Abstract
The enzyme α1,3-galactosyltransferase (α1,3GT or GCTA1) synthesizes α1,3-galactose (α1,3Gal) epitopes (Galα1,3Galβ1,4GlcNAc-R), which are the major xenoantigens causing hyperacute rejection in pig-to-human xenotransplantation. Complete removal of α1,3Gal from pig organs is the critical step toward the success of xenotransplantation. We reported earlier the targeted disruption of one allele of the α1,3GT gene in cloned pigs. A selection procedure based on a bacterial toxin was used to select for cells in which the second allele of the gene was knocked out. Sequencing analysis demonstrated that knockout of the second allele of the α1,3GT gene was caused by a T-to-G single point mutation at the second base of exon 9, which resulted in inactivation of the α1,3GT protein. Four healthy α1,3GT double-knockout female piglets were produced by three consecutive rounds of cloning. The piglets carrying a point mutation in the α1,3GT gene hold significant value, as they would allow production of α1,3Gal-deficient pigs free of antibiotic-resistance genes and thus have the potential to make a safer product for human use.
The enzyme α1,3-galactosyltransferase (α1,3GT or GGTA1) synthesizes α1,3Gal epitopes (Galα1,3Galβ1,4GlcNAc-R) on the cell surface of almost all mammals with the exception of humans, apes, and Old World monkeys (1). α1,3Gal epitopes are the major xenoantigens causing hyperacute rejection (HAR) in pig-to-human xenotransplantation (2–4). Many reports have also indicated that α1,3Gal epitopes are involved in acute vascular rejection (AVR) of xenografts (4–6). Piglets with α1,3GT heterozygous knockout have been cloned by our group (7) and another team (8) in the last year. To produce homozygous α1,3GT knockout piglets by natural breeding, assuming both male and female heterozygous knockout pigs are available at the same time and are fertile, is feasible but takes up to 12 months. However, by using a second-round knockout and cloning strategy, we could save up to 6 months and all cloned piglets would be α1,3GT double knockout (DKO). We have selected and enriched for α1,3GT DKO cells by using a bacterial toxin, toxin A from Clostridium difficile, which binds with high affinity to α1,3Gal epitopes and produces a cytotoxic effect on cells that are α1,3Gal-positive (9). Toxin A uses α1,3Gal epitopes as a cell surface receptor and causes “rounding” and lifting of the α1,3Gal-positive cells from the surface of the growth vessel (10, 11).
Heterozygous α1,3GT knockout fetal fibroblasts, 657A-I11 1-6 cells, were isolated from a day-32 pregnancy as described in (7). To avoid using a second antibiotic-resistance gene as a selection marker, we constructed an ATG (start codon)-targeting α1,3GT knockout vector, pPL680 (12), which also contains a neo gene, to knock out the second allele of the α1,3GT gene. 657A-I11 1-6 cells were transfected by electroporation with pPL680 and selected for the α1,3Gal-negative phenotype with purified C. difficile toxin A (13). One colony (680B1) was isolated and expanded after toxin A selection. When the 680B1 cells were stained with a fluorescein-labeled α1,3Gal-specific lectin, GS-IB4, about 80% of the cells were found to be α1,3Gal-negative. The fact that fewer than 100% of the cells in the colony were negative with GS-IB4 staining indicated that this colony contained a mixture of α1,3Gal-negative and -positive cells. We used 680B1 cells for somatic cell nuclear transfer (cloning) as described in (7). We transferred embryos to five recipient gilts, and three initial pregnancies were established, of which only one went beyond day 35 of gestation.
To determine whether all the fetuses cloned from 680B1 cells were α1,3GT DKO, we terminated the remaining pregnancy at day 39 and recovered four normal-sized fetuses. Fibroblast cell lines (680B1-1 to B1-4) were isolated from each of these four fetuses, and fluorescence-activated cell sorting (FACS) analysis with GS-IB4 staining showed that B1-1, B1-2, and B1-4 cells were α1,3Gal-negative, whereas B1-3 cells were positive for α1,3Gal (Fig. 1). Normal human serum (NHS) contains preformed antibodies to α1,3Gal and complement proteins, which together cause rapid lysis of cells that are α1,3Gal-positive. A complement lysis assay on these cells showed that B1-1, B1-2, and B1-4 cells were resistant to lysis by NHS, but B1-3 cells were lysed by NHS at the same rate (about 40% of cells lysed) as control wild-type pig cells (Fig. 2). Analysis of genomic DNA from these fetal cells by polymerase chain reaction (PCR) and Southern blot analysis indicated that none of the three α1,3Gal-negative cell lines had the expected restriction fragment pattern predicted for targeted disruption of the second α1,3GT allele with the pPL680 knockout vector. Instead, these fetal cells appeared to have the same allele pattern (one targeted allele and one wild-type allele) as their parent 657A-I11 1-6 cells, which contained only one disrupted α1,3GT allele. Northern blot analysis of the four cell lines (B1-1 to B1-4) showed that they expressed two mRNAs of similar size to those seen in the 657A-I11 1-6 cells (fig. S1).
Fig. 1.
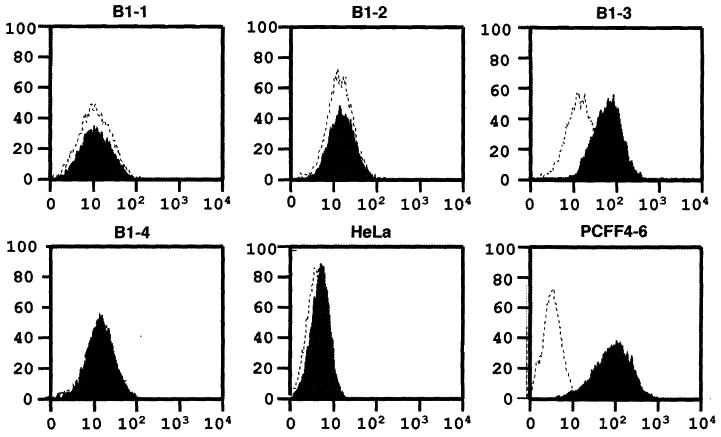
Flow cytometry analysis of 680B1-1 to B1-4 cells with GS-IB4 lectin staining. Horizontal and vertical axes denote intensity of fluorescence and number of events, respectively. Dotted line represents unstained cells analyzed by a fluorescence-activated cell sorter (Becton-Dickenson, Franklin, NJ). Shadow represents cells stained with fluorescein isothiocyanate–labeled GS-I-B4 lectin (EY Laboratories, Inc., San Mateo, CA). B1-1, B1-2, B1-3, and B1-4 are fetal fibroblasts derived from four day-39 fetuses. HeLa cells, a human cell line, were used as the negative control and PCFF4-6 cells, which were the parent cells for heterozygous and DKO of the α1,3GT gene, were used as the positive control.
Fig. 2.
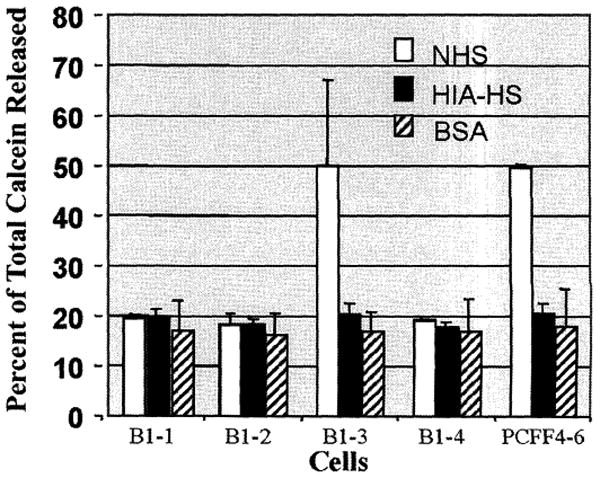
Complement lysis assay for DKO fetal fibroblasts and wild-type pig cells. Results are the average of three individual assays. Open box represents NHS. Solid box represents heat-inactivated human serum (HIA-HS) as the negative control, and hatched box represents bovine serum albumin (BSA) as the reagent control. About 20% calcein release (no cells lysed) is the base value for the negative control (HIA-HS) and the reagent control (BSA). For quality control and reproducibility purposes, we did not use fresh human serum for the assay, which usually gives about 90% calcein release. About 50% calcein release (about 40% of cells lysed) from wild-type pig cells is typical with commercial serum (frozen and lyophilized) from Sigma.
The 3.8-kb band corresponds in size to the wild-type α1,3GT transcript and the shorter 2.5-kb band is the same size expected for the truncated transcript of the first knockout allele (fig. S1). Because of the nature of the toxin A selection method, α1,3 Gal-negative cells are selected, regardless of whether inactivation of the second α1,3GT allele was caused by targeted disruption via the pPL680 vector or by any other mechanism. The fact that one normal-sized allele was observed (instead of two shorter knockout alleles) indicated that knockout of the second α1,3GT allele was due to mechanisms other than targeted homologous recombination-mediated disruption, promoter dysfunction, or mRNA missplicing and instability.
To identify the nature of the inactivation event for the second allele, we subcloned and sequenced α1,3GT cDNAs from all four cell lines (B1-1, B1-2, B1-3, and B1-4). Sequencing results revealed that there was a T-to-G transversion at the second base pair of exon 9 in the nontargeted α1,3GT allele of B1-1, B1-2, and B1-4 cells, but not in B1-3 cells or in the first knockout allele of all four cell lines. This T-to-G transversion in the α1,3GT coding region caused a single amino acid change from tyrosine to aspartic acid in the α1,3GT protein (Fig. 3). Although this mutation has not been observed in the inactivated α1,3GT gene of humans or higher primates (14), it is likely that the change of tyrosine, a hydrophobic amino acid, to aspartic acid, a hydrophilic amino acid, could disrupt α1,3GT function. Crystal structure analysis of bovine α1,3GT protein supports this speculation and shows that this tyrosine is at the center of the catalytic domain of bovine α1,3GT protein and is involved in uridine 5′-diphosphate–Gal binding (15, 16).
Fig. 3.
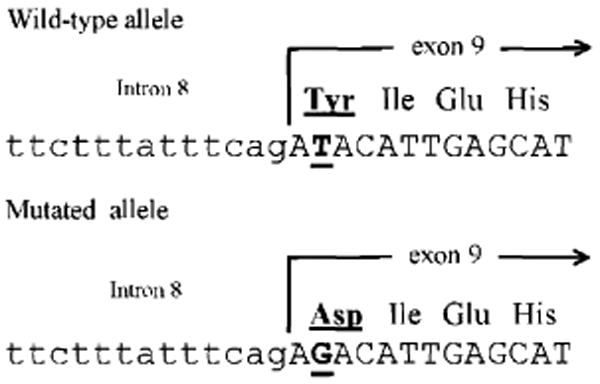
Sequencing analysis of the α1,3GT gene from wild-type pig cells and DKO porcine fetal fibroblasts. Upper and lower alignment show nucleotide sequence of the α1,3GT intron 8–exon 9 boundary from wild-type pigs and the second allele of the DKO pig fetuses (B1-1, B1-2, and B1-4), respectively. Small letters and capital letters denote intron and exon sequences, respectively. Underlined capital letters indicate the nucleotide where the point mutation occurred. Amino acids deduced from the correspondent mutated and wild-type DNA sequence are underlined. No other mutations were found in the coding region of the α1,3GT gene from the second allele of the DKO pig fetuses in our genomic and reverse transcriptase–PCR libraries.
To further confirm that the mutated cDNA cannot make functional α1,3GT protein, we cloned α1,3GT cDNAs from the nontargeted allele of B1-1 to B1-4 cells and wild-type pig cells into an expression vector and transfected them into human HeLa cells, which normally do not express α1,3GT protein. HeLa cells transfected with cDNA expression vectors from B1-1, B1-2, and B1-4 cells were negative for GS-IB4 lectin staining, indicating that the transfected pig cDNA from these cells did not make functional α1,3GT protein. In contrast, HeLa cells transfected with the cDNA from B1-3 cells and wild-type pig cells were positive with GS-IB4 staining. These results verified that the point mutation in cDNA from the second allele of the α1,3GT gene in B1-1, B1-2, and B1-4 cells resulted in synthesis of a defective α1,3GT protein. Although toxin A selection was repeated several times on 657A-I11 1-6 cells, with or without pPL680 vector transfection, no additional toxin A–resistant colonies were detected.
We performed somatic cell nuclear transfer (cloning) with all three DKO cell lines as described in (7). We transferred cloned embryos into 16 estrus-synchronized recipient gilts. Ten initial pregnancies were established, only two of which went to term. Two pregnancies were lost before day 30, five were lost between day 30 and day 40, and one was lost by day 60. The first five female α1,3GT DKO piglets (761-1 to 761-5), cloned from 680B1-2 cells, were born on 25 July 2002. One piglet (761-1) died shortly after birth, and necropsy revealed an enlarged tongue and unusually large kidneys. We have observed this phenotype in a few other α1,3Gal-positive cloned pigs, and it appears to be a function of the cloning process (incomplete reprogramming) and not the α1,3GT gene knockout per se. The other four DKO piglets were of normal size and healthy. Aorta endothelial cells and muscle and tail fibroblasts isolated from the dead piglet (761-1) were negative with GS-IB4 lectin staining. FACS analysis of muscle fibroblasts from piglet 761-1 also showed a negative result for GS-IB4 binding. Neonatal tail fibroblasts isolated from the four healthy piglets, when analyzed by FACS with GS-IB4, were all negative (fig. S2). Tissue sections of liver, kidney, spleen, skin, intestine, muscle, brain, heart, pancreas, lung, aorta, tongue, umbilicus, and tail obtained from piglet 761-1 were all negative with GS-IB4 staining, indicating a complete lack of detectable cell surface α1,3Gal epitopes. The GS-IB4 staining results for liver sections from a newborn wild-type piglet and from piglet 761-1 are shown in fig. S3. Southern blot and sequencing analysis of DNA samples from all five piglets confirmed the targeted disruption of the first allele of the α1,3GT gene and the T-to-G point mutation in the second base of exon 9 in the second allele of the α1,3GT gene. It has been reported that α1,3GT DKO mice developed cataracts at 4 to 6 weeks of age (17). Physical examination of the four piglets at 7 weeks of age did not reveal any abnormalities or cataracts. We will continue to monitor the piglets for the presence of cataracts.
We performed an in vivo immunogenicity test with α1,3GT-knockout mice. We injected islet-like cell clusters (ICCs) isolated from the pancreas of piglet 761-1 intraperitoneally into α1,3GT knockout mice. We used ICCs from a neonatal wild-type piglet as a control. As shown in fig. S4, no increase in the titer of immunoglobulin M (IgM) to α1,3Gal was observed in α1,3GT knockout mice after injection with ICCs from the α1,3GT DKO piglet, in contrast to significant IgM titer increases observed in those mice injected with wild-type piglet ICCs. This result clearly demonstrates that the DKO piglet cells do not make any α1,3Gal epitopes.
Thus, we have successfully produced four α1,3GT-deficient piglets by a toxin A–mediated selection method. Although our intent was to knock out the second allele of the α1,3GT gene by homologous recombination, this did not occur. Instead, because we used this powerful selection method, which allows us to isolate any event that results in loss of α1,3GT activity, we discovered a mutation in the second allele of the α1,3GT gene. Had we used standard selection methods with puromycin or hygromycin, we would not have found the mutation. Although the rate of spontaneous mutation in the pig genome is very low [about 4 × 10−8 for a spontaneous mutation per replication (18) in a mammalian gene similar in size to the α1,3GT gene], toxin A selection still enabled us to detect this crucial mutation. Clearly inactivation of the α1,3GT protein by this point mutation is a better outcome than by gene targeting with the pPL680 vector. It provides the opportunity to produce α1,3GT-deficient pigs without any antibiotic-resistance genes or other foreign DNA sequences, which should facilitate regulatory approval and, potentially, make a safer product for human use. It is certain that this point mutation will be maintained in the genome of these DKO pigs and their offspring, just as the few critical point mutations in the α1,3GT gene of humans and higher primates have been maintained over 20 million years (14). This genomic stability is not only due to the rarity of a reverse mutation event [about 5 × 10−11 per replication (18) for mammals] but, more importantly, the strong selection pressure against α1,3Gal-positive cells by the presence of antibodies to α1,3Gal in α1,3Gal-negative animals. Our results have demonstrated that removal of α1,3Gal epitopes on pig cells did not preclude development in utero, even though pig cells express up to 500 times the number of α1,3Gal epitopes as do mouse cells (4, 19). In addition, three consecutive rounds of cloning with rederived fetal cells did not appear to have a major detrimental effect on the overall development or health of the cloned pigs in this study. Analysis of tissues and organs from these α1,3GT DKO pigs in nonhuman primate models should provide clear indications of the involvement of α1,3Gal in HAR, AVR, and chronic rejection.
Supplementary Material
Acknowledgments
Supported in part by a grant from the National Institute of Standards and Technology Advanced Technology Program to PPL Inc. and by National Institutes of Health grant DK29961 (T.E.S). We thank B. Gragg, W. Lucero, T. Akers, H. Bishop, and J. McPherson for technical contributions to embryo transfer and animal husbandry; J. Cowell-Lucero, J. Hencke, and V. Marshall for help in cell culture and transfection; D. Innes for help in complement lysis assay; J. Profozich and T. Libert at the University of Pittsburgh for technical support; and staff at the Virginia-Maryland Regional College of Veterinary Medicine for performing cesarean delivery and physical examination of piglets. All animal work was done following a protocol approved by the PPL, Inc. (Blacksburg, VA) institutional animal care and use committee.
Footnotes
www.sciencemag.org/cgi/content/full/1078942/DC1
Materials and Methods
References and Notes
- 1.Galili U, et al. J Biol Chem. 1988;263:17755. [PubMed] [Google Scholar]
- 2.Good AH, et al. Transplant Proc. 1992;24:559. [PubMed] [Google Scholar]
- 3.Cooper DK, Koren E, Oriol R. Lancet. 1993;342:682. doi: 10.1016/0140-6736(93)91791-j. [DOI] [PubMed] [Google Scholar]
- 4.Galili U. Biochimie. 2001;83:557. doi: 10.1016/s0300-9084(01)01294-9. [DOI] [PubMed] [Google Scholar]
- 5.Sandrin MS, Mckenzie IFC. Curr Opin Immunol. 1999;11:527. doi: 10.1016/s0952-7915(99)00011-4. [DOI] [PubMed] [Google Scholar]
- 6.Logan JS. Curr Opin Immunol. 2000;12:563. doi: 10.1016/s0952-7915(00)00139-4. [DOI] [PubMed] [Google Scholar]
- 7.Dai Y, et al. Nature Biotechnol. 2002;20:251. doi: 10.1038/nbt0302-251. [DOI] [PubMed] [Google Scholar]
- 8.Lai L, et al. Science. 2002;295:1089. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- 9.Clark GF, Krivan HC, Wilkins TD, Smith DF. Arch Biochem Biophys. 1987;257:217. doi: 10.1016/0003-9861(87)90561-3. [DOI] [PubMed] [Google Scholar]
- 10.Kushnaryov VM, Sedmak JJ, Markwald RR, Faculjak ML, Loo PM. Cytobios. 1990;64:181. [PubMed] [Google Scholar]
- 11.Just I, Selzer J, von Eichel-Streiber C, Aktories K. J Clin Invest. 1995;95:1026. doi: 10.1172/JCI117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.pPL680 was made from three parts: a 1.8-kb PCR-generated fragment of α1,3GT intron 2 to exon 4 ended at the start codon ATG as the 5′ arm; Neo/pA coding region fused into the ATG of the 5′ arm in-frame; a 9.6-kb PCR-generated fragment of α1,3GT exon 4 to exon 7 as the 3′ arm.
- 13.657A-I11 1-6 cells were electroporated with pPL680 and cultured for 5 days. Cells were exposed to C. difficile toxin A (2 μg/ml) (Techlab, Blacksburg, VA) in growth medium for 2 hours. Toxin A medium was then replaced with fresh growth medium. Medium was changed 7 and 11 days posttransfection to remove the detached cells. One colony (680B1) was harvested 13 days posttransfection for expansion and cryopreservation.
- 14.Koike C, et al. J Biol Chem. 2002;277:10114. doi: 10.1074/jbc.M110527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gastinel LN, et al. EMBO J. 2001;20:638. doi: 10.1093/emboj/20.4.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohan R, Tvaroska I. Protein. 2001;44:428. doi: 10.1002/prot.1108. [DOI] [PubMed] [Google Scholar]
- 17.Tearle RG, et al. Transplantation. 1996;61:13. doi: 10.1097/00007890-199601150-00004. [DOI] [PubMed] [Google Scholar]
- 18.Drake JW, Charlesworth B, Charlesworth D, Crow J. Genetics. 1998;148:1667. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanemura M, Maruyama S, Galili U. Transplantation. 2000;69:187. doi: 10.1097/00007890-200001150-00034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


