Abstract
Repair of DNA double-strand breaks (DSBs) is critical for cell survival and for maintaining genome stability in eukaryotes. In Schizosaccharomyces pombe, the Mre11-Rad50-Nbs1 (MRN) complex and Ctp1 cooperate to perform the initial steps that process and repair these DNA lesions via homologous recombination (HR). While Ctp1 is recruited to DSBs in an MRN-dependent manner, the specific mechanism of this process remained unclear. We recently found that Ctp1 is phosphorylated on a domain rich in putative Casein kinase 2 (CK2) phosphoacceptor sites that resembles the SDTD repeats of Mdc1. Furthermore, phosphorylation of this motif is required for interaction with the FHA domain of Nbs1 that localizes Ctp1 to DSB sites. Here, we review and discuss these findings, and we present new data that further characterize the cellular consequences of mutating CK2 phosphorylation motifs of Ctp1, including data showing that these sites are critical for meiosis.
The integrity of the genome is constantly threatened by a variety of genotoxic insults, with one of the most detrimental being DNA double-strand breaks (DSBs). The heterotrimeric Mre11-Rad50-Nbs1 (MRN) complex is a critical element in the detection, signaling, and homologous recombination (HR) repair of DSBs.1 The MRN complex consists of Mre11 nuclease, Rad50 scaffolding, and Nbs1 regulatory subunits. The N-terminus of Nbs1 contains a forkhead-associated (FHA) domain and proximal tandem Brca1 C-terminal (BRCT) repeats while the C-terminus of Nbs1 harbors interaction motifs for Mre11 and ATM. Both the FHA domain and BRCT repeats are phosphopeptide-binding motifs, often involved in regulation of the cell cycle and DNA damage response by participating in phosphorylation-dependent protein-protein interactions.2, 3 While Mre11 and Rad50 have been thoroughly characterized structurally and functionally, the activities of Nbs1, especially those tied to the N-terminus, are not well understood. While it was initially reported that the N-terminal FHA-BRCT domains of Nbs1 enabled phosphorylation-dependent interaction with γH2AX at DSBs,4 this association was later shown to require mediator of the DNA-damage checkpoint 1 (Mdc1).5 Multiple groups then demonstrated in mammalian cells that the FHA-BRCT domains of Nbs1 interact with diphosphorylated SDTD repeats of Mdc1 and that this binding facilitates a positive feedback loop of ATM signaling from DSBs.6-9 Interestingly, this process is likely initiated through phosphorylation of Mdc1 SDTD sites by Casein kinase 2 (CK2). CK2 is a highly conserved serine/threonine protein kinase that is constitutively active, ubiquitously distributed in eukaryotes, and participates in an abundance of cellular processes.10 In particular, CK2 has recently been implicated in multiple DNA damage-response pathways, primarily through generating phosphorylation-dependent binding interfaces at sites of DNA damage that are recognized by FHA domain-containing DNA repair factors.11-18
The MRN protein complex performs its roles in HR through collaboration with the DNA end-processing factor Ctp1. Ctp1 is an ortholog of the budding yeast Sae2 and human CtIP proteins,19 and is phosphorylated both basally and in a DNA damage dependent manner.20 In S. pombe, Ctp1 and Nbs1 are genetically epistatic,19 and Ctp1 functions as a multi-copy suppressor of the DNA damage sensitivity caused by nbs1-s10, an FHA domain point mutant.20 Furthermore, in human cells, CtIPCtp1 interacts with the MRN complex,21 and coprecipitates specifically with Nbs1 when both proteins are overexpressed.22 From these data and the fact that fungi lack an Mdc1 homolog, we surmised that Nbs1 might associate with Ctp1 through a mechanism similar to the Nbs1-Mdc1 interaction. In support of this idea, our group and others identified a motif spanning amino acids 74-94 of Ctp1 that resembles the SDTD repeats of Mdc1, which we termed the SXT domain.23, 24 We observed that a ctp1-5A (ctp1-S77A T78A T79A S87A T89A) mutant, in which the five potential CK2 phosphoacceptor sites of the SXT domain were changed to alanine (Fig. 1A), exhibited slow growth and extensive DNA damage sensitivity. However, this mutation caused no alteration of Ctp1 protein stability or subcellular localization. Most importantly, we found that this mutation altered phosphorylation of Ctp1 and inhibited recruitment of Ctp1 to a defined DSB as observed by chromatin immunoprecipitation (ChIP).
Figure 1.
Basal phosphorylation of Ctp1 is carried out by Casein kinase 2. (A) Schematic of Ctp1 depicting conserved N-terminal coiled coil domain (CC), C-terminal core homology region (CXXC/RHR), and a CK2 phosphoacceptor motif (SXT). Motif enlargement shows residue substitutions in the ctp1-5A, ctp1-3A, ctp1-CK, and ctp1-3D strains. (B) ctp1-CK and ctp1-3D cells are sensitive to exogenous DNA damaging agents, similar to ctp1-3A cells. (C) Basal phosphorylation is equally altered in Ctp1-3A and Ctp1-CK relative to wild-type.
Remarkably, while the SXT domain of Ctp1 exhibits high conservation to the SDTD repeats of Mdc1, our additional mutational analyses revealed that a ctp1-3A (ctp1-T78A T79A T89A) mutant harboring combined alteration of SXT threonines alone (Fig. 1A) is sufficient for causing the severe DNA damage sensitivity observed in ctp1-5A. In addition, cells harboring individual or combined mutation of the SXT domain serines (Ser77/Ser87) exhibited no genotoxin sensitivity (data not shown). In contrast, the Mdc1-Nbs1 interaction depends on phosphorylation of both the serine and threonine of an individual SDTD motif.6-9
Mutation of SXT domain CK2 sites causes impedes basal phosphorylation of Ctp1 and causes DNA damage sensitivity
CK2 typically phosphorylates the minimum sequence S/T-X-X-D/E,25 a consensus that fits each threonine in the SXT domain of Ctp1. Although another recent study made use of a phosphospecific antibody to show that Thr89 of Ctp1 is constitutively phosphorylated in vivo,24 no specific evidence implicating CK2 in this process has been provided. To further investigate whether the basal phosphorylation of Ctp1 is likely catalyzed by CK2 or a protein kinase with a similar preferred phosphorylation site motif, we created a ctp1-CK (ctp1-E81A D82A E92A) mutant, in which the phosphoacceptor threonine residues of the SXT domain remain intact, while the acidic determinant (n+3) residue of each putative CK2 site was changed to alanine (Fig. 1A). This mutant exhibited slow growth and DNA damage sensitivity identical to the ctp1-3A mutant, suggesting a specific requirement for CK2 in the basal phosphorylation of Ctp1 (Fig. 1B). We previously demonstrated the importance of the SXT domain with regard to basal phosphorylation of Ctp1 through an increase in the gel mobility of Ctp1-5A protein. Here we show that this phenomenon is mirrored in ctp1-3A and ctp1-CK, as these mutants both run with identical faster mobility than wild-type (Fig. 1C). Since loss of the acidic determinant residues of each CK2 site alters gel mobility of Ctp1 to the same extent as loss of phosphoacceptor residues, this result suggests a specific requirement for CK2 in this process. Furthermore, these results corroborate loss of phosphorylation on the Ctp1 SXT domain with the extreme DNA damage sensitivity observed in the ctp1-3A and ctp1-CK strains (Fig. 1B). In addition, these mutants caused a dramatic increase in Ctp1 protein stability, which is known to occur in response to DNA damage.26 We further attempted to verify the role of CK2 in basal phosphorylation of Ctp1 through employing the temperature sensitive CK2 mutant orb5-19.27 However, we found that Ctp1 was extremely abundant at the restrictive temperature (data not shown), indicating that loss of multiple CK2-dependent processes leads to a large increase in spontaneous DNA damage.
We also generated a phospho-mimetic Ctp1 mutant, ctp1-3D (ctp1-T78D T79D T89D), in which the SXT domain threonines were exchanged for aspartic acid (Fig. 1A). Interestingly, this mutant showed enhanced DNA damage sensitivity similar to ctp1-3A (Fig. 1B), indicating that the significance of SXT domain phosphorylation is more extensive than increase in negative charge, such as providing a specific recognition platform for an FHA domain.
The SXT domain is required for Ctp1 activity
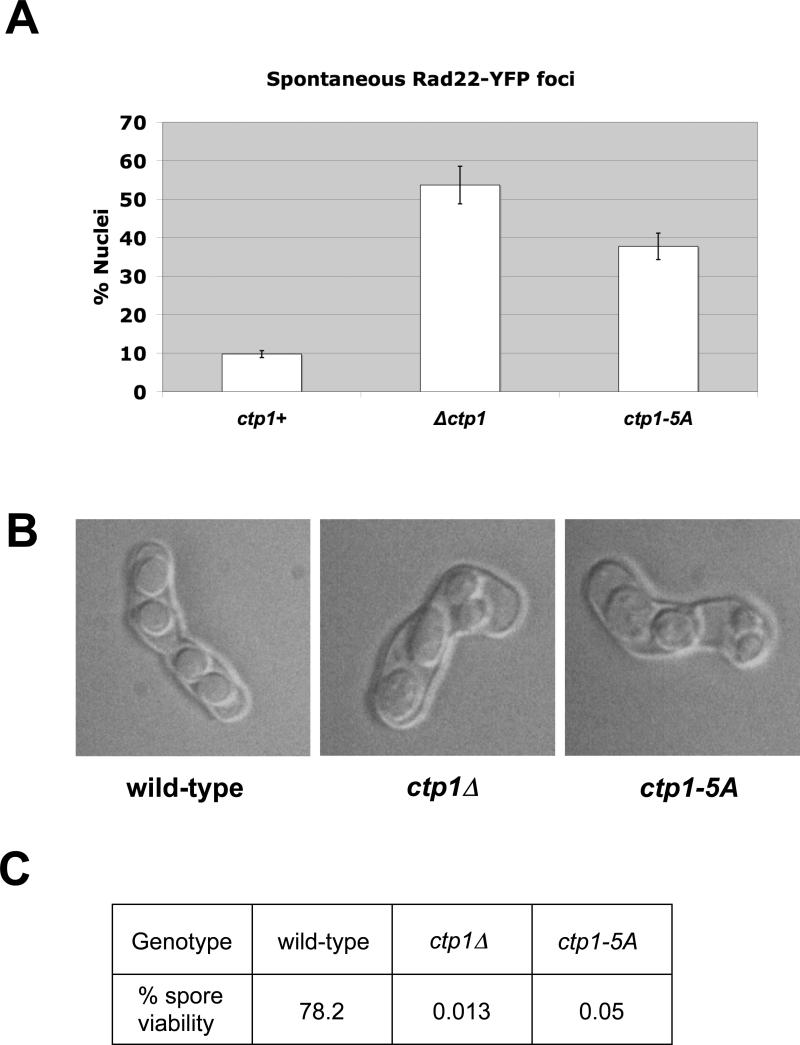
The similar phenotypes of ctp1Δ and ctp1-5A mutants indicated that the SXT domain is critical for Ctp1 activity. To further characterize the cellular consequences of inhibiting phosphorylation of Ctp1, we counted Rad22Rad52-YFP foci in ctp1-5A cells. Rad22 is essential for homologous recombination in fission yeast. We found that ctp1-5A cells accumulate a dramatically higher amount of spontaneous Rad22Rad52-YFP foci (38%) than ctp1+(11%), and that this is similar to what is observed in ctp1Δ (53%) (Fig. 2A). Since ctp1Δ cells are largely compromised for HR repair of DSBs (Limbo), a similar quantity of persistent Rad22Rad52-YFP foci suggests a loss of HR repair activity in the ctp1-5A mutant as well.
Figure 2.
The ctp1-5A mutant phenocopies cellular defects observed in ctp1Δ. (A) ctp1-5A cells accumulate increased levels of spontaneous Rad22-YFP foci. (B) Asci from a ctp1-5A × ctp1 5A mating are abnormal. (C) Spore viability in ctp1-5A × ctp1-5A is very low.
Ctp1 plays an essential role in the repair of programmed DSBs during meiosis.19 To examine the importance of the Ctp1 SXT domain in meiotic processes, we mated ctp1-5A haploids. Similar to what we observed in ctp1Δ × ctp1Δ crosses, the asci generated by a ctp1-5A × ctp1-5A mating are grossly defective in both spore size and number (Fig. 2B). Furthermore, the viability of the spores generated from the mating of ctp1-5A haploids is extremely poor (0.05%), similar to ctp1Δ (0.013%) (Fig. 2C). These results indicate that an intact SXT domain is essential for Ctp1 activity in both meiotic and mitotic cells. Taken together with our previous results, these data suggest that the ctp1-5A mutant essentially phenocopies the cellular defects observed in ctp1Δ cells.
Phosphorylation of Ctp1 mediates its association with the MRN complex through binding to the FHA domain of Nbs1
Although we were able to establish by ChIP that recruitment of Ctp1 to a defined DSB is Nbs1-dependent, isolation of Nbs1-Ctp1 complexes proved difficult, suggesting that Nbs1-Ctp1 binding might be short-lived or salt sensitive. By using an mre11-H134S nuclease-dead background,28 Ctp1-Nbs1 complexes were stabilized in vivo, and could be observed by co-immunoprecipitation. Furthermore, this association is dependent on the phosphopeptide-binding pocket of the Nbs1 FHA domain. Here, we utilized the ctp1-5A mutant to examine the effect of SXT domain phosphorylation on association of Ctp1 with Nbs1. Despite the fact that ctp1-5A induces a dramatic increase in the amount of total Ctp1 protein, the amount of Nbs1 associated with Ctp1-5A following TAP-pulldown was quite low, resembling the background binding of Nbs1 to IgG Sepharose observed in the negative control (Fig. 3A). This implies that phosphorylation of the SXT domain is critical for association with Nbs1.
Figure 3.
CK2-mediated phosphorylation of Ctp1 facilitates interaction with Nbs1. (A) Ctp1 and Nbs1 associate in a manner dependent on phosphorylation of the Ctp1 SXT motif and the Nbs1 FHA domain. Strains were generated that express TAP-tagged Ctp1 and FLAG-tagged Nbs1 in an mre11-H134S background. IgG Sepharose beads were used to precipitate TAP-tagged Ctp1 and any associated proteins. (B) Yeast-two hybrid assays show that Ctp1 associates with Nbs1. Titrating increasing amounts of 3-AT reveals a specific interation between Ctp1 and Nbs1 that is FHA domain-dependent.
We also verified association of Ctp1 and Nbs1 by yeast two-hybrid analyses. Although we had previously been unable to observe interaction of Nbs1 with Ctp1 under high-stringency (-LWHA) conditions,19 by titrating in increasing amounts of 3-AT, a competitive inhibitor of the HIS3 gene product, we detected a specific Ctp1-Nbs1 interaction that was greater than the autonomous activation of the HIS3 reporter gene by the pGBKT7-Ctp1 plasmid (Fig. 3B). Consistent with our previous findings, this interaction was impeded by point mutations (R27A, K45A, RKAA) in the FHA phosphopeptide-binding pocket of Nbs1 (Fig. 3B). Since the association of Ctp1 with Nbs1 appears to be strictly phosphorylation dependent, the low level of specific binding observed by two-hybrid assay may reflect an inability of Ctp1 to be robustly phosphorylated in an S. cerevisiae background. Together these results further substantiate the conclusion that the primary function of Ctp1 SXT domain phosphorylation is to promote association with Nbs1.
Discussion
Here we have provided additional evidence linking CK2 to the phospho-dependent interaction between Ctp1 and Nbs1 that is essential for HR repair of DSBs. and that this process represents overlapping functional conservation on at least two levels. First, CK2-dependent promotion of DNA damage response processes. Clear roles for CK2 in generating phosphorylation-dependent protein-protein interactions have been demonstrated in both non-homologous end joining (NHEJ) and single-strand break (SSB) repair.11-18 Our data now suggests that CK2 may also promote HR repair of DSBs by promoting association of Ctp1 with the MRN complex. Second, phosphorylated SDTD-like motifs serve as preferred docking sites for the N-terminal phosphopeptide-binding domains of Nbs1. In addition to the aforementioned interactions with Ctp1 and Mdc1 that facilitate HR and signal amplification from DSBs, respectively, Xrs2Nbs1 associates with phosphorylated Lif1XRCC4 SETD sites in an FHA domain-dependent manner to facilitate NHEJ in budding yeast.29 We infer from these observations that the N-terminus of Nbs1 likely performs a variety of signaling and DNA repair functions within the cellular DNA damage response.
In contrast to our results regarding N-terminal phosphorylation of Ctp1, recruitment of Sae2 and CtIP to DSBs requires CDK-mediated phosphorylation at a positionally conserved C-terminal site in budding yeast and humans, respectively.30, 31 It is unknown how this phosphorylation regulates Sae2 or CtIP recruitment to damaged DNA, but it is still interesting that phosphorylation of Ctp1/Sae2/CtIP is critical for the targeting and activity of each protein, even if manner in which this occurs may have diverged. Although basal phosphorylation of Ctp1 appears to be at least partially performed by CDKs, combined mutation of the ST/P sites in Ctp1 yields no phenotype,20 implying that CDK-mediated phosphorylation of Ctp1 has little or no role in its DNA repair-associated activities. Interestingly, while loss of CDK activity can be rescued using phosphomimetic Sae2/CtIP mutants,30, 31 a phospho-mimetic Ctp1 has deleterious effects on genotoxin sensitivity (Fig. 1B). This suggests that although the C-terminal CDK-mediated phosphorylation of CtIP/Sae2 contributes to the targeting and activity of these proteins at a DSB, it may not do so in the same manner as N-terminal CK2-mediated phosphorylation of Ctp1 (through generating an interaction with Nbs1/Xrs2). In support of this notion, a recent report demonstrates that while both the N- and C-termini of CtIP interact with the MRN complex, it is the extreme N-terminus of CtIP that specifically enables binding to Nbs1.32 A model that may unite these scenarios is one in which independent modifications of the N- and C-termini of CtIP/Sae2/Ctp1 could occur, each facilitating its own contact with the MRN complex.
Ctp1 is also phosphorylated in a DNA damage-dependent manner, a process that requires Mre11, Rad3ATR, Tel1ATM,20 as well as Ctp1 SXT domain phosphorylation and the FHA domain of Nbs123, 24. This suggests a mechanism in which localization of Ctp1 at a DSB is required for its DNA damage-induced phosphorylation by Rad3ATR/Tel1ATM. While PIKK-mediated DNA damage-induced phosphorylation of budding yeast Sae2Ctp1 is important for its repair and recombination functions,33 combined mutation of the PIKK-preferred ST/Q sites in Ctp1 fails to alter its phosphorylation or sensitivity in response to DNA damage.20 This suggests that either another kinase downstream of Rad3ATR/Tel1ATM performs this modification, or that PIKK-induced phosphorylation of Ctp1 occurs at a deviant non-S/TQ site, the latter of which has been observed in a few substrates such as BRCA1 and ATM itself.34, 35 Future studies will be necessary to map the specific site(s) where DNA damage-induced phosphorylation of Ctp1 occurs and to separate its functional significance from the initial priming phosphorylation event that is likely performed by CK2.
Materials and Methods
General Methods
Strains used in this study are listed in Table 1. Spore viability assays were performed exactly as previously described.19 Survival assays in response to chronic genotoxin exposure were performed by resuspending midlog phase cultures to 1 × 107 cells/ml, serially diluting the cultures fivefold, and spotting the dilutions onto YES agar plates containing the indicated amounts of CPT or HU.
S. pombe strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| PR109 | h- leu1-32 ura4-D18 | Lab stock |
| PR110 | h+ leu1-32 ura4-D18 | Lab stock |
| OL4121 | h- leu1-32 ura4-D18 ctp1::kanM×6 | Limbo, et al., 2007 |
| OL4122 | h+ leu1-32 ura4-D18 ctp1::kanM×6 | Limbo, et al., 2007 |
| YYY4181 | h? leu1-32 ura4-D18 his3-D1 arg3::HO site(kanM×6) ars1:nmt-(HO endonuclease):ampR:his3+:ars1 mre11-H134S-13myc:kanM×6 nbs1-5flag:kanM×6 ctp1-TAP:hph | Williams, et al., 2008 |
| YYY4182 | h? leu1-32 ura4-D18 his3-D1 arg3::HO site(kanM×6) ars1:nmt-(HO endonuclease):ampR:his3+:ars1 mre11-H134S-13myc:kanM×6 nbs1-5flag:kanM×6 | Williams, et al., 2008 |
| YYY4447 | h? leu1-32 ura4-D18 his3-D1 arg3::HO site(kanM×6) ars1:nmt-(HO endonuclease):ampR:his3+:ars1 mre11-H134S-13myc:kanM×6 nbs1-R27A K45A-5flag:kanM×6 ctp1-TAP:hph | Williams, et al., 2009 |
| GD4588 | h+ leu1-32 ura4-D18 ctp1-S77A T78A T79A S87A T89A-TAP:kanM×6 | Williams, et al., 2009 |
| OL4589 | h- leu1-32::ctp1-3flag:leu+ ura4-D18 ctp1::hphM×6 | Limbo, et al., 2007 |
| GD 4834 | h- leu1-32 ura4-D18 ctp1-S77A T78A T79A S87A T89A-TAP:kanM×6 | This study |
| GD 4835 | h? leu1-32 ura4-D18 his3-D1 arg3::HO site(kanM×6) ars1:nmt-(HO endonuclease):ampR:his3+:ars1 mre11-H134S-13myc:kanM×6 nbs1-5flag:kanM×6 ctp1-S77A T78A T79A S87A T89A-TAP:kanM×6 | This study |
| GD 4836 | h- leu1-32 ura4-D18 ctp1-T78D T79D T89D-TAP:kanM×6 | This study |
| GD 4837 | h? leu1-32 ura4-D18 ctp1-S77A T78A T79A S87A T89A-TAP:kanM×6 rad22-YFP:kanM×6 | This study |
| GD 4838 | h- leu1-32::ctp1-E81A D82A E92A-3flag:leu+ ura4-D18 ctp1::hphM×6 | This study |
| GD 4839 | h- leu1-32::ctp1-T78A T79A T89A-3flag:leu+ ura4-D18 ctp1::hphM×6 | This study |
| GD 4840 | h? leu1-32 ura4-D18 ctp1::hphM×6 rad22-YFP:kanM×6 | This study |
| GD 4841 | h? leu1-32 ura4-D18 ctp1-TAP:kanM×6 rad22-YFP:kanM×6 | This study |
Ctp1 immunoprecipitation and Western blotting
Following preparation of whole-cell extracts in lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 2.5 mM EDTA, 0.002% NP-40, 50 mM NaF, protease inhibitor tablet [Roche, complete Mini]), Ctp1-FLAG was immunoprecipitated with α-FLAG antibody (Sigma) and then subjected to western blotting as previously described.20
Microscopy
Flourescent microscopy of Rad22-YFP expressing strains was performed as previously described.36 For meiosis experiments, cells of opposite mating types were mixed and incubated for 3 days at 25°C on SSA plates to obtain zygotic asci. Images were taken with a Nikon Eclipse E800 microscope, Photometrics Quantix CCD camera, and IPlab Spectrum software.
Nbs1-Ctp1 Coimmunoprecipitation
Soluble whole-cell extracts were made from exponentially growing cells by disrupting in low-salt lysis buffer (50 mM Tris [pH 8.0], 50 mM NaCl, 2.5 mM EDTA, 0.002% NP-40, 50 mM NaF, protease inhibitor tablet [Roche, complete Mini]) with a bead beater. Ctp1-TAP and associated proteins were precipitated from extracts with a 50% suspension of IgG Sepharose (GE Healthcare), washed three times with low-salt lysis buffer, resolved on 8% SDS-PAGE, and examined by western blotting.
Yeast Two-Hybrid Analysis
Indicated proteins were fused to Gal4 activating domain and DNA binding domain in pGADT7 and pGBKT7, respectively, and expressed in S. cerevisiae strain AH109 (Clontech Matchmaker system). Strains positive for cotransformation with the indicated pair of plasmids were assayed by growth on Dex-WL plates (minimal glucose media lacking tryptophan and leucine). Positive interactions were assayed by growth on medium-stringency Dex-WLH plates (minimal glucose media lacking tryptophan, leucine, and histidine) and high-stringency Dex-WLHA (minimal glucose media lacking tryptophan, leucine, histidine, and adenine). The stringency of Dex-WLH plates was increased by addition of 3-AT.
Acknowledgements
We thank Scott Williams and John Tainer for previous collaborations on this project. G.E.D. is supported by a National Research Service Award from the National Institutes of Health. This work was supported by NIH grants awarded to P.R. CA77325 and John Tainer/P.R. CA117638.
References
- 1.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–20. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 2.Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–94. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, et al. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–51. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- 5.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–83. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–26. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, et al. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–40. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. PNAS USA. 2008;105:11200–5. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan JS, Litchfield DW. Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim Biophys Acta. 2008;1784:33–47. doi: 10.1016/j.bbapap.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, et al. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 12.Bekker-Jensen S, Fugger K, Danielsen JR, Gromova I, Sehested M, Celis J, et al. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J Biol Chem. 2007;282:19638–43. doi: 10.1074/jbc.C700060200. [DOI] [PubMed] [Google Scholar]
- 13.Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn SH, Foley M, et al. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol. 2005;15:656–60. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 14.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol. 2007;27:3793–803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch CA, Agyei R, Galicia S, Metalnikov P, O'Donnell P, Starostine A, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–85. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, et al. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 17.Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, Leng M, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol Cell Biol. 2004;24:8356–65. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA. APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 2008;7:292–302. doi: 10.1016/j.dnarep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–46. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, et al. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol. 2008;28:3639–51. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–20. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 23.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–11. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–68. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 26.Watson A, Mata J, Bahler J, Carr A, Humphrey T. Global gene expression responses of fission yeast to ionizing radiation. Mol Biol Cell. 2004;15:851–60. doi: 10.1091/mbc.E03-08-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snell V, Nurse P. Genetic analysis of cell morphogenesis in fission yeast--a role for casein kinase II in the establishment of polarized growth. EMBO J. 1994;13:2066–74. doi: 10.1002/j.1460-2075.1994.tb06481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmbos PL, Wu D, Daley JM, Wilson TE. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics. 2008;180:1809–19. doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–92. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–65. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J, Chen J. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem. 2009;284:31746–52. doi: 10.1074/jbc.M109.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol. 2004;24:4151–65. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006;25:3504–14. doi: 10.1038/sj.emboj.7601231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–6. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 36.Dovey CL, Russell P. Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe. Genetics. 2007;177:47–61. doi: 10.1534/genetics.107.077255. [DOI] [PMC free article] [PubMed] [Google Scholar]