase 4 (PP4), protein phosphatase 5 (PP5) and protein phosphatase 6 (PP6). Among them, PP1 is ubiquitously expressed and highly conserved. Members of this subfamily often contain multiple subunits, including the catalytic subunit and regulatory/targeting subunits that guide the catalytic subunit to the right place at the right time. The founding member of the FCP/SCP subfamily is represented by FCP1, which is a phosphatase that dephosphorylates the carboxy-terminal domain of RNA polymerase II [63].
Phosphatases and TGF-β receptor activity
The phosphorylation cascade from receptor to Smad proteins plays an important role in the activation of TGF-β signaling. Dephosphorylation of receptors and Smad proteins contributes to the duration and intensity of TGF-β signaling. An increasing number of protein phosphatases have been reported to regulate TGF-β signaling at both receptor and Smad levels.
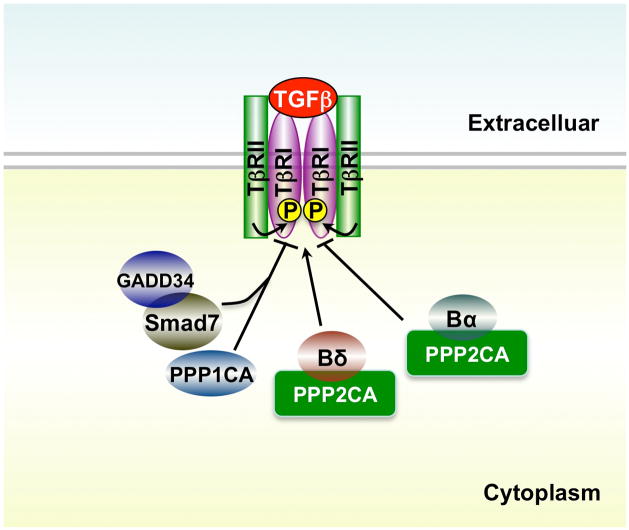
At the receptor level, several studies reported the involvement of PPP family phosphatases in modulating the receptor activity. Members of the PPP family contain the catalytic subunit and a highly diverse array of regulatory and/or targeting subunits, and therefore represent hundreds of protein serine/threonine phosphatases. For example, PP1 has at least 16 regulatory subunits, each of which may play a unique role in targeting the catalytic subunit of protein phosphatase 1 (PP1c) to its specific substrates in a tempo-spatial manner. A previous study in Drosophila melanogaster identified PP1c as a negative regulator of Dpp (Decapentaplegic) signaling. PP1c binds to SARA (Smad anchor for receptor activation) through a specific motif and is targeted to the Dpp receptor complexes [64]. Disruption of this PP1-SARA interaction leads to hyperphosphorylation of the Dpp type I receptor. Although there is no direct evidence supporting dephosphorylation of the type I receptor by PP1, SARA-recruited PP1c can reduce the phosphorylation level of the type I receptor and negatively regulate Dpp signaling [64]. Thus, SARA is a novel targeting subunit that recruits PP1c to the type I receptor.
Further studies suggest that PP1 protein also plays a role in regulating TGF-β type I receptor in various mammalian cell types. Shi et al. found that upon TGF-β stimulation, Smad7 interacts with growth arrest and DNA damage protein (GADD34), a regulatory/targeting subunit of the human PP1 holoenzyme. The formation of Smad7-GADD34 complex recruits the catalytic subunit of PP1 to dephosphorylate the type I TGF-β receptor in Mv1Lu mink lung epithelial cells, a process enhanced by SARA as in the case in Drosophila. The Smad7/GADD/PP1c complex significantly inhibits TGF-β-mediated cellular responses indicating that the formation of PP1 holoenzyme mediated by Smad7 functions as a negative feedback in TGF-β signaling pathway (Figure 2) [65]. Consistent with this finding, ectopic expression of Smad7 and PP1α (PP1c isoform α) potently inhibit TGF-β/ALK1-induced Smad1/5 phosphorylation in endothelial cells (ECs), while knockdown of either Smad7 or PP1α enhances Smad1/5 phosphorylation [33]. It is interesting to note that Smad7 and PP1α gene expression is up-regulated by TGF-β treatment in ECs through a non-canonical TGF-β/ALK1 pathway. These studies implicate that Smad7 may recruit PP1 to dephosphorylate the type I receptors, and consequently control receptor-induced downstream signaling events.
Figure 2. Regulation of TGF-β receptors by phosphatases.
PPP1CA, catalytic subunit of protein phosphatase 1; PPP2CA, catalytic subunit of protein phosphatase 2A; Bα, regulatory subunit of PP2A; Bδ, regulatory subunit of PP2A; GADD34, growth arrest and DNA damage 34.
In addition to PP1, PP2A is another well-known protein phosphatase that associates with TGF-β receptors [66–68]. PP2A is also a member of the PPP family, and this remarkably conserved phosphatase accounts for the majority of PS/TP activity in most cells. PP2A functions as heterotrimeric enzyme, which consists of catalytic C subunit, structural/scaffolding A subunit and a regulatory B-type subunit. The B-type subunits function as targeting factors, and hence confer specificity, diversity and regulation of the PP2A holoenzyme. The interaction between TGF-β receptors and PP2A subunits was first reported in 1998 [68], and a recent study suggested an opposite effect of two PP2A B-type regulatory subunits, Bα and Bδ, in TGF-β/Activin/Nodal signaling pathway [66] (Figure 2). In the latter study, Butat et al noticed that in Xenopus embryos the phenotype of Bα knockdown or Bδ overexpression is highly similar to those with loss of Nodal signaling. Further studies suggest that Bα enhances TGF-β/Activin/Nodal signaling by stabilizing the basal level of the type I receptor, whereas Bδ negatively modulates these pathways by restricting receptor activity [66]. Unexpectedly, there is no solid evidence supporting the role of PP2A in direct dephosphorylation of either the TGF-β receptors or Smad2. However, PP2A may dephosphorylate Smad3, but not Smad2, under hypoxic condition, suggesting a complicated regulatory function of PP2A phosphatase in the regulation of TGF-β signaling [69].
Phosphatases that target the SXS motif of R-Smads
Phosphorylation of R-Smad proteins in the SXS motif by type I receptors is a central event during TGF-β signal transduction activation. The SXS phosphorylation triggers a cascade of intracellular responses from R-Smad/Co-Smad complex formation in the cytoplasm to the transcriptional control in the nucleus (Figure 1). Since signal transduction pathways are regulated by dynamic interplay between protein kinases and phosphatases, it is reasonable to postulate that the SXS motif must be dephosphorylated by phosphatases to ensure proper balance of Smad signaling. Experimental evidence has shown that R-Smads shuttle between nucleus and cytoplasm. While persistent accumulation of the R-Smad/Smad4 complex in the nucleus requires the continuous TGF-β receptor activity, nuclear export of R-Smads has been shown to depend on their dephosphorylation and dissociation of Smad complexes [70–73]. This further suggests the existence of nuclear Smad phosphatases that target the SXS motif to terminate TGF-β-Smad signaling.
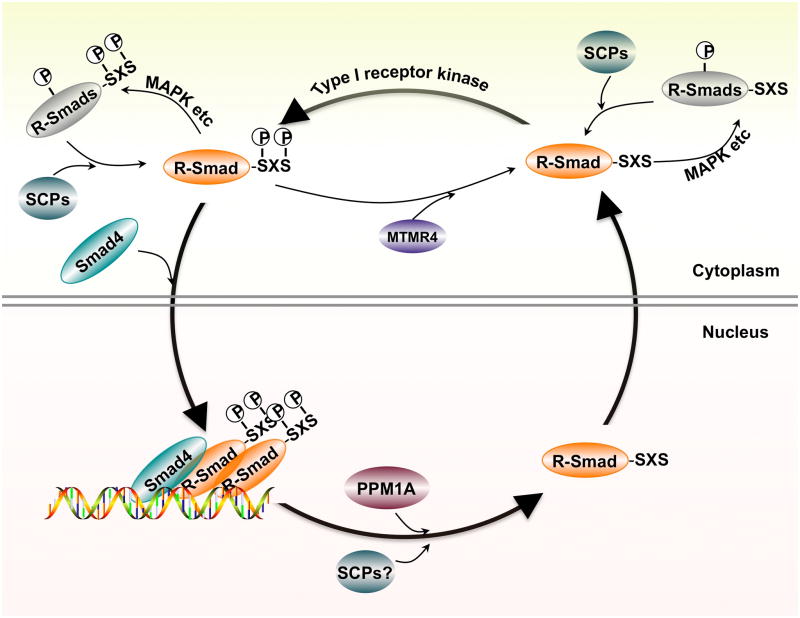
In the study by Lin et al. [74], a library of human serine/threonine phosphatases were screened for their activity towards dephosphorylating Smad2/3. Experimentally, the screen is rather simple by first co-expressing Smad2/3 (the substrate) and a phosphatase (the enzyme) in the same cell and then looking for those phosphatases that reduce or abolish phosphorylation of Smad2/3 that is induced by a constitutively active mutant of TGF-β type I receptor. Out of these thirty-nine phosphatases, only a nuclear localized protein phosphatase, PPM1A/PP2Cα, reduces the level of Smad2/3 phosphorylation that is induced by the activated TGF-β receptor [74]. Furthermore, RNAi-mediated knockdown of endogenous PPM1A expression increases the strength and duration of Smad2/3 SXS phosphorylation in the nucleus. A series of biochemical experiments further pinpoint PPM1A as a direct phosphatase towards Smad2/3 dephosphorylation rather than inactivation of the upstream type I receptor [74]. PPM1A can directly interact with phospho-Smad2/3, not unphosphorylated Smad2/3, and can dephosphorylate recombinant phospho-Smad2/3 proteins in vitro [74]. Importantly, PPM1A facilitates the interaction of dephosphorylated Smad2/3 with RanBP3, a nuclear export factor [75]. As a result, PPM1A-mediated dephosphorylation of Smad2/3 promotes nuclear export of Smad2/3 and shuts off TGF-β-induced anti-proliferative and transcriptional responses (Figure 3). Smad-antagonizing activity of PPM1A is also observed during Nodal-dependent early embryogenesis in zebrafish [74].
Figure 3. Regulation of R-Smad nucleocytoplasmic cycling by reversible phosphorylation.
R-Smads are phosphorylated by type I receptor kinase and then translocated into the nucleus. In the nucleus, Smads are dephosphorylated by phosphatases such as PPM1A, leading to their export back to the cytoplasm.
Despite these nuclear localized phosphatases that function to facilitate R-Smad export from nucleus to cytoplasm, a DUSP family phosphatase MTMR4 (myotubularin-related protein 4) also negatively regulates TGF-β signaling by interacting with and dephosphorylating activated R-Smads in the early endosome, and then blocking R-Smad nuclear translocation [76].
Similar to Smad2/3, Smad1/5/8 is also regulated through reversible phosphorylation. Duan et al. reported that PPM1A dephosphorylates the C-terminal SXS motif of phosphorylated Smad1/5/8, suggesting that PPM1A is a pan-Smad phosphatase [77]. Are there any phosphatases that specifically target the TGF-β or BMP branch? It has been reported that Small C-terminal Domain Phosphatases (SCPs) 1, 2 and 3, Pyruvate Dehydrogenase Phosphatase (PDP) were initially identified to dephosphorylate the SXS motif of Smad1 in Xenopus and Drosophila, respectively [78–79]. Furthermore, knockdown of SCP1/2 [78–79] or PDP [78–79] appears to increase the level of Smad1 phosphorylation in mammalian cells.
Reversible phosphorylation at the linker region of R-Smads
Besides phosphorylation at the C-terminal SXS motif of Smads by the TGF-β type I receptors that represents the key event in Smad activation, phosphorylation at additional sites also regulates the activity of Smads. In contrast to the conserved N-terminal MH1 domain and C-terminal MH2 domain, sequences in the linker region between the MH1 and MH2 domain in R-Smad proteins are very divergent. The linker region contains multiple sites of phosphorylation by a number of intracellular protein kinases that are activated by signals such as mitogenic growth factors, and therefore the linker integrates the activation of these protein kinases into the fine-tuning of Smad activation.
Proline-directed protein kinases such as mitogen-activated protein kinases (MAPKs) and cyclin-dependent kinase (CDKs) are major groups of protein kinases that exhibit preference for specific serine/threonine residues in the linker region [80–81]. Studies demonstrate that phosphorylation in the linker region of R-Smads plays both positive and negative roles in TGF-β signaling. Phosphorylation of Smad3 by CDK2/4 inhibits its transcriptional activity, reduces the anti-proliferative action of TGF-β, and serves as a novel means by which CDK2 promotes aberrant cell cycle progression and confers cancer cell resistance to the growth-inhibitory effects TGF-β [81]. While CDK-mediated R-Smad phosphorylation appears to play negative roles in TGF-β signaling, MAPK-mediated phosphorylation has a dual role in Smad2/3 regulation. For example, ERK-dependent Smad2 phosphorylation at Thr8 enhances its activity, but ERK-mediated Smad2 phosphorylation at Ser245/250/255 and Thr220 as well as Smad3 phosphorylation at Ser204/208 and Thr179 play an inhibitory role on Smad2/3 transcriptional activity [82]. In addition to CDKs and ERK, a number of intracellular protein kinases have been found to phosphorylate the linker region of R-Smads. p38, Rho kinase and c-Jun N-terminal kinase phosphorylate Smad2/3 at multiple sites and enhance their transcriptional activity [83–87]. Other kinases, e.g. CK1γ2, CK1ε, CaMKII, PKC, GRK2 and MEKK1, can also target R-Smads and regulate Smad-dependent transcriptional responses [88–89]. Like Smad2/3 in the TGF-β/activin pathway, Smad1 in BMP pathway can also be phosphorylated in its linker region by protein kinases. For example, Ser187, Ser195, Ser206 and Ser214 sites in the linker region of Smad1 are phosphorylated by ERK [90–91], while Ser193 and Ser210 sites are phosphorylated by GSK-3β [92–93]. Thus, the linker region is a critical regulatory platform in TGF-β superfamily signaling.
Phosphorylation in the linker regions serves an important function in regulating the activity, stability and transport of R-Smads. Linker phosphorylation of Smad2/3 by TGF-β facilitates the binding of NEDD4L to Smad2/3 and consequently results in Smad2/3 polyubiquitination and degradation [41]. Similarly, Smurf1 binding to the phosphorylated linker promotes Smad1 ubiquitination; Smurf1 also causes cytoplasmic retention of Smad1 through blocking Smad1-Nup214 interaction [92].
As increasing evidence indicating the importance of the linker phosphorylation in TGF-β signaling, protein phosphatases have been identified to reversely control this dynamic process. Specifically, phosphatases SCP1/2/3 dephosphorylate specific residues in the linker, but not in the SXS motif of Smad2/3 [94–95]. The activity of SCPs antagonizes the mitogen-mediated inhibition of TGF-β transcriptional responses. Interestingly, for Smad1, SCPs not only mediate dephosphorylation in its linker region, but also dephosphorylate its SXS motif [78,95]. However, SCPs do not dephosphorylate all phospho-residues in the linker and MH1 domain. It is of great interest to identify additional phosphatases that selectively dephosphorylate individual phospho-residues in these regions.
Conclusions and Perspectives
Accumulating evidence illustrates the important role of post-translational modification to regulate components in the TGF-β signaling pathway, and ultimately the critical cellular responses induced by TGF-β ligands. Recent advancements emphasize the importance of how reversible phosphorylation and dephosphorylation controls the physiological consequences in response to developmental or environmental cues. Notably, a few phosphatases have been identified that both negatively and positively regulate TGF-β superfamily signaling.
Despite the progress, the precise role of most of the phosphatases remains undisclosed, largely due to the complexity and pleiotrophy of both the TGF-β signaling and phosphatases. For example, the molecular mechanism for PPP phosphatases-mediated regulation of the receptor activity is still elusive. What are the direct substrates of PP1 or PP2A during receptor regulation? How and in which subcellular compartments are the individual subunits or the holoenzyme are recruited to the receptor? Even for nuclear phosphatases such as PPM1A, do they dephosphorylate the Smad activator complex on chromatin and dissociate the complex or only dephosphorylate phospho-R-Smads after the Smad/DNA dissociation? The answers to these questions are important in addressing the functions and mechanism for how phosphatases fine-tune the TGF-β signaling strength and duration during embryogenesis and other developmental processes.
Because the TGF-β superfamily is evolutionarily conserved, more exhaustive genetic screens in multiple species (e.g. Drosophila, C. elegans and RNAi screens in mammalian cells) are anticipated to search for additional protein phosphatases involved in the regulation of TGF-β signaling. Several features for these new phosphatases include: 1) they either directly dephosphorylate the receptors or Smads; 2) they act either for the entire TGF-β superfamily or in a pathway-restricted manner; 3) functionally, they may play either positive or negative roles, depending on the targeted residues of the receptors or Smads; 4) they may also function to regulate Smad nuclear entry, act to stabilize/dissociate the Smad transcription complex or to modulate Smad nuclear export. Identification and characterization of new phosphatases will not only help us to draw a clearer view for the complex regulation of TGF-β signaling, but also lead to a more complete understanding of signaling crosstalk and integration between the TGF-β signaling pathway and the diverse signaling pathways in cells.
Considering critical roles of TGF-β signaling in tumorigenesis, deregulation of phosphatases in the TGF-β pathway may be an important requirement for tumor progression. Since both PP2A complexes and PPM1A are implicated in tumorigenesis, it is plausible that these phosphatases exert their functions in tumor development through regulation of the TGF-β signaling. Further investigations are needed to elucidate the pathological roles of TGF-β receptor/Smad phosphatases in cancer and other human diseases (e.g. vascular diseases and autoimmune diseases) where TGF-β signaling is a critical player.
References
- 1.Hill CS. TGF-beta signalling pathways in early Xenopus development. Curr Opin Genet Dev. 2001;11:533–540. doi: 10.1016/s0959-437x(00)00229-x. [DOI] [PubMed] [Google Scholar]
- 2.von Bubnoff A, Cho KW. Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev Biol. 2001;239:1–14. doi: 10.1006/dbio.2001.0388. [DOI] [PubMed] [Google Scholar]
- 3.Whitman M. Smads and early developmental signaling by the TGFbeta superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 5.Gordon KJ, Blobe GC. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R, Feng XH. TGF-beta receptor signaling. Biochim Biophys Acta. 1997;1333:F105–150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- 7.Freeman JW, DeArmond D, Lake M, Huang W, Venkatasubbarao K, Zhao S. Alterations of cell signaling pathways in pancreatic cancer. Front Biosci. 2004;9:1889–1898. doi: 10.2741/1388. [DOI] [PubMed] [Google Scholar]
- 8.Han C, Demetris AJ, Liu Y, Shelhamer JH, Wu T. Transforming growth factor-beta (TGF-beta) activates cytosolic phospholipase A2alpha (cPLA2alpha)-mediated prostaglandin E2 (PGE)2/EP1 and peroxisome proliferator-activated receptor-gamma (PPAR-gamma)/Smad signaling pathways in human liver cancer cells. A novel mechanism for subversion of TGF-beta-induced mitoinhibition. J Biol Chem. 2004;279:44344–44354. doi: 10.1074/jbc.M404852200. [DOI] [PubMed] [Google Scholar]
- 9.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 10.Varga J. Antifibrotic therapy in scleroderma: extracellular or intracellular targeting of activated fibroblasts? Curr Rheumatol Rep. 2004;6:164–170. doi: 10.1007/s11926-004-0062-8. [DOI] [PubMed] [Google Scholar]
- 11.Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–29. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 12.Itoh S, ten Dijke P. Negative regulation of TGF-beta receptor/Smad signal transduction. Curr Opin Cell Biol. 2007;19:176–184. doi: 10.1016/j.ceb.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 14.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 15.Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- 16.Boesen CC, Radaev S, Motyka SA, Patamawenu A, Sun PD. The 1.1 A crystal structure of human TGF-beta type II receptor ligand binding domain. Structure. 2002;10:913–919. doi: 10.1016/s0969-2126(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 17.Galliher AJ, Schiemann WP. Src phosphorylates Tyr284 in TGF-beta type II receptor and regulates TGF-beta stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res. 2007;67:3752–3758. doi: 10.1158/0008-5472.CAN-06-3851. [DOI] [PubMed] [Google Scholar]
- 18.Luo K, Lodish HF. Positive and negative regulation of type II TGF-beta receptor signal transduction by autophosphorylation on multiple serine residues. EMBO J. 1997;16:1970–1981. doi: 10.1093/emboj/16.8.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawler S, Feng XH, Chen RH, Maruoka EM, Turck CW, Griswold-Prenner I, Derynck R. The type II transforming growth factor-beta receptor autophosphorylates not only on serine and threonine but also on tyrosine residues. J Biol Chem. 1997;272:14850–14859. doi: 10.1074/jbc.272.23.14850. [DOI] [PubMed] [Google Scholar]
- 20.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 21.Byfield SD, Roberts AB. Lateral signaling enhances TGF-beta response complexity. Trends Cell Biol. 2004;14:107–111. doi: 10.1016/j.tcb.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 23.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 24.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Liu F, Hata A, Baker JC, Doody J, Carcamo J, Harland RM, Massague J. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe M, Masuyama N, Fukuda M, Nishida E. Regulation of intracellular dynamics of Smad4 by its leucine-rich nuclear export signal. EMBO Rep. 2000;1:176–182. doi: 10.1093/embo-reports/kvd029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao Z, Liu X, Henis YI, Lodish HF. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc Natl Acad Sci U S A. 2000;97:7853–7858. doi: 10.1073/pnas.97.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu L, Chen YG, Massague J. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFbeta-dependent phosphorylation. Nat Cell Biol. 2000;2:559–562. doi: 10.1038/35019649. [DOI] [PubMed] [Google Scholar]
- 30.ten Dijke P, Miyazono K, Heldin CH. Signaling inputs converge on nuclear effectors in TGF-beta signaling. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- 31.Roberts AB. TGF-beta signaling from receptors to the nucleus. Microbes Infect. 1999;1:1265–1273. doi: 10.1016/s1286-4579(99)00258-0. [DOI] [PubMed] [Google Scholar]
- 32.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 33.Valdimarsdottir G, Goumans MJ, Itoh F, Itoh S, Heldin CH, ten Dijke P. Smad7 and protein phosphatase 1alpha are critical determinants in the duration of TGF-beta/ALK1 signaling in endothelial cells. BMC Cell Biol. 2006;7:16. doi: 10.1186/1471-2121-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoover LL, Kubalak SW. Holding their own: the noncanonical roles of Smad proteins. Sci Signal. 2008;1:pe48. doi: 10.1126/scisignal.146pe48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- 36.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99:2107–2112. doi: 10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 38.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, Imamura T. NEDD4–2 (neural precursor cell expressed, developmentally down-regulated 4–2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem J. 2005;386:461–470. doi: 10.1042/BJ20040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massague J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K, Miyazono K. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol Biol Cell. 2001;12:1431–1443. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xin H, Xu X, Li L, Ning H, Rong Y, Shang Y, Wang Y, Fu XY, Chang Z. CHIP controls the sensitivity of transforming growth factor-beta signaling by modulating the basal level of Smad3 through ubiquitin-mediated degradation. J Biol Chem. 2005;280:20842–20850. doi: 10.1074/jbc.M412275200. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Xin H, Xu X, Huang M, Zhang X, Chen Y, Zhang S, Fu XY, Chang Z. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol Cell Biol. 2004;24:856–864. doi: 10.1128/MCB.24.2.856-864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang M, Liang YY, Wrighton K, Ungermannova D, Wang XP, Brunicardi FC, Liu X, Feng XH, Lin X. Ubiquitination and proteolysis of cancer-derived Smad4 mutants by SCFSkp2. Mol Cell Biol. 2004;24:7524–7537. doi: 10.1128/MCB.24.17.7524-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan M, Cao X, Wu Y, Bai S, Wu L, Shi X, Wang N. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3:171–176. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280:22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 48.Moren A, Hellman U, Inada Y, Imamura T, Heldin CH, Moustakas A. Differential ubiquitination defines the functional status of the tumor suppressor Smad4. J Biol Chem. 2003;278:33571–33582. doi: 10.1074/jbc.M300159200. [DOI] [PubMed] [Google Scholar]
- 49.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 50.Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH. Activation of transforming growth factor-beta signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J Biol Chem. 2003;278:18714–18719. doi: 10.1074/jbc.M302243200. [DOI] [PubMed] [Google Scholar]
- 51.Lee PS, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem. 2003;278:27853–27863. doi: 10.1074/jbc.M301755200. [DOI] [PubMed] [Google Scholar]
- 52.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 53.Tajima Y, Goto K, Yoshida M, Shinomiya K, Sekimoto T, Yoneda Y, Miyazono K, Imamura T. Chromosomal region maintenance 1 (CRM1)-dependent nuclear export of Smad ubiquitin regulatory factor 1 (Smurf1) is essential for negative regulation of transforming growth factor-beta signaling by Smad7. J Biol Chem. 2003;278:10716–10721. doi: 10.1074/jbc.M212663200. [DOI] [PubMed] [Google Scholar]
- 54.Itoh S, Landstrom M, Hermansson A, Itoh F, Heldin CH, Heldin NE, ten Dijke P. Transforming growth factor beta1 induces nuclear export of inhibitory Smad7. J Biol Chem. 1998;273:29195–29201. doi: 10.1074/jbc.273.44.29195. [DOI] [PubMed] [Google Scholar]
- 55.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikauchi Y, Imamura T, Miyazono K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J Biol Chem. 2002;277:39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- 57.Wrighton KH, Lin X, Feng XH. Critical regulation of TGFbeta signaling by Hsp90. Proc Natl Acad Sci U S A. 2008;105:9244–9249. doi: 10.1073/pnas.0800163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 59.Simonsson M, Heldin CH, Ericsson J, Gronroos E. The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem. 2005;280:21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- 60.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 61.Gallego M, Virshup DM. Protein serine/threonine phosphatases: life, death, and sleeping. Curr Opin Cell Biol. 2005;17:197–202. doi: 10.1016/j.ceb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 63.Majello B, Napolitano G. Control of RNA polymerase II activity by dedicated CTD kinases and phosphatases. Front Biosci. 2001;6:D1358–1368. doi: 10.2741/majello. [DOI] [PubMed] [Google Scholar]
- 64.Bennett D, Alphey L. PP1 binds Sara and negatively regulates Dpp signaling in Drosophila melanogaster. Nat Genet. 2002;31:419–423. doi: 10.1038/ng938. [DOI] [PubMed] [Google Scholar]
- 65.Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164:291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Batut J, Schmierer B, Cao J, Raftery LA, Hill CS, Howell M. Two highly related regulatory subunits of PP2A exert opposite effects on TGF-beta/Activin/Nodal signalling. Development. 2008;135:2927–2937. doi: 10.1242/dev.020842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petritsch C, Beug H, Balmain A, Oft M. TGF-beta inhibits p70 S6 kinase via protein phosphatase 2A to induce G(1) arrest. Genes Dev. 2000;14:3093–3101. doi: 10.1101/gad.854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griswold-Prenner I, Kamibayashi C, Maruoka EM, Mumby MC, Derynck R. Physical and functional interactions between type I transforming growth factor beta receptors and Balpha, a WD-40 repeat subunit of phosphatase 2A. Mol Cell Biol. 1998;18:6595–6604. doi: 10.1128/mcb.18.11.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heikkinen PT, Nummela M, Leivonen SK, Westermarck J, Hill CS, Kahari VM, Jaakkola PM. Hypoxia-activated Smad3-specific dephosphorylation by PP2A. J Biol Chem. 285:3740–3749. doi: 10.1074/jbc.M109.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- 71.Schmierer B, Hill CS. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor beta-dependent nuclear accumulation of Smads. Mol Cell Biol. 2005;25:9845–9858. doi: 10.1128/MCB.25.22.9845-9858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu L, Kang Y, Col S, Massague J. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. Mol Cell. 2002;10:271–282. doi: 10.1016/s1097-2765(02)00586-5. [DOI] [PubMed] [Google Scholar]
- 73.Lin X, Chen Y, Meng A, Feng X. Termination of TGF-beta superfamily signaling through SMAD dephosphorylation--a functional genomic view. J Genet Genomics. 2007;34:1–9. doi: 10.1016/S1673-8527(07)60001-0. [DOI] [PubMed] [Google Scholar]
- 74.Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng XH. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dai F, Lin X, Chang C, Feng XH. Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-beta signaling. Dev Cell. 2009;16:345–357. doi: 10.1016/j.devcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu J, Pan L, Qin X, Chen H, Xu Y, Chen Y, Tang H. MTMR4 attenuates TGF{beta} signaling by dephosphorylating R-Smads in endosomes. J Biol Chem. 2010 doi: 10.1074/jbc.M109.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duan X, Liang YY, Feng XH, Lin X. Protein serine/threonine phosphatase PPM1A dephosphorylates Smad1 in the bone morphogenetic protein signaling pathway. J Biol Chem. 2006;281:36526–36532. doi: 10.1074/jbc.M605169200. [DOI] [PubMed] [Google Scholar]
- 78.Knockaert M, Sapkota G, Alarcon C, Massague J, Brivanlou AH. Unique players in the BMP pathway: small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc Natl Acad Sci U S A. 2006;103:11940–11945. doi: 10.1073/pnas.0605133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen HB, Shen J, Ip YT, Xu L. Identification of phosphatases for Smad in the BMP/DPP pathway. Genes Dev. 2006;20:648–653. doi: 10.1101/gad.1384706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 81.Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 82.Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Funaba M, Zimmerman CM, Mathews LS. Modulation of Smad2-mediated signaling by extracellular signal-regulated kinase. J Biol Chem. 2002;277:41361–41368. doi: 10.1074/jbc.M204597200. [DOI] [PubMed] [Google Scholar]
- 84.Engel ME, McDonnell MA, Law BK, Moses HL. Interdependent SMAD and JNK signaling in transforming growth factor-beta-mediated transcription. J Biol Chem. 1999;274:37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 85.Mori S, Matsuzaki K, Yoshida K, Furukawa F, Tahashi Y, Yamagata H, Sekimoto G, Seki T, Matsui H, Nishizawa M, Fujisawa J, Okazaki K. TGF-beta and HGF transmit the signals through JNK-dependent Smad2/3 phosphorylation at the linker regions. Oncogene. 2004;23:7416–7429. doi: 10.1038/sj.onc.1207981. [DOI] [PubMed] [Google Scholar]
- 86.Kamaraju AK, Roberts AB. Role of Rho/ROCK and p38 MAP kinase pathways in transforming growth factor-beta-mediated Smad-dependent growth inhibition of human breast carcinoma cells in vivo. J Biol Chem. 2005;280:1024–1036. doi: 10.1074/jbc.M403960200. [DOI] [PubMed] [Google Scholar]
- 87.Yamagata H, Matsuzaki K, Mori S, Yoshida K, Tahashi Y, Furukawa F, Sekimoto G, Watanabe T, Uemura Y, Sakaida N, Yoshioka K, Kamiyama Y, Seki T, Okazaki K. Acceleration of Smad2 and Smad3 phosphorylation via c-Jun NH(2)-terminal kinase during human colorectal carcinogenesis. Cancer Res. 2005;65:157–165. [PubMed] [Google Scholar]
- 88.Wrighton KH, Feng XH. To (TGF)beta or not to (TGF)beta: fine-tuning of Smad signaling via post-translational modifications. Cell Signal. 2008;20:1579–1591. doi: 10.1016/j.cellsig.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wrighton KH, Lin X, Feng XH. Phospho-control of TGF-beta superfamily signaling. Cell Res. 2009;19:8–20. doi: 10.1038/cr.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389:618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 91.Aubin J, Davy A, Soriano P. In vivo convergence of BMP and MAPK signaling pathways: impact of differential Smad1 phosphorylation on development and homeostasis. Genes Dev. 2004;18:1482–1494. doi: 10.1101/gad.1202604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 93.Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wrighton KH, Willis D, Long J, Liu F, Lin X, Feng XH. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-beta signaling. J Biol Chem. 2006;281:38365–38375. doi: 10.1074/jbc.M607246200. [DOI] [PubMed] [Google Scholar]
- 95.Sapkota G, Knockaert M, Alarcon C, Montalvo E, Brivanlou AH, Massague J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J Biol Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]